Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Development and application of ubiquitin-based chemical probes†
Xin
Sui
 ab,
Yu
Wang
a,
Yun-Xiang
Du
b,
Lu-Jun
Liang
b,
Qingyun
Zheng
b,
Yi-Ming
Li
*a and
Lei
Liu
ab,
Yu
Wang
a,
Yun-Xiang
Du
b,
Lu-Jun
Liang
b,
Qingyun
Zheng
b,
Yi-Ming
Li
*a and
Lei
Liu
 *b
*b
aSchool of Food and Biological Engineering, Key Laboratory of Metabolism and Regulation for Major Diseases of Anhui Higher Education Institutes, Hefei University of Technology, Hefei 230009, China. E-mail: ymli@hfut.edu.cn
bTsinghua-Peking Center for Life Sciences, Ministry of Education Key Laboratory of Bioorganic Phosphorus Chemistry and Chemical Biology, Center for Synthetic and Systems Biology, Department of Chemistry, Tsinghua University, Beijing 100084, China. E-mail: lliu@mail.tsinghua.edu.cn
First published on 4th August 2020
Abstract
Protein ubiquitination regulates almost every process in eukaryotic cells. The study of the many enzymes involved in the ubiquitination system and the development of ubiquitination-associated therapeutics are important areas of current research. Synthetic tools such as ubiquitin-based chemical probes have been making an increasing contribution to deciphering various biochemical components involved in ubiquitin conjugation, recruitment, signaling, and deconjugation. In the present minireview, we summarize the progress of ubiquitin-based chemical probes with an emphasis on their various structures and chemical synthesis. We discuss the utility of the ubiquitin-based chemical probes for discovering and profiling ubiquitin-dependent signaling systems, as well as the monitoring and visualization of ubiquitin-related enzymatic machinery. We also show how the probes can serve to elucidate the molecular mechanism of recognition and catalysis. Collectively, the development and application of ubiquitin-based chemical probes emphasizes the importance and utility of chemical protein synthesis in modern chemical biology.
1. Introduction
Protein post-translational modifications (e.g., methylation, acetylation, phosphorylation, glycosylation, and ubiquitination) regulate various biological processes in all eukaryotic cells, and dysregulation of the associated enzymes gives rise to diverse pathologies. Ubiquitination, the post-translational attachment of a 76-residue protein named ubiquitin (Ub), is orchestrated by the actions of four enzyme classes.1–3 The Ub activating enzyme E1 catalyses the formation of an E1-Ub thioester at the expense of ATP (Fig. 1a). Then, the active Cys of the conjugating enzyme E2 attacks the E1-Ub thioester to produce an E2-Ub thioester, and the Ub ligase E3 transfers the Ub from the active Cys of E2 to the Lys of the substrate protein. The reverse of this process is accomplished by deubiquitinating enzymes (DUBs), which catalyse the cleavage of the isopeptide bond.4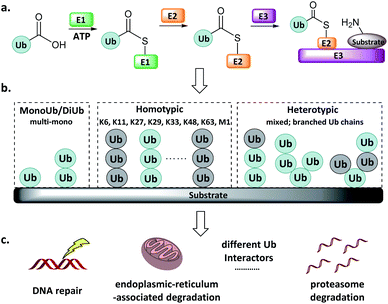 | ||
| Fig. 1 Ub system. (a) Actions of E1, E2, and E3. (b) Mono-, di- and poly-Ub chains. (c) Distinct Ub chains are recognized by different interactors. | ||
Ub can be attached to substrate proteins as a single unit or in the form of Ub chains, wherein successive Ubs are connected at Ub's M1, K6, K11, K27, K29, K33, K48, and K63 amino groups. In addition to the homotypic Ub chains, heterotypic and branched Ub chains have also been discovered5 (Fig. 1b). These Ubs adopt distinct conformations, which differentially influence the fate of the protein to which they are attached in a manner reminiscent of a code. For example, K48-linked Ubs signal proteasome degradation, while K63-linked Ubs regulate the innate immune signalling pathways.1–3,6,7 In addition, Ub-like (UbL) modifications such as Nedd8, SUMO, and ISG15 have also been identified.8–10 Defects in components of the Ub/UbL processes influence disease pathogenesis, especially cancer and neurodegeneration. A more detailed understanding of Ub/UbL modifications in cellular processes such as DNA repair and immune response remains to be acquired (Fig. 1c).
Genetic and proteomic methods have been developed to study ubiquitination. For instance, proteomic studies using an antibody targeting Lys-ε-Gly–Gly have revealed >50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ubiquitination sites in human cells.11 More recently, a Ub clipping method was developed to map Ubs in vivo, showing that branched Ubs account for 10–20% of the total Ub abundance.12 Despite these advances, understanding of ubiquitination processes is far from complete, and currently available tools are inadequate to fill in the gaps. For example, elucidation of the E2, E3, or DUB responsible for a specific reaction remains difficult using genetic mutations or small interfering RNA; and monitoring of the dynamics of the reversible ubiquitination processes requires higher levels of spatiotemporal resolution.
000 ubiquitination sites in human cells.11 More recently, a Ub clipping method was developed to map Ubs in vivo, showing that branched Ubs account for 10–20% of the total Ub abundance.12 Despite these advances, understanding of ubiquitination processes is far from complete, and currently available tools are inadequate to fill in the gaps. For example, elucidation of the E2, E3, or DUB responsible for a specific reaction remains difficult using genetic mutations or small interfering RNA; and monitoring of the dynamics of the reversible ubiquitination processes requires higher levels of spatiotemporal resolution.
To supplement canonical methods for the study of ubiquitination, Ub-based chemical probes have been developed to capture or monitor Ub-related enzymes and interactors either covalently or non-covalently.13 Ub-based chemical probes usually comprise a reactive group, a reporting group, and a Ub conjugate module (Fig. 2). The Ub conjugate module contains either a monomeric Ub, Ub chain, or ubiquitinated substrate protein. The reactive group can be used to capture or enrich the Ub enzymes and interactors. The reporting group is used for visualization and/or identification.
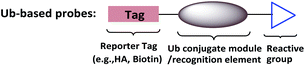 | ||
| Fig. 2 Ub-based chemical probes usually comprise a Ub conjugate module, a reactive group and a reporting group. | ||
Ub-based chemical probes have been demonstrated to be effective tools for discovering and monitoring Ub-related enzymes and interactors.14–16 They can also be used to study the mechanism of the ubiquitination or deubiquitination event.17–19 Further development of Ub-based chemical probes with expanded functions and enhanced sensitivity is an important area in Ub research. This article aims to review the different classes of Ub-based chemical probes and their applications.
2. Designs of Ub-based chemical probes
2.1 Probes capturing enzyme active sites
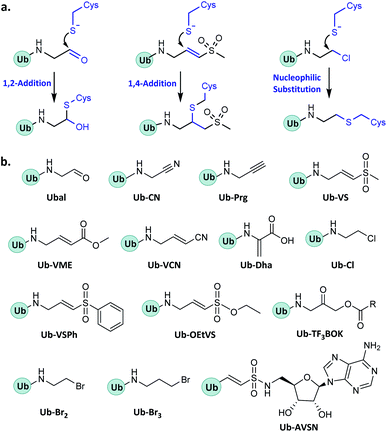
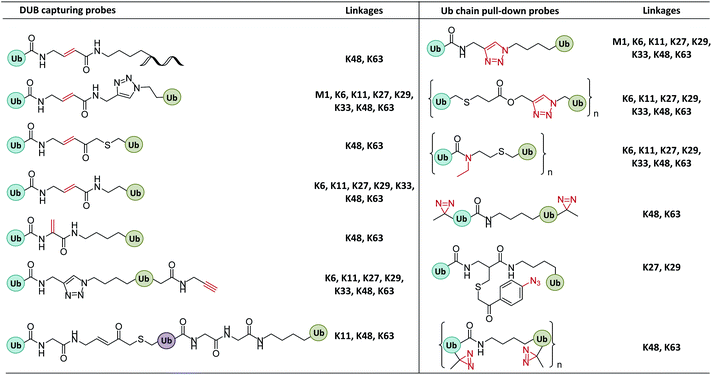
In enzymatic ubiquitination and deubiquitination processes, the catalytic site is usually an active Cys. Strategies to capture the active Cys commonly rely on nucleophilic addition or substitution reactions (Fig. 3).2.2 Probes capturing Ub interactors
The recruitment and binding of different Ubs by the Ub interactors is usually non-covalent in nature. The development of Ub-based chemical probes to capture or enrich Ub interactors can inform the study of these recognition events.3. Synthesis of Ub-based chemical probes
Synthesis of the Ub-based chemical probes usually involves two steps: first, synthesis of Ub conjugate module; second, incorporation of the active group. Ub conjugates are usually difficult to obtain through direct recombinant expression, and therefore need to be synthesized through chemical means.3.1 Total chemical synthesis of Ub conjugate module
In the total chemical synthesis of Ub conjugates, the target Ub conjugate is usually divided into separate segments at a site close to the isopeptide. Ub conjugates are generally synthesized from a donor Ub thioester and a substrate protein bearing a “Cys-like” auxiliary group at or close to the isopeptide Lys. After their ligation, the “Cys-like” auxiliary is removed from the ligated product to yield the target Ub conjugate45 (Fig. 5).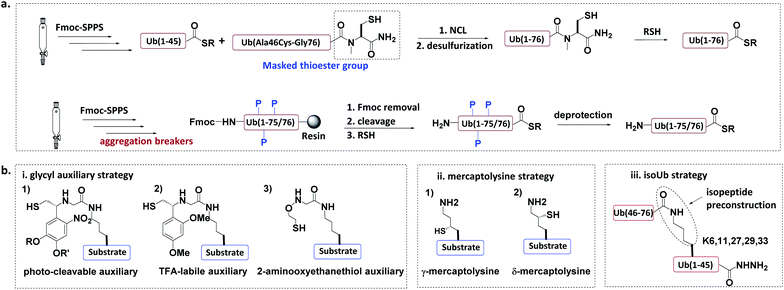 | ||
| Fig. 5 Chemical synthesis of Ub conjugate. (a) Total chemical synthesis of Ub conjugate. (b) Construction of isopeptide bonds. | ||
Another approach for the synthesis of long peptides on the solid support is to incorporate “aggregation breakers” such as pseudoproline and dimethoxybenzyl dipeptides,62 both of which disrupt the aggregation of the nascent peptide on the resin surface. Using this strategy, Ovaa et al. successfully synthesized Ub thioesters and even SUMO2 (92 amino acids).63
3.1.2.1 Glycyl auxiliary strategy. A photo-cleavable thiol-containing glycyl auxiliary can be introduced to the Lys of the substrate protein through SPPS.64 The auxiliary reacts with the Ub-thioester through NCL, and then the auxiliary can be selectively removed via photolysis to generate the desired ubiquitinated substrate. Two alternative auxiliaries are 1-(2,4-dimethoxyphenyl)-2-mercaptoethyl, which can be removed by trifluoroacetic acid after ligation;63,65 and a 2-aminooxy-ethanethiol auxiliary, which exhibits faster ligation.66
3.1.2.2 Mercaptolysine strategy. Brik and Liu groups designed δ- and γ-mercaptolysine and incorporated them into substrate proteins.67,68 Both δ- and γ-mercaptolysine can be ligated with a Ub-thioester, leading to the formation of an isopeptide bond after desulfurization. Compared to the glycyl-auxiliary strategy, the mercaptolysine uses a primary amino group to participate in the ligation and has a faster ligation speed.
3.1.2.3 IsoUb strategy.69. This strategy employs the 76-residue isoUb unit, which is made from two adjacent Ub segments, each of which contains an N-terminal Cys and a C-terminal hydrazide to facilitate ligation. Multiple isoUb units can be assembled through sequential hydrazide-based NCL. Unlike native Ub, the isoUb unit does not aggregate, and can therefore be readily synthesized. Furthermore, the ligation of isoUb takes place at Cys with less hydrolysis by-products. Using the isoUb strategy, a K11/K48-branched hexa-Ub (456 amino acids) was synthesized.
3.2 Protein semi-synthesis of Ub conjugate module
The total chemical synthesis may present technical challenges to some biochemistry-oriented laboratories. Protein semi-synthesis methods may be a useful alternate.70–74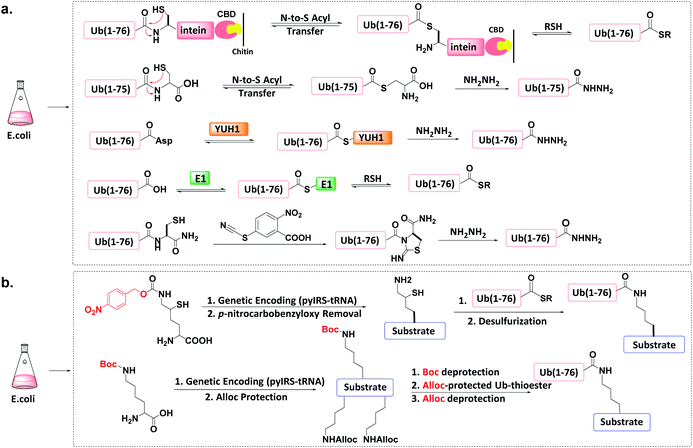 | ||
| Fig. 6 Protein semi-synthesis of Ub conjugate. (a) Recombinant production of Ub thioesters. (b) Recombinant introduction of isopeptide bonds. | ||
Other methods can produce recombinant Ub-hydrazide (as a Ub thioester equivalent). First, Macmillan et al. discovered that peptidyl C-terminal Cys can undergo N-to-S acyl transfer to generate a transient thioester.77 After adding hydrazine to Ub(1-76C), N-to-S acyl transfer generates Ub(1-75)-hydrazide with a yield of ca. 50 mg L−1 of LB medium. Second, Liu et al. discovered that the Ub hydrolase YUH1 can hydrolyse Ub analogues to form thioester intermediates.78 By appending Asp to the Ub C-terminal and treating Ub(1-77D) with YUH1 and hydrazine, Ub(1-76)-hydrazide was obtained in a high yield (ca. 30–40 mg L−1 of LB medium). Third, the transesterification of E1 and Ub to form an active E1-Ub thioester can be intercepted by the addition of thiol, leading to the formation of Ub (1-76)-thioester in good yields (e.g., 50 mg L−1).36,79,80 Finally, Liu et al. found that a small molecule cyanylating reagent (2-nitro-5-thiocyanatobenzoic acid) can modify a recombinant protein at its C-terminus via nucleophilic acyl substitution, generating a protein hydrazide if the nucleophile is hydrazine.81
An unnatural amino acid strategy has also been used to introduce a side-chain Boc-protected Lys into the substrate protein.83 The remaining free amines on the substrate protein and on the donor Ub-thioester are chemically masked by another protecting group such as allyloxycarbonyl (Alloc). After Boc deprotection, Ub-thioester was reacted with the substrate protein forming an isopeptide bond through Ag-catalysed thioester-amine condensation. Finally, the removal of Alloc led to the formation of the target Ub conjugate.
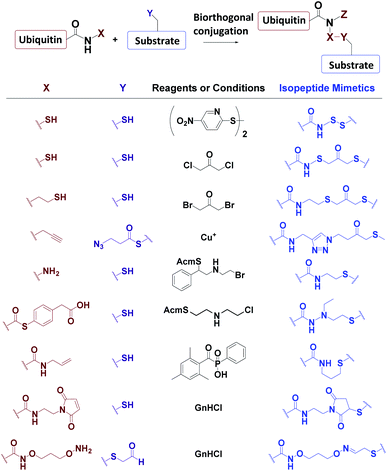
3.3 Biorthogonal synthesis of mimic Ub conjugates
Ub conjugates incorporating isopeptide bond mimetics can also be synthesized from the expressed Ub70–73,84,85 (Fig. 7).3.4 Incorporation of chemically reactive groups
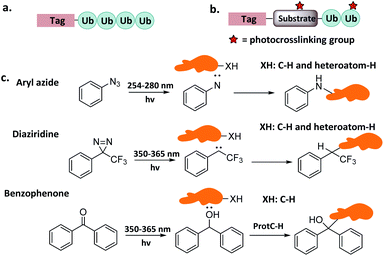
Ub-based chemical probes incorporate two types of active groups: one to capture the active Cys residue, and the other to capture protein-interacting interface amino acids.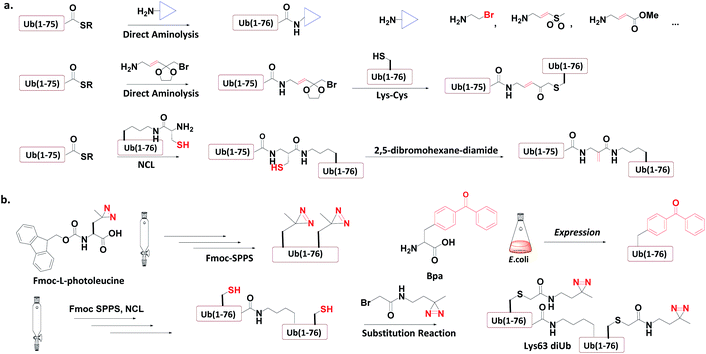 | ||
| Fig. 8 Incorporation of chemically reactive groups. (a) Strategies to capture the Cys residue. (b) Strategies to incorporate photocrosslinking groups. | ||
3.4.1.1 Direct aminolysis. Kessler et al. used the protein semi-synthesis method to prepare an HA-labelled Ub-thioester and then introduced the active group to the C-terminus of Ub through direct aminolysis.22 This approach was used to make a series of monoUb active probes including Ub-Michael acceptor and Ub-halides. Zhuang et al. attached a bifunctional linker containing a masked Michael acceptor at the C-terminus of the remote Ub thioester.96 The ketal group in the linker was then deprotected to generate an α-bromo ketone. After a thiol substitution reaction was carried out with the α-bromo ketone intermediate, a diUb probe was obtained.
3.4.1.2 NCL. Brik et al. mutated the remote Ub's Gly76 to Cys and loaded it onto the Lys side chain amino group of the receptor Ub through SPPS.24 After ligation with the remote Ub thioester through NCL, 2,5-dibromohexanediamide was used to convert the Cys76 residue to Dha. In this manner, a diUb probe was obtained. Ovaa et al. designed an NCL ligation handle, which allows the ligation of Ub onto a protein. This building block was loaded on the proximal Ub and then reacted with Ub(1-75)-thioester to construct via NCL. Through in situ selective thiol elimination, a diUb probe was constructed.
3.4.2.1 Direct SPPS. Ovaa et al. introduced photoleucine containing a photocrosslinking group into the sequence of Ub through SPPS, and then enzymatically assembled it into a Ub chain to generate a photocrosslinking polyUb probe.97
3.4.2.2 Insertion of unnatural amino acids. Virdee et al. used an evolved engineered pair of tRNA and aminoacyl-tRNA synthetase to embed p-benzoyl-L-phenylalanine (Bpa) into the sequence of Ub.41 This Ub mutant was covalently put onto the active Cys residue-to-Lys mutated E2 to construct an E2-Ub photocrosslinking probe.
3.4.3.3 Post-ligation thiol modification. Tian et al. mutated Ub's Ile44 to Cys and obtained a diUb conjugate through hydrazide based NCL.98 The two Cys residues of the synthetic conjugate were then modified with photocrosslinking groups (diazirine or aryl azide) through selective substitution reaction, leading to the generation of a diUb photocrosslinking probe.
4. Applications of Ub-based chemical probes
4.1 MonoUb/UbL probes
The covalent product of Ub-CN and DUB was not compatible with the reducing conditions of SDS/PAGE. In 2001, Ploegh et al. developed Ub-VS that can form irreversible adducts.15 This probe reacted efficiently with UCH-L3, and also showed high activity with an array of Ub carboxyl terminal hydrolases such as Ubp1, Ubp2, Ubp6, Ubp15, and Yuh1. Using this probe, a unique DUB, i.e. USP14, was identified in the mammalian 26S proteasome. In 2002, more monoUb probes were developed, including HA-Ub-Cl, HA-Ub-Br2, HA-Ub-Br3, HA-Ub-VS, HA-Ub-VME, HA-Ub-VSPh, and HA-Ub-VCN.22 These probes exhibited distinct DUB profiling, enabling the identification of DUBs by affinity-tag based mass proteomics (AP-MS). A total of 23 active DUBs were identified in EL4 cells. HSPC263, a protein labelled by HA-Ub-Br2, had no sequence homology with known DUB and was determined to belong to a new OUT family of DUB.
To overcome false positives caused by non-covalent interactions and non-selective capturing, Wertz et al. developed a ‘reactive-site-centric chemoproteomics’ method for the detection of probe-labelled residues by enhanced monoUb-probes. A C-terminal alkyne was added and attached with a cleavable biotin-azide tag via click reaction.102 Using the new probe, a previously unannotated DUB, ZUFSP with high Lys63-linked specificity was identified.103
Finally, mono-Ub probes can be used for the identification of DUBs in defined cellular compartments.104 For instance, HA-Ub-VS was used to study the function of USP7 during lipogenesis. Transcriptional coregulator Tip60, whose expression is regulated by polyubiquitination on multiple sites, plays a key role in adipocyte differentiation. Combining activity monitoring using Ub-VS and further in vitro and in vivo experiments, it was found that early adipogenesis is regulated through USP7-mediated de-ubiquitination of the Tip60.105
Another study elucidated the extent to which activation and inhibition of UCH-L5 was tuned by its two adaptor RPN13 and INO80. Both adaptors bind to UCH-L5 as demonstrated by the crystal structures.18 However, the consequences of binding are different: RNP13 exerts activation activity, whereas INO80 inhibits activity. To explain regulatory mechanisms, Ub-Prg was used to capture the catalytic conformation by which RPN13DEUBAD activates UCH-L5; and the catalytic conformation that truncated INO80DEUBAD lacks to inhibit UCH-L5. On the basis of multiple intermediate structures, it was found that RPN13DEUBAD activates UCH-L5 by positioning its domains while INO80DEUBAD inhibits UCH-L5 by blocking Ub binding.18
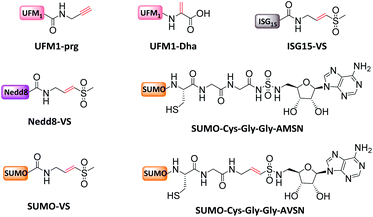
Finally, monoUb probes can also be employed to study the mechanism for activation and thioester bond formation in E1s. Two mono-Ub probes were used to capture E1 intermediates,31 including Ub-AMSN probe to mimic the adenylate intermediate, and Ub-AVSN to mimic the tetrahedral intermediate. These two structures have shown significant conformational changes, from an open conformation before release of pyrophosphate to a closed conformation required for thioester bond formation.19 Similar probes have also been applied to capture Sumo E1's active intermediates and study structural mechanism.19
Mootz et al. prepared a SUMO-1 photo-crosslinking probe by introducing Bpa at the Arg50 of Sumo-1.113 The active site-independent probe captured the known effector protein RanBP2 in complex cell lysate, portending its use for profiling the sumo-related pathways of real cell systems.
4.2 Ub/UbL chain probes
To circumvent the steric hindrance of the diUb probes, other diUb probes incorporating a Michael acceptor that more closely resemble native diUbs were developed. For example, Ovaa et al. used K11 and K48 diUb probes to label Cezanne (a K11-specific DUB that regulates cellular inflammation, NF-κB signalling and T cell activation) and found that Cezanne can be exclusively labelled by K11 diUb active probes.62 Brik et al. found that IsoT, USP2, OTUB1, and OTUB2 hydrolyse K48 diUb based on dehyrdoalanine (Dha) probe.24
Nonetheless, activity-based diUb probes cannot readily capture K27-active DUBs due to the steric hindrance of the K27 isopeptide bond. Li et al. reported a photocrosslinking K27 diUb probe bearing an aryl azide group,115 which probe captured K27-selective DUBs from cell lysates (OTUD2 and USP13). A similar photocrosslinking K29-diUb probe also captured K29 specific DUBs (ZRANB1 and OTUD2).
Severe acute respiratory syndrome coronavirus papain-like protease (SARS PLpro) is a DUB which recognizes K48 polyUb chains via at least two binding sites of S2–S1, rather than S1–S1′.116 To elucidate the structural basis of this selectivity, a distal diUbK48 probe was developed to crosslink SARS PLpro, and the covalent complex crystalized and solved at 2.85 Å.117,118 It was found that at least three Ub-binding sites cooperated to cleave K48 Ub. The S1 and S2 sites of PLpro remodelled K48 diUb to an extended conformation, resulting in K48 specificity.
Finally, a triUb probe with well-defined cross-linking site can be used to investigate whether a DUB performs endo- and exo-Ub chain cleavage.35 For example, USP9X, a DUB regulating multiple important cellular processes including apoptosis and stem cell self-renewal, displays multiple Ub-binding sites and selectivity for K11, K48, and K63 Ubs. In order to investigate its cleavage mode, Zhuang et al. developed K11, K48, and K63 tri-Ub probes, in which the active group is located at the isopeptide bond between the distal and middle Ubs. Experiments with these probes revealed that USP9X cleaved the K11, K48, and K63 Ubs in different modes (i.e. endo, exo, and mixed).
In another study of Ub interactors, Stengel et al. developed polyUb probes bearing up to ten Ub units.38 These probes were applied to proteomic detection, from which 70, 44, and 37 proteins were found to be interactors of K27, K29, and K33 Ub, respectively. The authors also used gel eluted liquid fraction entrapment electrophoresis to separate the Ubs into Ub2, Ub4, Ub6+ to examine their interactors separately.39 Significant differences were observed in the interactors of Ubs with different lengths. For example, Ub-associated domain-containing protein 1 (Ubac1), RING finger 123 (RNF123), and USP15 only interact with long Ub chains (Ub4, Ub6+).
In addition, triazole-free DUB-resistant Ub probes were developed containing N-ethyl isopeptide bonds.36 These probes were used to profile the interactome of K29 Ubs, which captured some new interactors not identified by the triazole probes. The main limitation of the Ub pull-down probes is that weak interactors cannot be captured. Tian et al. reported diazirine-based photoaffinity probes that can capture K48- and K63-Ub interactors in cell lysates.98 Meanwhile, Glickman et al. developed photo-crosslinking polyUb probes for detecting proteasome subunits that interact with Ubs.97
4.3 Ub/UbL substrate probes
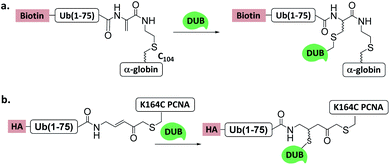 | ||
| Fig. 11 Ub/UbL substrate probes used to capture and/or study DUBs. (a) α-globin-Ub-Dha. (b) K164C Ub-PCNA-MAL. | ||
Another Ub-substrate probe, Ub-PCNA-MAL, was developed by Zhuang et al.40 (Fig. 11b). Two ubiquitination sites on yeast PCNA (K107 and K164) were examined for the probes. Pull-down experiments in yeast lysate and quantitative mass spectrometry analysis showed that both probes could capture a variety of DUBs. Ubp3 and Ubp10 were found to be specific for K164. This finding indicates that DUBs can distinguish ubiquitination sites on a substrate protein.
 | ||
| Fig. 12 Ub/UbL substrate probes. (a) E2-Ub-AVS. (b) E2-Ub-Dha. (c) E2-SUMO. (d) E2-Ub photocrosslinking probe. (e) Ub-nucleosome probe. | ||
More recently, a variant of the probe E2-Ub-AVS was developed, which showed improved labelling efficiency and was applied to profiling HECT or RBR E3s in neuroblastoma SH-SY5Y cell extracts. About 40 HECT/RBR E3s were profiled, consisting of 80% known HECT/RBR ligase. Notably, 33 RING ligases were also enriched, of which a novel RING ligase MYCBP2 was discovered to utilizes a unique RING-Cys relay (RCR) mechanism mediating the transfer of Ub onto the substrate threonine and serine residues. In vitro and in vivo experiments verified NMNAT2 (nicotinamide mononucleotide adenyltransferase) was the substrate of MYCBP2.29
Shi et al. reported an E2-Ub-Dha probe (Fig. 12b), containing UBE2D2, an Ub moiety and a Dha reacting group.121In vitro labelling assay showed that the probe can react with HECT E3 NEDD4 and UBE3C efficiently. In vivo profiling using HeLa cells identified several HECT E3s including NEDD4, UBE3C and HUWE1. Moreover, two RBR E3 (ARIH1, ARIH2) and several RING E3s were also enriched, similar to E2-Ub-AVS probe.
Other Ub substrate probes have been designed to capture RING-type E3s. Unlike HECT/RBR E3s that can be capture by activity-based probes, RING-type E3s do not bear a catalytic Cys. Bode et al. developed a photo-reactive E2-SUMO probe to trap RING type SUMO E3 ligase.42 C93 of SUMO E2 Ubc9 was mutated to 2,3-diaminopropionic acid to form a stable amide-linked E2-SUMO conjugate (Fig. 12c), and diazirine was introduced to Ubc9 F22 to trap E3 ligases. When the probe was incubated with cell lysate, SUMO E3 ligase RanBP2 can be enriched. Moreover, Virdee et al. developed an E2-Ub photo-reactive probe (Fig. 12d), in which Bpa was genetically introduced. The activity of the probe was demonstrated by crosslinking with RNF4 in a SUMO chain dependent manner.
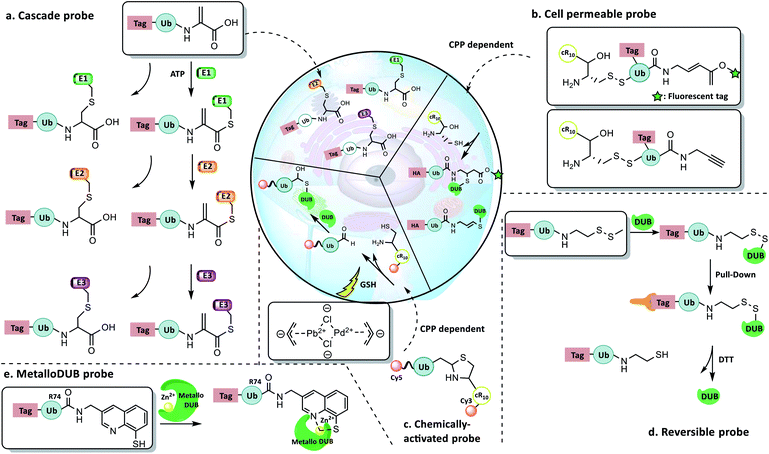
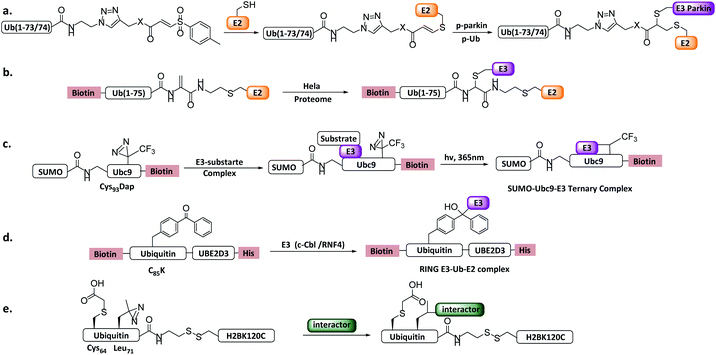
4.4 New concept probes
5. Summary and perspective
This article reviews the development and application of the state-of-the-art Ub-based chemical probes, highlighting the need for innovative technologies and novel concepts to study the ever-increasingly complex facets of Ub biology. Special attention has been given to the use of chemical probes to capture and monitor the enzymes or Ub interactors previously not targeted by conventional probe designs. Progress in this regard will enable a more in-depth dissection of the many enigmatic aspects of ubiquitination in the cells, and elucidation of the mechanisms behind the complex ubiquitination machinery. Furthermore, it should be pointed out that proteomic identification of trapped proteins by the synthetic and semisynthetic probes is still not easy and has become the bottleneck in the field. Also, chemical probes capable of in vivo profiling are also increasingly needed. New Ub-based chemical probes will continue to emerge, inspired both by the innovation and progress of chemical protein synthesis, and the fascinating biology and clinical importance of ubiquitination.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by the National Key R&D Program of China (2017YFA0505200), NSFC (91753205, 21532004, 81621002, and 21621003 for L.L.; 21877024 for Y.-M.L.), and the Fundamental Research Funds for the Central Universities (No. JZ2019HGPB0105).Notes and references
- D. Komander and M. Rape, Annu. Rev. Biochem., 2012, 81, 203–229 CrossRef CAS PubMed.
- K. N. Swatek and D. Komander, Cell Res., 2016, 26, 399–422 CrossRef CAS PubMed.
- A. Hershko and A. Ciechanover, Annu. Rev. Biochem., 1998, 67, 425–479 CrossRef CAS PubMed.
- T. E. T. Mevissen and D. Komander, Annu. Rev. Biochem., 2017, 86, 159–192 CrossRef CAS PubMed.
- D. L. Haakonsen and M. Rape, Trends Cell Biol., 2019, 29, 704–716 CrossRef CAS PubMed.
- R. Yau and M. Rape, Nat. Cell Biol., 2016, 18, 579–586 CrossRef CAS PubMed.
- E. Oh, D. Akopian and M. Rape, Annu. Rev. Cell Biol., 2018, 34, 137–162 CrossRef CAS PubMed.
- R. I. Enchev, B. A. Schulman and M. Peter, Nat. Rev. Mol. Cell Biol., 2015, 16, 30–44 CrossRef CAS PubMed.
- X. Zhao, Mol. Cell, 2018, 71, 409–418 CrossRef CAS PubMed.
- K. I. Kim and D.-E. Zhang, Biochem. Biophys. Res. Commun., 2003, 307, 431–434 CrossRef CAS PubMed.
- W. Kim, E. J. Bennett, E. L. Huttlin, A. Guo, J. Li, A. Possemato, M. E. Sowa, R. Rad, J. Rush, M. J. Comb, J. W. Harper and S. P. Gygi, Mol. Cell, 2011, 44, 325–340 CrossRef CAS PubMed.
- K. N. Swatek, J. L. Usher, A. F. Kueck, C. Gladkova, T. E. T. Mevissen, J. N. Pruneda, T. Skern and D. Komander, Nature, 2019, 572, 533–537 CrossRef CAS PubMed.
- K. F. Witting, M. P. C. Mulder and H. Ovaa, J. Mol. Biol., 2017, 429, 3388–3394 CrossRef CAS PubMed.
- Y. A. Lam, W. Xu, G. N. DeMartino and R. E. Cohen, Nature, 1997, 385, 737–740 CrossRef CAS PubMed.
- A. Borodovsky, B. M. Kessler, R. Casagrande, H. S. Overkleeft, K. D. Wilkinson and H. L. Ploegh, EMBO J., 2001, 20, 5187–5196 CrossRef CAS PubMed.
- J. Hemelaar, A. Borodovsky, B. M. Kessler, D. Reverter, J. Cook, N. Kolli, T. Gan-Erdene, K. D. Wilkinson, G. Gill, C. D. Lima, H. L. Ploegh and H. Ovaa, Mol. Cell. Biol., 2004, 24, 84–95 CrossRef CAS PubMed.
- S. Misaghi, P. J. Galardy, W. J. Meester, H. Ovaa, H. L. Ploegh and R. Gaudet, J. Biol. Chem., 2005, 280, 1512–1520 CrossRef CAS PubMed.
- D. D. Sahtoe, W. J. van Dijk, F. El Oualid, R. Ekkebus, H. Ovaa and T. K. Sixma, Mol. Cell, 2015, 57, 887–900 CrossRef CAS PubMed.
- S. K. Olsen, A. D. Capili, X. Lu, D. S. Tan and C. D. Lima, Nature, 2010, 463, 906–912 CrossRef CAS PubMed.
- R. Ekkebus, S. I. van Kasteren, Y. Kulathu, A. Scholten, I. Berlin, P. P. Geurink, A. de Jong, S. Goerdayal, J. Neefjes, A. J. Heck, D. Komander and H. Ovaa, J. Am. Chem. Soc., 2013, 135, 2867–2870 CrossRef CAS.
- S. Sommer, N. D. Weikart, U. Linne and H. D. Mootz, Bioorg. Med. Chem., 2013, 21, 2511–2517 CrossRef CAS PubMed.
- A. Borodovsky, H. Ovaa, N. Kolli, T. Gan-Erdene, K. D. Wilkinson, H. L. Ploegh and B. M. Kessler, Chem. Biol., 2002, 9, 1149–1159 CrossRef CAS PubMed.
- S. D. Whedon, N. Markandeya, A. Rana, N. A. Senger, C. E. Weller, F. Turecek, E. R. Strieter and C. Chatterjee, J. Am. Chem. Soc., 2016, 138, 13774–13777 CrossRef CAS PubMed.
- N. Haj-Yahya, H. P. Hemantha, R. Meledin, S. Bondalapati, M. Seenaiah and A. Brik, Org. Lett., 2014, 16, 540–543 CrossRef CAS PubMed.
- M. Jbara, S. Laps, M. Morgan, G. Kamnesky, G. Mann, C. Wolberger and A. Brik, Nat. Commun., 2018, 9, 3154 CrossRef PubMed.
- J. Yu, C. Li, B. Li, X. Zhu, R. Zhang, L. Ji, D. Tang, A. M. Asiri, X. Sun, Q. Li, S. Liu and Y. Luo, Chem. Commun., 2019, 55, 6401–6404 RSC.
- K. R. Love, R. K. Pandya, E. Spooner and H. L. Ploegh, ACS Chem. Biol., 2009, 4, 275–287 CrossRef PubMed.
- K. C. Pao, M. Stanley, C. Han, Y. C. Lai, P. Murphy, K. Balk, N. T. Wood, O. Corti, J. C. Corvol, M. M. Muqit and S. Virdee, Nat. Chem. Biol., 2016, 12, 324–331 CrossRef CAS PubMed.
- K. C. Pao, N. T. Wood, A. Knebel, K. Rafie, M. Stanley, P. D. Mabbitt, R. Sundaramoorthy, K. Hofmann, D. M. F. van Aalten and S. Virdee, Nature, 2018, 556, 381–385 CrossRef CAS PubMed.
- R. Meledin, S. M. Mali, O. Kleifeld and A. Brik, Angew. Chem., Int. Ed., 2018, 57, 5645–5649 CrossRef CAS PubMed.
- Z. S. Hann, C. Ji, S. K. Olsen, X. Lu, M. C. Lux, D. S. Tan and C. D. Lima, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 15475–15484 CrossRef CAS.
- V. E. Albrow, E. L. Ponder, D. Fasci, M. Bekes, E. Deu, G. S. Salvesen and M. Bogyo, Chem. Biol., 2011, 18, 722–732 CrossRef CAS PubMed.
- A. Iphofer, A. Kummer, M. Nimtz, A. Ritter, T. Arnold, R. Frank, J. van den Heuvel, B. M. Kessler, L. Jansch and R. Franke, ChemBioChem, 2012, 13, 1416–1420 CrossRef.
- T. E. T. Mevissen, Y. Kulathu, M. P. C. Mulder, P. P. Geurink, S. L. Maslen, M. Gersch, P. R. Elliott, J. E. Burke, B. D. M. van Tol, M. Akutsu, F. E. Oualid, M. Kawasaki, S. M. V. Freund, H. Ovaa and D. Komander, Nature, 2016, 538, 402–405 CrossRef CAS PubMed.
- P. Paudel, Q. Zhang, C. Leung, H. C. Greenberg, Y. Guo, Y. H. Chern, A. Dong, Y. Li, M. Vedadi, Z. Zhuang and Y. Tong, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 7288–7297 CrossRef CAS.
- Q. Zheng, T. Wang, G. C. Chu, C. Zuo, R. Zhao, X. Sui, L. Ye, Y. Yu, J. Chen, X. Wu, W. Zhang, H. Deng, J. Shi, M. Pan, Y. M. Li and L. Liu, Angew. Chem., Int. Ed., 2020 DOI:10.1002/anie.202002974.
- X. Zhang, A. H. Smits, G. B. van Tilburg, P. W. Jansen, M. M. Makowski, H. Ovaa and M. Vermeulen, Mol. Cell, 2017, 65, 941–955.e948 CrossRef CAS PubMed.
- X. Zhao, J. Lutz, E. Hollmuller, M. Scheffner, A. Marx and F. Stengel, Angew. Chem., Int. Ed., 2017, 56, 15764–15768 CrossRef CAS PubMed.
- J. Lutz, E. Hollmuller, M. Scheffner, A. Marx and F. Stengel, Angew. Chem., Int. Ed., 2020, 59, 12371–12375 CrossRef PubMed.
- P. Gong, G. A. Davidson, W. Gui, K. Yang, W. P. Bozza and Z. Zhuang, Chem. Sci., 2018, 9, 7859–7865 RSC.
- S. Mathur, A. J. Fletcher, E. Branigan, R. T. Hay and S. Virdee, Cell Chem. Biol., 2020, 27, 74–82.e76 CrossRef CAS PubMed.
- Y. Zhang, T. Hirota, K. Kuwata, S. Oishi, S. G. Gramani and J. W. Bode, J. Am. Chem. Soc., 2019, 141, 14742–14751 CrossRef CAS PubMed.
- L. Zhou, M. T. Holt, N. Ohashi, A. Zhao, M. M. Muller, B. Wang and T. W. Muir, Nat. Commun., 2016, 7, 10589 CrossRef CAS PubMed.
- D. P. Murale, S. C. Hong, M. M. Haque and J. S. Lee, Proteome Sci., 2016, 15, 14 CrossRef PubMed.
- R. Yang and C. F. Liu, Top. Curr. Chem., 2015, 362, 89–106 CrossRef CAS.
- R. Ingenito, E. Bianchi, D. Fattori and A. Pessi, J. Am. Chem. Soc., 1999, 121, 11369–11374 CrossRef CAS.
- J. B. Blanco-Canosa and P. E. Dawson, Angew. Chem., Int. Ed., 2008, 47, 6851–6855 CrossRef CAS PubMed.
- A. P. Tofteng, K. K. Sorensen, K. W. Conde-Frieboes, T. Hoeg-Jensen and K. J. Jensen, Angew. Chem., Int. Ed., 2009, 48, 7411–7414 CrossRef CAS PubMed.
- J. S. Zheng, H. N. Chang, F. L. Wang and L. Liu, J. Am. Chem. Soc., 2011, 133, 11080–11083 CrossRef CAS PubMed.
- K. S. Kumar, L. Spasser, L. A. Erlich, S. N. Bavikar and A. Brik, Angew. Chem., Int. Ed., 2010, 49, 9126–9131 CrossRef CAS PubMed.
- M. Pan, S. Gao, Y. Zheng, X. Tan, H. Lan, X. Tan, D. Sun, L. Lu, T. Wang, Q. Zheng, Y. Huang, J. Wang and L. Liu, J. Am. Chem. Soc., 2016, 138, 7429–7435 CrossRef CAS PubMed.
- S. Gao, M. Pan, Y. Zheng, Y. Huang, Q. Zheng, D. Sun, L. Lu, X. Tan, X. Tan, H. Lan, J. Wang, T. Wang, J. Wang and L. Liu, J. Am. Chem. Soc., 2016, 138, 14497–14502 CrossRef CAS PubMed.
- G. M. Fang, Y. M. Li, F. Shen, Y. C. Huang, J. B. Li, Y. Lin, H. K. Cui and L. Liu, Angew. Chem., Int. Ed., 2011, 50, 7645–7649 CrossRef CAS PubMed.
- G. M. Fang, J. X. Wang and L. Liu, Angew. Chem., Int. Ed., 2012, 51, 10347–10350 CrossRef CAS PubMed.
- J.-S. Zheng, S. Tang, Y.-K. Qi, Z.-P. Wang and L. Liu, Nat. Protoc., 2013, 8, 2483 CrossRef CAS PubMed.
- J. Li, Y. Li, Q. He, Y. Li, H. Li and L. Liu, Org. Biomol. Chem., 2014, 12, 5435–5441 RSC.
- Y. M. Li, M. Y. Yang, Y. C. Huang, Y. T. Li, P. R. Chen and L. Liu, ACS Chem. Biol., 2012, 7, 1015–1022 CrossRef CAS PubMed.
- M. Pan, Q. Zheng, S. Ding, L. Zhang, Q. Qu, T. Wang, D. Hong, Y. Ren, L. Liang, C. Chen, Z. Mei and L. Liu, Angew. Chem., Int. Ed., 2019, 58, 2627–2631 CrossRef CAS PubMed.
- H. Lan, K. Wu, Y. Zheng, M. Pan, Y. C. Huang, S. Gao, Q. Y. Zheng, J. S. Zheng, Y. M. Li, B. Xiao and L. Liu, J. Pept. Sci., 2016, 22, 320–326 CrossRef CAS PubMed.
- H. Ai, Y. Guo, D. Sun, S. Liu, Y. Qi, J. Guo, Q. Qu, Q. Gong, S. Zhao, J. Li and L. Liu, ChemBioChem, 2019, 20, 221–229 CrossRef CAS PubMed.
- M. Pan, Q. Zheng, S. Gao, Q. Qu, Y. Yu, M. Wu, H. Lan, Y. Li, S. Liu, J. Li, D. Sun, L. Lu, T. Wang, W. Zhang, J. Wang, Y. Li, H.-G. Hu, C. Tian and L. Liu, CCS Chem., 2019, 1, 476–489 CrossRef CAS.
- M. P. Mulder, F. El Oualid, J. ter Beek and H. Ovaa, ChemBioChem, 2014, 15, 946–949 CrossRef CAS PubMed.
- M. P. C. Mulder, R. Merkx, K. F. Witting, D. S. Hameed, D. El Atmioui, L. Lelieveld, F. Liebelt, J. Neefjes, I. Berlin, A. C. O. Vertegaal and H. Ovaa, Angew. Chem., Int. Ed., 2018, 57, 8958–8962 CrossRef CAS PubMed.
- C. Chatterjee, R. K. McGinty, J. P. Pellois and T. W. Muir, Angew. Chem., Int. Ed., 2007, 46, 2814–2818 CrossRef CAS PubMed.
- R. Yang, X. Bi, F. Li, Y. Cao and C. F. Liu, Chem. Commun., 2014, 50, 7971–7974 RSC.
- C. E. Weller, W. Huang and C. Chatterjee, ChemBioChem, 2014, 15, 1263–1267 CrossRef CAS PubMed.
- K. S. Ajish Kumar, M. Haj-Yahya, D. Olschewski, H. A. Lashuel and A. Brik, Angew. Chem., Int. Ed., 2009, 48, 8090–8094 CrossRef CAS.
- R. Yang, K. K. Pasunooti, F. Li, X. W. Liu and C. F. Liu, J. Am. Chem. Soc., 2009, 131, 13592–13593 CrossRef CAS PubMed.
- S. Tang, L. J. Liang, Y. Y. Si, S. Gao, J. X. Wang, J. Liang, Z. Mei, J. S. Zheng and L. Liu, Angew. Chem., Int. Ed., 2017, 56, 13333–13337 CrossRef CAS PubMed.
- Y.-K. Qi, Y.-Y. Si, S.-S. Du, J. Liang, K.-W. Wang and J.-S. Zheng, Sci. China: Chem., 2019, 62, 299–312 CrossRef CAS.
- R. E. Thompson and T. W. Muir, Chem. Rev., 2020, 120, 3051–3126 CrossRef CAS PubMed.
- S. M. Mali, S. K. Singh, E. Eid and A. Brik, J. Am. Chem. Soc., 2017, 139, 4971–4986 CrossRef CAS PubMed.
- X. Bi, K. K. Pasunooti and C.-F. Liu, Sci. China: Chem., 2018, 61, 251–265 CrossRef CAS.
- Y. Si, L. Liang, S. Tang, Y. Qi, Y. Huang and L. Liu, Sci. China: Chem., 2018, 61, 412–417 CrossRef CAS.
- T. W. Muir, D. Sondhi and P. A. Cole, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 6705–6710 CrossRef CAS PubMed.
- J. Shi and T. W. Muir, J. Am. Chem. Soc., 2005, 127, 6198–6206 CrossRef CAS PubMed.
- A. L. Adams, B. Cowper, R. E. Morgan, B. Premdjee, S. Caddick and D. Macmillan, Angew. Chem., Int. Ed., 2013, 52, 13062–13066 CrossRef CAS PubMed.
- G. C. Chu, M. Pan, J. Li, S. Liu, C. Zuo, Z. B. Tong, J. S. Bai, Q. Gong, H. Ai, J. Fan, X. Meng, Y. C. Huang, J. Shi, H. Deng, C. Tian, Y. M. Li and L. Liu, J. Am. Chem. Soc., 2019, 141, 3654–3663 CrossRef CAS PubMed.
- O. N. Burchak, M. Jaquinod, C. Cottin, L. Mugherli, K. Iwai, F. Chatelain and M. Y. Balakirev, ChemBioChem, 2006, 7, 1667–1669 CrossRef CAS PubMed.
- F. El Oualid, R. Merkx, R. Ekkebus, D. S. Hameed, J. J. Smit, A. de Jong, H. Hilkmann, T. K. Sixma and H. Ovaa, Angew. Chem., Int. Ed., 2010, 49, 10149–10153 CrossRef CAS PubMed.
- Y. Qiao, G. Yu, K. C. Kratch, X. A. Wang, W. W. Wang, S. Z. Leeuwon, S. Xu, J. S. Morse and W. R. Liu, J. Am. Chem. Soc., 2020, 142, 7047–7054 CrossRef CAS PubMed.
- S. Virdee, P. B. Kapadnis, T. Elliott, K. Lang, J. Madrzak, D. P. Nguyen, L. Riechmann and J. W. Chin, J. Am. Chem. Soc., 2011, 133, 10708–10711 CrossRef CAS PubMed.
- C. Castaneda, J. Liu, A. Chaturvedi, U. Nowicka, T. A. Cropp and D. Fushman, J. Am. Chem. Soc., 2011, 133, 17855–17868 CrossRef CAS PubMed.
- Y. K. Qi, H. S. Ai, Y. M. Li and B. Yan, Front. Chem., 2018, 6, 19 CrossRef PubMed.
- L. Lu, Y. Guo, T. Wang, L. Liang, S. Zhao, F. Wang and L. Liu, Sci. China: Chem., 2020, 63, 237–243 CrossRef CAS.
- C. Chatterjee, R. K. McGinty, B. Fierz and T. W. Muir, Nat. Chem. Biol., 2010, 6, 267–269 CrossRef CAS PubMed.
- J. Chen, Y. Ai, J. Wang, L. Haracska and Z. Zhuang, Nat. Chem. Biol., 2010, 6, 270–272 CrossRef CAS PubMed.
- L. Yin, B. Krantz, N. S. Russell, S. Deshpande and K. D. Wilkinson, Biochemistry, 2000, 39, 10001–10010 CrossRef CAS PubMed.
- M. T. Morgan, M. Haj-Yahya, A. E. Ringel, P. Bandi, A. Brik and C. Wolberger, Science, 2016, 351, 725–728 CrossRef CAS PubMed.
- Y. E. Lewis, T. Abeywardana, Y. H. Lin, A. Galesic and M. R. Pratt, ACS Chem. Biol., 2016, 11, 931–942 CrossRef CAS PubMed.
- S. Eger, M. Scheffner, A. Marx and M. Rubini, J. Am. Chem. Soc., 2010, 132, 16337–16339 CrossRef CAS PubMed.
- V. H. Trang, E. M. Valkevich, S. Minami, Y. C. Chen, Y. Ge and E. R. Strieter, Angew. Chem., Int. Ed., 2012, 51, 13085–13088 CrossRef CAS PubMed.
- E. M. Valkevich, R. G. Guenette, N. A. Sanchez, Y. C. Chen, Y. Ge and E. R. Strieter, J. Am. Chem. Soc., 2012, 134, 6916–6919 CrossRef CAS PubMed.
- H. P. Hemantha, S. N. Bavikar, Y. Herman-Bachinsky, N. Haj-Yahya, S. Bondalapati, A. Ciechanover and A. Brik, J. Am. Chem. Soc., 2014, 136, 2665–2673 CrossRef CAS PubMed.
- S. K. Singh, I. Sahu, S. M. Mali, H. P. Hemantha, O. Kleifeld, M. H. Glickman and A. Brik, J. Am. Chem. Soc., 2016, 138, 16004–16015 CrossRef CAS PubMed.
- G. Li, Q. Liang, P. Gong, A. H. Tencer and Z. Zhuang, Chem. Commun., 2014, 50, 216–218 RSC.
- M. Chojnacki, W. Mansour, D. S. Hameed, R. K. Singh, F. El Oualid, R. Rosenzweig, M. A. Nakasone, Z. Yu, F. Glaser, L. E. Kay, D. Fushman, H. Ovaa and M. H. Glickman, Cell Chem. Biol., 2017, 24, 443–457.e446 CrossRef CAS PubMed.
- J. Liang, L. Zhang, X. L. Tan, Y. K. Qi, S. Feng, H. Deng, Y. Yan, J. S. Zheng, L. Liu and C. L. Tian, Angew. Chem., Int. Ed., 2017, 56, 2744–2748 CrossRef CAS PubMed.
- A. Hershko and I. A. Rose, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 1829–1833 CrossRef CAS PubMed.
- T. Li, N. I. Naqvi, H. Yang and T. S. Teo, Biochem. Biophys. Res. Commun., 2000, 272, 270–275 CrossRef CAS PubMed.
- H. Holzl, B. Kapelari, J. Kellermann, E. Seemuller, M. Sumegi, A. Udvardy, O. Medalia, J. Sperling, S. A. Muller, A. Engel and W. Baumeister, J. Cell Biol., 2000, 150, 119–130 CrossRef CAS PubMed.
- D. S. Hewings, J. Heideker, T. P. Ma, A. P. AhYoung, F. El Oualid, A. Amore, G. T. Costakes, D. Kirchhofer, B. Brasher, T. Pillow, N. Popovych, T. Maurer, C. Schwerdtfeger, W. F. Forrest, K. Yu, J. Flygare, M. Bogyo and I. E. Wertz, Nat. Commun., 2018, 9, 1162 CrossRef PubMed.
- P. Haahr, N. Borgermann, X. Guo, D. Typas, D. Achuthankutty, S. Hoffmann, R. Shearer, T. K. Sixma and N. Mailand, Mol. Cell, 2018, 70, 165–174.e166 CrossRef CAS PubMed.
- M. Y. Balakirev, S. O. Tcherniuk, M. Jaquinod and J. Chroboczek, EMBO Rep., 2003, 4, 517–522 CrossRef CAS PubMed.
- Y. Gao, A. Koppen, M. Rakhshandehroo, I. Tasdelen, S. F. van de Graaf, J. van Loosdregt, O. van Beekum, N. Hamers, D. van Leenen, C. R. Berkers, R. Berger, F. C. Holstege, P. J. Coffer, A. B. Brenkman, H. Ovaa and E. Kalkhoven, Nat. Commun., 2013, 4, 2656 CrossRef PubMed.
- L. Kategaya, P. Di Lello, L. Rouge, R. Pastor, K. R. Clark, J. Drummond, T. Kleinheinz, E. Lin, J. P. Upton, S. Prakash, J. Heideker, M. McCleland, M. S. Ritorto, D. R. Alessi, M. Trost, T. W. Bainbridge, M. C. M. Kwok, T. P. Ma, Z. Stiffler, B. Brasher, Y. Tang, P. Jaishankar, B. R. Hearn, A. R. Renslo, M. R. Arkin, F. Cohen, K. Yu, F. Peale, F. Gnad, M. T. Chang, C. Klijn, E. Blackwood, S. E. Martin, W. F. Forrest, J. A. Ernst, C. Ndubaku, X. Wang, M. H. Beresini, V. Tsui, C. Schwerdtfeger, R. A. Blake, J. Murray, T. Maurer and I. E. Wertz, Nature, 2017, 550, 534–538 CrossRef CAS PubMed.
- A. P. Turnbull, S. Ioannidis, W. W. Krajewski, A. Pinto-Fernandez, C. Heride, A. C. L. Martin, L. M. Tonkin, E. C. Townsend, S. M. Buker, D. R. Lancia, J. A. Caravella, A. V. Toms, T. M. Charlton, J. Lahdenranta, E. Wilker, B. C. Follows, N. J. Evans, L. Stead, C. Alli, V. V. Zarayskiy, A. C. Talbot, A. J. Buckmelter, M. Wang, C. L. McKinnon, F. Saab, J. F. McGouran, H. Century, M. Gersch, M. S. Pittman, C. G. Marshall, T. M. Raynham, M. Simcox, L. M. D. Stewart, S. B. McLoughlin, J. A. Escobedo, K. W. Bair, C. J. Dinsmore, T. R. Hammonds, S. Kim, S. Urbe, M. J. Clague, B. M. Kessler and D. Komander, Nature, 2017, 550, 481–486 CrossRef CAS PubMed.
- G. Gavory, C. R. O'Dowd, M. D. Helm, J. Flasz, E. Arkoudis, A. Dossang, C. Hughes, E. Cassidy, K. McClelland, E. Odrzywol, N. Page, O. Barker, H. Miel and T. Harrison, Nat. Chem. Biol., 2018, 14, 118–125 CrossRef CAS PubMed.
- L. C. Dang, F. D. Melandri and R. L. Stein, Biochemistry, 1998, 37, 1868–1879 CrossRef CAS PubMed.
- U. Hassiepen, U. Eidhoff, G. Meder, J. F. Bulber, A. Hein, U. Bodendorf, E. Lorthiois and B. Martoglio, Anal. Biochem., 2007, 371, 201–207 CrossRef CAS PubMed.
- Z. R. Zhou, Y. H. Zhang, S. Liu, A. X. Song and H. Y. Hu, Biochem. J., 2012, 441, 143–149 CrossRef CAS PubMed.
- K. F. Witting, G. J. van der Heden van Noort, C. Kofoed, C. Talavera Ormeno, D. El Atmioui, M. P. C. Mulder and H. Ovaa, Angew. Chem., Int. Ed., 2018, 57, 14164–14168 CrossRef CAS PubMed.
- K. F. Taupitz, W. Dorner and H. D. Mootz, Chem.–Eur. J., 2017, 23, 5978–5982 CrossRef CAS PubMed.
- J. F. McGouran, S. R. Gaertner, M. Altun, H. B. Kramer and B. M. Kessler, Chem. Biol., 2013, 20, 1447–1455 CrossRef CAS PubMed.
- X. D. Tan, M. Pan, S. Gao, Y. Zheng, J. Shi and Y. M. Li, Chem. Commun., 2017, 53, 10208–10211 RSC.
- M. Bekes, W. Rut, P. Kasperkiewicz, M. P. Mulder, H. Ovaa, M. Drag, C. D. Lima and T. T. Huang, Biochem. J., 2015, 468, 215–226 CrossRef CAS PubMed.
- D. Flierman, G. J. van der Heden van Noort, R. Ekkebus, P. P. Geurink, T. E. Mevissen, M. K. Hospenthal, D. Komander and H. Ovaa, Cell Chem. Biol., 2016, 23, 472–482 CrossRef CAS PubMed.
- M. Bekes, G. J. van der Heden van Noort, R. Ekkebus, H. Ovaa, T. T. Huang and C. D. Lima, Mol. Cell, 2016, 62, 572–585 CrossRef CAS PubMed.
- Y. Ye, G. Blaser, M. H. Horrocks, M. J. Ruedas-Rama, S. Ibrahim, A. A. Zhukov, A. Orte, D. Klenerman, S. E. Jackson and D. Komander, Nature, 2012, 492, 266–270 CrossRef CAS PubMed.
- R. Byrne, T. Mund and J. D. F. Licchesi, ChemBioChem, 2017, 18, 1415–1427 CrossRef CAS PubMed.
- L. Xu, J. Fan, Y. Wang, Z. Zhang, Y. Fu, Y. M. Li and J. Shi, Chem. Commun., 2019, 55, 7109–7112 RSC.
- S. D. Briggs, T. Xiao, Z. W. Sun, J. A. Caldwell, J. Shabanowitz, D. F. Hunt, C. D. Allis and B. D. Strahl, Nature, 2002, 418, 498 CrossRef CAS PubMed.
- R. K. McGinty, J. Kim, C. Chatterjee, R. G. Roeder and T. W. Muir, Nature, 2008, 453, 812–816 CrossRef CAS PubMed.
- M. P. Mulder, K. Witting, I. Berlin, J. N. Pruneda, K. P. Wu, J. G. Chang, R. Merkx, J. Bialas, M. Groettrup, A. C. Vertegaal, B. A. Schulman, D. Komander, J. Neefjes, F. El Oualid and H. Ovaa, Nat. Chem. Biol., 2016, 12, 523–530 CrossRef CAS PubMed.
- W. Gui, C. A. Ott, K. Yang, J. S. Chung, S. Shen and Z. Zhuang, J. Am. Chem. Soc., 2018, 140, 12424–12433 CrossRef CAS PubMed.
- D. S. Hameed, A. Sapmaz, L. Gjonaj, R. Merkx and H. Ovaa, ChemBioChem, 2018, 19, 2553–2557 CrossRef CAS PubMed.
- G. Mann, G. Satish, R. Meledin, G. B. Vamisetti and A. Brik, Angew. Chem., Int. Ed., 2019, 58, 13540–13549 CrossRef CAS PubMed.
- A. de Jong, K. Witting, R. Kooij, D. Flierman and H. Ovaa, Angew. Chem., Int. Ed., 2017, 56, 12967–12970 CrossRef CAS PubMed.
- D. S. Hameed, A. Sapmaz, L. Burggraaff, A. Amore, C. J. Slingerland, G. J. P. van Westen and H. Ovaa, Angew. Chem., Int. Ed., 2019, 58, 14477–14482 CrossRef CAS PubMed.
Footnote |
| † Dedicated to the memory of Professor Huib Ovaa. |
| This journal is © The Royal Society of Chemistry 2020 |