Open Access Article
Open Access ArticleAtomic differentiation of silver binding preference in protein targets: Escherichia coli malate dehydrogenase as a paradigm†
Haibo
Wang
 *a,
Xinming
Yang
a,
Minji
Wang
*a,
Xinming
Yang
a,
Minji
Wang
 ab,
Menglong
Hu
c,
Xiaohan
Xu
a,
Aixin
Yan
d,
Quan
Hao
c,
Hongyan
Li
a and
Hongzhe
Sun
ab,
Menglong
Hu
c,
Xiaohan
Xu
a,
Aixin
Yan
d,
Quan
Hao
c,
Hongyan
Li
a and
Hongzhe
Sun
 *a
*a
aDepartment of Chemistry, CAS-HKU Joint Laboratory of Metallomics on Health and Environment, The University of Hong Kong, Pokfulam Road, Hong Kong, P. R. China. E-mail: haibo_wang@connect.hku.hk; hsun@hku.hk
bSchool of Chemistry and Molecular Engineering, East China Normal University, No. 3663 Zhongshan Road North, Shanghai, 200062, P. R. China
cSchool of Biomedical Sciences, The University of Hong Kong, Laboratory Block, 21 Sassoon Road, Pokfulam, Hong Kong, P. R. China
dSchool of Biological Sciences, The University of Hong Kong, Pokfulam Road, Hong Kong, P. R. China
First published on 10th September 2020
Abstract
Understanding how metallodrugs interact with their protein targets is of vital importance for uncovering their molecular mode of actions as well as overall pharmacological/toxicological profiles, which in turn facilitates the development of novel metallodrugs. Silver has been used as an antimicrobial agent since antiquity, yet there is limited knowledge about silver-binding proteins. Given the multiple dispersed cysteine residues and histidine–methionine pairs, Escherichia coli malate dehydrogenase (EcMDH) represents an excellent model to investigate silver coordination chemistry as well as its targeting sites in enzymes. We show by systematic biochemical characterizations that silver ions (Ag+) bind EcMDH at multiple sites including three cysteine-containing sites. By X-ray crystallography, we unravel the binding preference of Ag+ to multiple binding sites in EcMDH, i.e., Cys113 > Cys251 > Cys109 > Met227. Silver exhibits preferences to the donor atoms and residues in the order of S > N > O and Cys > Met > His > Lys > Val, respectively, in EcMDH. For the first time, we report the coordination of silver to a lysine in proteins. Besides, we also observed argentophilic interactions (Ag⋯Ag, 2.7 to 3.3 Å) between two silver ions coordinating to one thiolate. Combined with site-directed mutagenesis and an enzymatic activity test, we unveil that the binding of Ag+ to the site IV (His177-Ag-Met227 site) plays a vital role in Ag+-mediated MDH inactivation. This work stands as the first unusual and explicit study of silver binding preference to multiple binding sites in its authentic protein target at the atomic resolution. These findings enrich our knowledge on the biocoordination chemistry of silver(I), which in turn facilitates the prediction of the unknown silver-binding proteins and extends the pharmaceutical potentials of metal-based drugs.
Introduction
Despite the wide application of metallodrugs for the treatment of various diseases clinically,1–5 the molecular targets mediating their biological effects remain largely unknown.3,6,7 It is very important to understand how metal-based drugs interact with their molecular targets, which defines the overall pharmacological and toxicological profiles of metallo-medicines.8–11 However, there is a paucity of detailed structural information on the metallodrug–protein adducts so far, which hinders the exploration of metallodrug targeting sites in proteins and their pharmacological potentials.Silver ions and silver nanoparticles (AgNPs) are nowadays being extensively used as antimicrobial agents in healthcare and food industry owing to their broad-spectrum antimicrobial properties and less chance of resistance, yet their molecular targets remain largely unknown.4,12–18 Proteins are deemed to be vital targets of silver and the widespread assumption of silver toxicity is that silver binds to the thiolate or thioester of cysteine or methionine residues and subsequently substitutes the cognate ions in the catalytic sites of metalloenzymes.16,19,20 Particularly, being isoelectronic with Cu+, Ag+ has been reported to mimic Cu+ at its native binding sites in copper proteins by adopting various coordination numbers and geometries.21–23 However, to date, the binding of Ag+ to proteins, especially the authentic silver-binding targets, has not been structurally extensively investigated. To the best of our knowledge, azurin,24 multicopper oxidase CueO,21 urease,25 and trypanothione reductase26 are the only silver inhibited proteins in vitro with available silver-bound crystal structures.
Considering the diversity of silver-binding proteins and the multifaceted factors defining the silver–protein interaction, the silver targeting sites in proteins should be richly varied and exquisitely attuned to different proteins and residues.3,16,27 However, limited information on silver-binding proteins restrained the thorough examination, particularly at the atomic level, on which sites or residues of proteins are targeted by silver and what is the preferred coordination geometry of silver in proteins. In the case of multiple sites present, how silver differentiates the binding priority to these sites and what is the driving force for the binding preferences. These long-standing issues remain to be addressed.16
Until very recently, we resolved and identified 34 silver-binding proteins (Ag+-proteome) in E. coli using our home-made two-dimensional approach of liquid chromatography coupled with GE-ICP-MS (LC-GE-ICP-MS).27,28 We unveiled that silver eradicates E. coli through targeting multiple proteins and disrupting multiple pathways, including glycolysis, TCA cycle, glyoxylate shunt and ROS defense system. E. coli malate dehydrogenase (EcMDH or MDH) is the only enzyme involved in both TCA cycle and glyoxylate shunt, indicating its vital role as a silver target.
EcMDH contains three cysteine residues and several methionine–histidine pairs, while all these cysteine sites are far away from the active sites. These properties of E. coli MDH render it as an exceptional model system for investigating the long-standing issues between silver and its protein target. In this work, we found that silver binds to MDH at multiple sites based on systematic characterizations. Subsequently, we demonstrate a diversified coordination behavior of Ag+ to MDH by using an array of Ag-bound MDH X-ray structures. For the first time, we unravel the Ag+ binding preference to its multiple binding sites of its protein target at the atomic level. This work highlights the first successful attempt to capture the binding preference of silver to its protein target, enriching the bio-coordination chemistry of metals.
Results and discussions
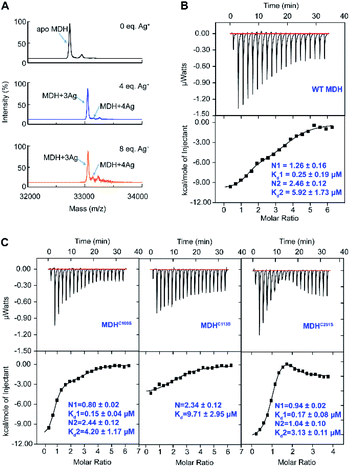
To study how Ag+ binds to MDH in vitro, we overexpressed and purified the MDH protein (Tables S1 and 2†). The purity was confirmed by size exclusion chromatography (Fig. S1A†) and SDS-PAGE (Fig. S1B†). Binding of Ag+ to MDH (monomer) was examined by matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). The peak at m/z of 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 732.4 was assignable to native MDH monomer, while an intense peak and a weak peak at m/z of 33
732.4 was assignable to native MDH monomer, while an intense peak and a weak peak at m/z of 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 053.6 and 33
053.6 and 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 160.4 (calcd m/z of 33
160.4 (calcd m/z of 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 052.6 and 33
052.6 and 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 159.5 respectively) were assignable to MDH with three and four Ag+ ions bound respectively (Fig. 1A). Further increasing Ag+ concentration to 8 eq. led to neither higher intensity of the peak at m/z of 33
159.5 respectively) were assignable to MDH with three and four Ag+ ions bound respectively (Fig. 1A). Further increasing Ag+ concentration to 8 eq. led to neither higher intensity of the peak at m/z of 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 160.4 nor more peak at higher m/z (Fig. 1A), verifying that the binding stoichiometry of Ag/MDH is 4
160.4 nor more peak at higher m/z (Fig. 1A), verifying that the binding stoichiometry of Ag/MDH is 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The MALDI-TOF MS data indicate that the first three Ag+ ions might bind MDH with much higher affinity than the fourth one.
1. The MALDI-TOF MS data indicate that the first three Ag+ ions might bind MDH with much higher affinity than the fourth one.
We therefore further measured the binding affinity of Ag+ towards MDH by isothermal titration calorimetry (ITC). ITC data exhibited two binding modes and were fitted with a two-set binding model, which gave rise to apparent dissociation constants (Kd) of 0.25 ± 0.09 and 5.92 ± 1.73 μM for the first (N1 = 1.26 ± 0.16) and second (N2 = 2.46 ± 0.12) Ag+-binding modes respectively (Fig. 1B and Table S3†). These values generally agree well with our previous studies on the binding affinity between silver ions and cysteines (at sub-micro molar level).29,30 Given that silver is highly thiophilic, we hypothesize that cysteine residues are involved in silver binding and constructed three MDH mutants with cysteines being mutated into serine and then examined the binding affinity of Ag+ to MDH mutants under identical conditions. Two binding modes were still observed for MDHC109S and MDHC251S. In line with the wild-type MDH, MDHC109S and MDHC251S retained the high binding affinity to Ag+ with the binding capacity of ca. 0.97 (Kd = 0.15 ± 0.04 μM) and 0.94 (Kd = 0.17 ± 0.08 μM) respectively, indicating that Cys109 and Cys251 are not the strongest binding sites (Fig. 1C and Table S3†). While for the second binding mode, MDHC109S and MDHC251S bound ca. 1.44 and 0.94 eq. of Ag+ with the Kd of 4.20 ± 0.17 and 3.13 ± 0.11 μM respectively (Fig. 1C and Table S3†). The loss of one weak binding site of MDHC109S and MDHC251S demonstrates that Ag+ binds to Cys109 or Cys251 with relatively lower affinity. While MDHC113S bound 2.34 ± 0.12 molar equivalents of Ag+ with the Kd of 9.71 ± 2.75 μM, and the loss of the strong binding site in this mutant suggests that Ag+ binds to Cys113 with relatively higher affinity (Fig. 1C and Table S3†). Collectively, these data demonstrate that Ag+ binds to the three cysteine sites of MDH, with Cys113 exhibiting the highest affinity, followed by Cys109 and Cys251.
Our combined data from MALDI-TOF MS and ITC show that silver ions bind to MDH mainly via cysteine residues and each cysteine site binds one silver ion under physiological condition. Nevertheless, the coordination geometries as well as the forces driving the binding preference of silver ions to the three cysteine sites and the fourth site remain unclear.
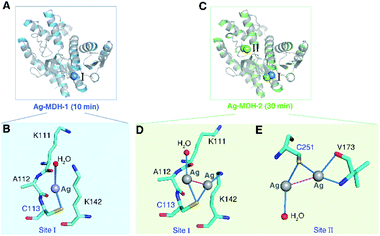
To further explore the local coordination geometry of silver in its binding sites at the atomic level, we attempted to crystallize the apo-form MDH. Tetragonal-shaped single crystals were formed in three days and reached a size suitable for X-ray diffraction after one week (Fig. S2A†) and the structure was solved at the resolution of 1.54 Å (Fig. S2B†). To obtain the Ag+-bound form crystals, we soaked the crystals into cryo-protectant solutions containing 2 mM Ag+ for 10 min (Ag-MDH-1), 30 min (Ag-MDH-2), and 1 h (Ag-MDH-3), yielding Ag-bound MDH crystals loaded with various number of Ag+ at the resolution from 2.06 Å to 2.55 Å (Table S4†). The crystallographic results showed no significant overall conformational changes among the Ag-bound MDHs and apo-form (RMSD of 0.233 Å, 0.312 Å, and 0.309 Å when superimposing Ag-MDH-1, Ag-MDH-2, and Ag-MDH-3 (Chain A) to apo-MDH, respectively). For all Ag-MDHs, the identification of Ag was made on the significant positive peaks (≥20σ) in the mFo-DFc (difference Fourier) map (Table S5†).
In the crystal structure of Ag-MDH-1, only one Ag+ bound to the Cys113 site (Site I), with Ag+ coordinating to the side chain of the Sγ of Cys113 and a water molecule (Fig. 2A and B). The bond lengths of Ag+ to Cys113-Sγ or H2O are the same (2.60 Å) with a pseudo-linear S–Ag–O angle of 159.1° (Tables S6 and S7†). When the crystal soaking duration was extended to 30 min (Ag-MDH-2), four silver ions were observed at Cys113 site (Site I) and Cys251 site (Site II) with two silver ions observed at each site (Fig. 2C). Unlike Ag-MDH-1, the side-chain of Cys113 bound two silver ions with Ag⋯Ag distance of 2.7 Å in Ag-MDH-2 (Fig. 2D). The first silver ion (Ag401) at this site further coordinated to a water molecule whilst the other (Ag402) to the Nζ of Lys142. Superimposing the Chain A of the two structures showed that the silver bound to Cys113 of Ag-MDH-1 is located in the center of the two silver ions of Ag-MDH-2 (Fig. S3A†). In Ag-MDH-2, Cys251 also bound two silver ions (Ag⋯Ag distance of 3.1 Å) with one being further coordinated by a water molecule and another by main-chain nitrogen and oxygen of Val173 respectively (Fig. 2E). For the silver ions observed in Ag-MDH-1 and Ag-MDH-2, most of them share a similar quasi-linear coordination geometry with the angle of ligand-Ag-ligand ranging from 153.8° to 177.7° except Ag403 (in Ag-MDH-2) (Table S7†), which has three coordinations with Cys251 and Val173 in nearly trigonal planar geometry.
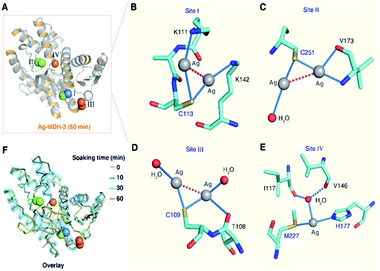
In the structure of Ag-MDH-3, four Ag+-binding sites with seven silver ions were identified in each chain of MDH. Consistent with the biochemical studies, three of the Ag+-binding sites contain Cys residues, i.e., Cys113 (Site I), Cys251 (Site II), and Cys109 (Site III), with two silver ions found in each thiolate of three cysteine residues (Fig. 3A). Similarly, argentophilic interactions were observed between the two silver ions that bound to the thiolate group of the same cysteine (Ag⋯Ag, 2.8–3.3 Å) (Fig. 3B–D and Table 1).31 In addition, each silver ion bound to a nitrogen atom from either a lysine side-chain (Site I) or a valine backbone (Site II), or an additional water molecule (Site III), with most of them forming quasi-linear coordination geometry (Fig. 3B–D and Table 2). In the Cys109 and Cys251 sites, a backbone oxygen atom formed an additional coordination to one of the silver atoms (L–Ag–S, 102–104°). The four silver ions bound to Cys113 and Cys251 in Ag-MDH-3 overlap with those in Ag-MDH-2, albeit that one of the silver ion bound to Cys113 in Ag-MDH-3 is coordinated by the Nζ of Lys111 rather than a water molecule (Fig. S3A and B†).
| Metal | Ligand | Distance (Å) |
|---|---|---|
| a Only weak interactions exist. | ||
| Ag401 | Cys109-Sγ | 2.5 |
| Ag402 | Cys109-Sγ | 2.4 |
| Ag402 | Thr108-O | 2.9 |
| Ag401 | HOH512 | 2.9 |
| Ag402 | HOH511 | 2.3 |
| Ag401 | Ag402 | 3.0a |
| Ag403 | Cys113-Sγ | 2.9 |
| Ag404 | Cys113-Sγ | 2.5 |
| Ag403 | Lys111-Nζ | 2.6 |
| Ag404 | Lys142-Nζ | 2.5 |
| Ag403 | Ag404 | 2.8a |
| Ag405 | Cys251-Sγ | 2.3 |
| Ag406 | Cys251-Sγ | 2.4 |
| Ag405 | Val173-N | 2.2 |
| Ag405 | Val173-O | 2.5 |
| Ag406 | HOH513 | 2.9 |
| Ag405 | Ag406 | 3.3 |
| Ag407 | Met227-Sδ | 2.7 |
| Ag407 | His177-Nε2 | 2.5 |
| Ag407 | HOH509 | 2.7 |
| Atom1 | Atom2 | Atom3 | Angle (°) |
|---|---|---|---|
| a All Ag atoms and water molecules are assigned their universal chain IDs. The measurements are based on the atomic coordination for the polymeric Chain A and Ag atoms or water molecules. | |||
| HOH512 | Ag401 | Cys109-Sγ | 136.3 |
| HOH511 | Ag402 | Cys109-Sγ | 145.5 |
| Thr108-O | Ag402 | Cys109-Sγ | 104.6 |
| Lys111-Nζ | Ag403 | Cys113-Sγ | 154.0 |
| Lys142-Nζ | Ag404 | Cys113-Sγ | 151.7 |
| Val173-N | Ag405 | Cys251-Sγ | 168.9 |
| Val173-O | Ag405 | Cys251-Sγ | 102.7 |
| HOH513 | Ag406 | Cys251-Sγ | 170.3 |
| His177-Nε2 | Ag407 | Met227-Sδ | 146.6 |
| HOH509 | Ag407 | His177-Nε2 | 88.3 |
| HOH509 | Ag407 | Met227-Sδ | 89.3 |
As a soft metal, Ag+ exhibits higher affinity to nitrogen than oxygen atoms, the higher binding affinity of Ag+ to the Cys113 and Cys251 sites than the other sites could be ascribed to the coordination of Ag+ to the adjacent nitrogen atoms of the lysine side chains (Lys111 and Lys142) and the nitrogen atom of the valine main chain (Val173), respectively (Fig. 3B and C). At the fourth silver-binding site, Met227, His177 and a water molecule (HOH 509) formed a trigonal pyramidal geometry (the S–Ag–N angle of 146.6°) for Ag+ binding with lower occupancy than the previous three Ag+-binding sites (Fig. 3E). Additionally, the water molecule was further stabilized by two hydrogen bonds (O⋯O, 2.8–3.0 Å) from the backbone oxygen atoms of Ile117 and Val146.
To verify these four sites are indeed silver binding sites, we further constructed a triple cysteine mutant MDH3CS as well as two variants with four residues mutated (MDH3CS/M227S and MDH3CS/H177S) and examined their silver binding capability by ITC and MALDI-TOF MS. For the triple Cys mutant MDH3CS, a weak signal of Ag-bound MDH3CS was detected by MALDI-TOF MS (MDH3CS, m/z = 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 681.2; Ag-MDH3CS, m/z = 32
681.2; Ag-MDH3CS, m/z = 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 787.6) (Fig. S4A†), whereas, no obvious Ag+ binding was observed by ITC (Fig. S4B†), indicating that the affinity of the fourth Ag+-binding might be too weak to be detected by ITC. Consistently, no peak corresponding to Ag+-binding protein was observed for MDH3CS after treatment with 4 eq. of Ag+ by GE-ICP-MS, for which the weakly bound metal ions are dissociated during the gel electrophoresis separation (Fig. S4C†). For MDH3CS/M227S and MDH3CS/H177S, no MW change was observed for the former one upon incubation with silver ions, whereas a weak signal for one Ag+ bound to MDH3CS/H177S was observed in MALDI-TOF MS (Fig. S5A and B†), indicating that Ag+ binds to Met227 with higher affinity than His177. Taken together, these data demonstrate that mutation of the major silver-binding residues could completely abrogate silver binding to MDH, verifying the authenticity of the silver-binding sites in MDH resolved by crystallographic structures.
787.6) (Fig. S4A†), whereas, no obvious Ag+ binding was observed by ITC (Fig. S4B†), indicating that the affinity of the fourth Ag+-binding might be too weak to be detected by ITC. Consistently, no peak corresponding to Ag+-binding protein was observed for MDH3CS after treatment with 4 eq. of Ag+ by GE-ICP-MS, for which the weakly bound metal ions are dissociated during the gel electrophoresis separation (Fig. S4C†). For MDH3CS/M227S and MDH3CS/H177S, no MW change was observed for the former one upon incubation with silver ions, whereas a weak signal for one Ag+ bound to MDH3CS/H177S was observed in MALDI-TOF MS (Fig. S5A and B†), indicating that Ag+ binds to Met227 with higher affinity than His177. Taken together, these data demonstrate that mutation of the major silver-binding residues could completely abrogate silver binding to MDH, verifying the authenticity of the silver-binding sites in MDH resolved by crystallographic structures.
Compared with MALDI-TOF MS and ITC results (vide supra), our crystallographic structures collectively illustrated identical silver-binding sites and a more detailed silver-binding preference in MDH. Despite only one Ag+ binds to each cysteine site in solution, two Ag+ were observed in each thiolate with weak argentophilic interactions and seven Ag+ were found in one MDH molecule in the Ag-MDH-3 crystal structure. The observation of two Ag+ bound to each cysteine site in the crystal structure is likely due to the use of a high concentration of AgNO3 (2 mM) and a long duration (1 h) for crystal soaking. Argentophilic interactions between silver ions in the form of di-nuclear,25,26 tetra-nuclear32 and even hepta-nuclear33 were observed in urease, ferredoxin and metallothionein as well. Nevertheless, the observed two Ag+ ions in each thiolate indeed provide two potential silver binding modes at each cysteine site, which may represent novel silver shuttle systems under physiological conditions.
To investigate the effect of Ag+-binding on the enzyme function, we further measured the activity of MDH after incubation with different amounts of Ag+. The result showed a dose-dependent inhibition of Ag+ on MDH activity with 3 eq. and 6 eq. of Ag+ inhibiting over 50% and 90% of MDH activity respectively (Fig. 4A). To uncover the binding of Ag+ to which site contribute to MDH inhibition, we compared the structures of Ag-MDH-3 and previously reported structure of EcMDH in ternary complex with citrate (as substrate analog) and cofactor NAD+ (PDB: 1EMD).34 Superimposition of 1EMD with Ag-MDH-3 demonstrates that Ag+-binding site IV is the one being most adjacent to the active site (Fig. 4B and C). A close look at the comparison of Ag-MDH-3 and NAD-bound MDH shows that the binding of silver to His177 and Met227 form a steric hindrance to the nicotinamide carboxamide of NAD+ (Fig. 4D). In comparison with citrate-bound MDH, the side chain of His177 in Ag-MDH-3 rotates 30.4° to the opposite direction of citrate for silver binding. Such a rotation results in 2.3 Å shift of the His177-Nε2 away from citrate binding (Fig. 4E).
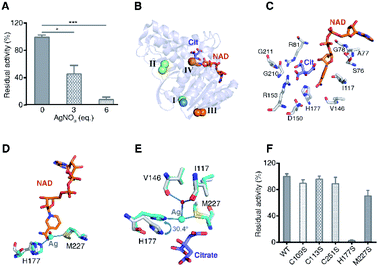 | ||
| Fig. 4 Binding of Ag+ to site IV (His177-Met227 site) contributes to Ag+-mediated MDH inactivation. (A) Dose dependent inhibition of MDH by Ag+ (n = 3). (B) Superimposition of native MDH (PDB: 1EMD) with Ag-bound MDH (Ag-MDH-3). (C) Active site of MDH with key residues shown in sticks (PDB: 1EMD). (D) An overlay image comparing the relative position of Ag+ (cyan sphere) with the cofactor NAD+ (PDB: 1EMD). (E) An overlay image comparing the relative position of Ag+ (cyan sphere) with the substrate analog citrate (PDB: 1EMD). (F) Normalized residual activity of WT MDH and MDH mutants (n = 3). The results are shown as mean ± SEM (A, F). *P < 0.05, **P < 0.01 and ***P < 0.001. Structural alignment was done over Cα residues using DaliLite (B–E). | ||
To further unveil which residue is pivotal for Ag+ inhibition on MDH activity, we examined the enzyme activities of WT MDH and a series of MDH mutants, including MDHC109S, MDHC113S, MDHC251S, MDHH177S, and MDHM227S. As shown in Fig. 4F, individual site-directed mutation of Cys109, Cys113, and Cys251 to Ser led to no significant change of MDH activity. In contrast, the mutation of Met227 to Ser resulted in a decrease of ∼30% of MDH activity and mutation of His177 to Ser completely inactivated the enzyme (Fig. 4F), demonstrating that these two residues play critical roles for the catalytic activity of MDH. Indeed, previous studies reported that Met227 is involved in the binding of the ribose from NAD+34 and the His177–Asp150 pair acts as an essential proton relay system via hydrogen bonds during the catalytic reaction.35–37 Thus, the binding of Ag+ to the site IV as shown by the crystal structure could potentially disrupt the optimal orientation of NAD+ to the active site and abrogate the proton transfer capability of catalytic His177, leading to the MDH inactivation.
Taken together, the X-ray studies show that silver ions bind to four sites of MDH with higher affinity to the three Cys sites and relatively lower affinity to the fourth Met/His site. Although the three cysteine sites are all located at the solvent accessible surface, they exhibit different affinities to silver ions due to slightly different coordination spheres of silver ions, i.e., the nitrogen atoms of side-chains of Lys111/Lys142 and the main-chain of Val173 at Cys113 and Cys251 sites respectively. The distance between Val173/N and Ag405 is shorter (0.7 Å) than that between Thr108/O and Ag402, which might contribute to the higher affinity of silver ions to Cys251 than to Cys109. The crystallographic data are generally coherent with the biochemical results that Ag+ binds to four sites of MDH albeit the different stoichiometry. Importantly, the binding of Ag+ to the site IV (His177-Ag-Met227) plays a vital role in Ag+-mediated MDH inactivation. For the three cysteine sites, despite the long distance between each cysteine to the active site (>20 Å), whether silver binding to these sites contributes to MDH inhibition, e.g., through unusual allosteric regulation, warrants further investigation.
Conclusion
We unveil, at the atomic resolution, a process of silver ion binding to the multiple sites of MDH via a series of Ag-bound MDH structures. Consistent with the biochemical characterizations, Ag+ ions bind to the multiple sites of MDH with the binding preference in the order of Cys113 > Cys251 > Cys109 > Met227/His177. Generally, silver ions show a diversified coordination geometry through binding to S, N and O from Cys/Met, His/Lys/Val and Val/H2O respectively. Coordination of silver to Lys was observed for the first time in protein structures. The coordination numbers of silver generally range from 2 to 4 and angles from 130° to 178° with quasi-linear as preferred geometry, followed by trigonal planar. We found that, in the case of silver coordination to a single residue, it exhibits preferences to the atoms and residues in the order of S > N > O and Cys > Met > His > Lys > Val respectively. When silver coordinates to the sites containing the same dominant residue of cysteine, the flexibility of surrounding residues together with the solvent accessibility confer the site specific avidity for silver. Moreover, we observed argentophilic interactions between silver ions (Ag⋯Ag distance of 2.7–3.3 Å) coordinating to the same thiolate of each cysteine site, which could only be observed in soaked crystals (high concentration of Ag+ being used) but not in solutions. Importantly, combined with enzymatic activity test on the site-directed MDH mutants, we uncover that the binding of Ag+ to the site IV (His177-Ag-Met227 site) plays a pivotal role in Ag+-mediated MDH inactivation.Collectively, using MDH as a showcase, we demonstrate a diversified bio-coordination chemistry of silver to its authentic protein targets, which are exquisitely attuned to different sites and residues. Our work represents for the first unusual and explicit study of silver-binding to the multiple sites in its authentic protein target at the atomic resolution. Recently, we demonstrated that Ag+ targets Cys149 and His176 at the active site of GAPDH (an essential enzyme in glycolysis pathway without using metal ion as cofactor), leading to abolishment of its enzymatic activity via a mix-type inhibition.38 The diversity of silver-binding proteins and the multiple properties defining the silver–ligand interactions may provide novel insights into pharmacological and toxicological profiles of antimicrobial silver, thereby opening a new horizon for the study of the interactions between metallo-based medicines and their protein targets. The atomistic knowledge of silver–ligand interaction could also facilitate the development of novel metalloenzyme inhibitors as metallodrug candidates.39,40
Additional information
The coordinates and structure factors of Ag-MDH-1, Ag-MDH-2 and Ag-MDH-3 are deposit at Protein Data Bank (wwPDB) with accession codes of 5Z3W, 7CGC, and 7CGD, respectively.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge the Research Grants Council of Hong Kong (R7070-18, 17307017P, 1733616P and AoE/P-705/16), National Science Foundation of China (21671203), Norman & Cecilia Yip Foundation (HS) and The University of Hong Kong as well as a Hong Kong PhD Fellowship (HBW). We thank the Center for Genomic Sciences, Li Ka Shing Faculty of Medicine for the mass spectrometry facilities. The crystal diffraction data were collected at Shanghai Synchrotron Radiation Facility (SSRF), the Chinese Academy of Sciences. We thank the staff at BL17U1 beamline of SSRF for their generous help and Vivian WW Yam (HKU) for helpful discussion.Notes and references
- K. J. Franz and N. Metzler-Nolte, Chem. Rev., 2019, 119, 727–729 CrossRef CAS.
- J. J. Soldevila-Barreda and N. Metzler-Nolte, Chem. Rev., 2019, 119, 829–869 CrossRef CAS.
- J. A. Lemire, J. J. Harrison and R. J. Turner, Nat. Rev. Microbiol., 2013, 11, 371–384 CrossRef CAS.
- R. J. Turner, Microb. Biotechnol., 2017, 10, 1062–1065 CrossRef.
- Y. C. Ong, S. Roy, P. C. Andrews and G. Gasser, Chem. Rev., 2019, 119, 730–796 CrossRef CAS.
- N. P. E. Barry and P. J. Sadler, Chem. Commun., 2013, 49, 5106–5131 RSC.
- K. D. Mjos and C. Orvig, Chem. Rev., 2014, 114, 4540–4563 CrossRef CAS.
- H. Wang, Y. Zhou, X. Xu, H. Li and H. Sun, Curr. Opin. Chem. Biol., 2020, 55, 171–179 CrossRef CAS.
- H. Li, R. Wang and H. Sun, Acc. Chem. Res., 2019, 52, 216–227 CrossRef CAS.
- M. P. Sullivan, H. U. Holtkamp and C. G. Hartinger, Met. Ions Life Sci., 2018, 18, 351–386 CAS.
- C. G. Hartinger, M. Groessl, S. M. Meier, A. Casini and P. J. Dyson, Chem. Soc. Rev., 2013, 42, 6186–6199 RSC.
- R. J. Turner, Antibiotics, 2018, 7, 112 CrossRef.
- J. J. Harrison, H. Ceri and R. J. Turner, Nat. Rev. Microbiol., 2007, 5, 928–938 CrossRef CAS.
- J. R. Morones-Ramirez, J. A. Winkler, C. S. Spina and J. J. Collins, Sci. Transl. Med., 2013, 5, 190ra181 Search PubMed.
- J.-Y. Maillard and P. Hartemann, Crit. Rev. Microbiol., 2013, 39, 373–383 CrossRef CAS.
- S. Eckhardt, P. S. Brunetto, J. Gagnon, M. Priebe, B. Giese and K. M. Fromm, Chem. Rev., 2013, 113, 4708–4754 CrossRef CAS.
- S. Chernousova and M. Epple, Angew. Chem., Int. Ed., 2013, 52, 1636–1653 CrossRef CAS.
- K. M. Fromm, Nat. Chem., 2011, 3, 178 CrossRef CAS.
- S. Medici, M. Peana, G. Crisponi, V. M. Nurchi, J. I. Lachowicz, M. Remelli and M. A. Zoroddu, Coord. Chem. Rev., 2016, 327, 349–359 CrossRef.
- J. L. Clement and P. S. Jarrett, Met.-Based Drugs, 1994, 1, 467–482 CrossRef CAS.
- S. K. Singh, S. A. Roberts, S. F. McDevitt, A. Weichsel, G. F. Wildner, G. B. Grass, C. Rensing and W. R. Montfort, J. Biol. Chem., 2011, 286, 37849–37857 CrossRef CAS.
- M. Meury, M. Knop and F. P. Seebeck, Angew. Chem., Int. Ed., 2017, 56, 8115–8119 CrossRef CAS.
- F. Long, C. C. Su, M. T. Zimmermann, S. E. Boyken, K. R. Rajashankar, R. L. Jernigan and E. W. Yu, Nature, 2010, 467, 484–U140 CrossRef CAS.
- M. J. Panzner, S. M. Bilinovich, J. A. Parker, E. L. Bladholm, C. J. Ziegler, S. M. Berry and T. C. Leeper, J. Inorg. Biochem., 2013, 128, 11–16 CrossRef CAS.
- L. Mazzei, M. Cianci, A. Gonzalez Vara and S. Ciurli, Dalton Trans., 2018, 47, 8240–8247 RSC.
- P. Baiocco, A. Ilari, P. Ceci, S. Orsini, M. Gramiccia, T. Di Muccio and G. Colotti, ACS Med. Chem. Lett., 2011, 2, 230–233 CrossRef CAS.
- H. Wang, A. Yan, Z. Liu, X. Yang, Z. Xu, Y. Wang, R. Wang, M. Koohi-Moghadam, L. Hu, W. Xia, H. Tang, Y. Wang, H. Li and H. Sun, PLoS Biol., 2019, 17, e3000292 CrossRef.
- L. Hu, T. Cheng, B. He, L. Li, Y. Wang, Y.-T. Lai, G. Jiang and H. Sun, Angew. Chem., Int. Ed., 2013, 52, 4916–4920 CrossRef CAS.
- X. Liao, F. Yang, R. Wang, X. He, H. Li, R. Y. T. Kao, W. Xia and H. Sun, Chem. Sci., 2017, 8, 8061–8066 RSC.
- X. Liao, F. Yang, H. Li, P.-K. So, Z. Yao, W. Xia and H. Sun, Inorg. Chem., 2017, 8, 8061–8066 CAS.
- H. Schmidbaur and A. Schier, Angew. Chem., Int. Ed., 2015, 54, 746–784 CrossRef CAS.
- M. Martic, I. N. Jakab-Simon, L. T. Haahr, W. R. Hagen and H. E. Christensen, J. Biol. Inorg Chem., 2013, 18, 261–276 CrossRef CAS.
- C. W. Peterson, S. S. Narula and I. M. Armitage, FEBS Lett., 1996, 379, 85–93 CrossRef CAS.
- M. D. Hall and L. J. Banaszak, J. Mol. Biol., 1993, 232, 213–222 CrossRef CAS.
- J. K. Bell, H. P. Yennawar, S. K. Wright, J. R. Thompson, R. E. Viola and L. J. Banaszak, J. Biol. Chem., 2001, 276, 31156–31162 CrossRef CAS.
- M. Lemaire, M. Miginiac-Maslow and P. Decottignies, Eur. J. Biochem., 1996, 236, 947–952 CrossRef CAS.
- J. J. Birktoft and L. J. Banaszak, J. Biol. Chem., 1983, 258, 472–482 CAS.
- H. Wang, M. Wang, X. Yang, X. Xu, Q. Hao, A. Yan, M. Hu, R. Lobinski, H. Li and H. Sun, Chem. Sci., 2019, 10, 7193–7199 RSC.
- L. Riccardi, V. Genna and M. De Vivo, Nat. Rev. Chem., 2018, 2, 100–112 CrossRef CAS.
- R. Wang, T. P. Lai, P. Gao, H. Zhang, P. L. Ho, P. C. Woo, G. Ma, R. Y. Kao, H. Li and H. Sun, Nat. Commun., 2018, 9, 439 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc04151c |
| This journal is © The Royal Society of Chemistry 2020 |