Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Pinkment: a synthetic platform for the development of fluorescent probes for diagnostic and theranostic applications†
Maria
Weber‡
 ab,
Hai-Hao
Han‡
ab,
Hai-Hao
Han‡
 cd,
Bo-Han
Li‡
ce,
Maria L.
Odyniec
cd,
Bo-Han
Li‡
ce,
Maria L.
Odyniec
 a,
Charlotte E. F.
Jarman
a,
Yi
Zang
c,
Steven D.
Bull
a,
Charlotte E. F.
Jarman
a,
Yi
Zang
c,
Steven D.
Bull
 a,
Amanda B.
Mackenzie
gh,
Adam C.
Sedgwick
a,
Amanda B.
Mackenzie
gh,
Adam C.
Sedgwick
 *f,
Jia
Li
*ce,
Xiao-Peng
He
*f,
Jia
Li
*ce,
Xiao-Peng
He
 *d and
Tony D.
James
*d and
Tony D.
James
 *a
*a
aDepartment of Chemistry, University of Bath, Bath, BA2 7AY, UK. E-mail: t.d.james@bath.ac.uk
bCentre for Doctoral Training, Centre for Sustainable & Circular Technologies, University of Bath, Bath, BA2 7AY, UK
cNational Centre for Drug Screening, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 189 Guo Shoujing Rd., Shanghai 201203, PR China. E-mail: jli@simm.ac.cn
dKey Laboratory for Advanced Materials & Feringa Nobel Prize Scientist Joint Research Centre, School of Chemistry and Molecular Engineering, Frontiers Center for Materiobiology and Dynamic Chemistry, East China University of Science and Technology, 130 Meilong Rd., Shanghai 200237, PR China. E-mail: xphe@ecust.edu.cn
eUniversity of Chinese Academy of Sciences, No. 19A Yuquan Road, Beijing 100049, PR China
fDepartment of Chemistry, University of Texas at Austin, 105 East 24th Street A5300, Austin, Texas 78712-1224, USA. E-mail: a.c.sedgwick@utexas.edu
gDepartment of Pharmacy and Pharmacology, University of Bath, Bath, BA2 7AY, UK
hCentre for Therapeutic Innovation, University of Bath, Bath, BA2 7AY, UK
First published on 6th August 2020
Abstract
Reaction-based fluorescent-probes have proven successful for the visualisation of biological species in various cellular processes. Unfortunately, in order to tailor the design of a fluorescent probe to a specific application (i.e. organelle targeting, material and theranostic applications) often requires extensive synthetic efforts and the synthetic screening of a range of fluorophores to match the required synthetic needs. In this work, we have identified Pinkment-OH as a unique “plug-and-play” synthetic platform that can be used to develop a range of ONOO− responsive fluorescent probes for a variety of applications. These include theranostic-based applications and potential material-based/bioconjugation applications. The as prepared probes displayed an excellent sensitivity and selectivity for ONOO− over other ROS. In vitro studies using HeLa cells and RAW 264.7 macrophages demonstrated their ability to detect exogenously and endogenously produced ONOO−. Evaluation in an LPS-induced inflammation mouse model illustrated the ability to monitor ONOO− production in acute inflammation. Lastly, theranostic-based probes enabled the simultaneous evaluation of indomethacin-based therapeutic effects combined with the visualisation of an inflammation biomarker in RAW 264.7 cells.
Introduction
There is a growing need for new and effective diagnostic tools that can evaluate biomarkers involved in inflammatory based diseases.1–6 Inflammation is the innate defence mechanism of the body that recognises damaged cells, pathogens and infections. The inflammatory response often results in the generation of reactive oxygen species/reactive nitrogen species (ROS/RNS), which are involved in the functional regulation of M1 and M2 macrophages.7,8 The M1 pro-inflammatory phenotype is induced by lipopolysaccharide (LPS), which triggers the generation of ROS from NADPH using NADPH oxidase (NOX).9 This production of ROS regulates an array of cellular events including the activation of the nuclear factor kappa-B (NF-κB), the production of cytokines and cell survival whereas, high levels of ROS are associated with programmed cell death, i.e. apoptosis.7,10–14 The high sensitivity and high spatial and temporal resolution of fluorescent probes allow us to visualise these key cellular events. Our group and others have focused on the fluorescence-based detection of ROS/RNS such as ONOO−, H2O2 and HOCl.1,15–21 To achieve the selective detection of a particular ROS requires the careful consideration of both fluorophore and reactive motif. In this regard, resorufin is a particularly attractive fluorophore due to its red shifted fluorescence and easy to functionalise scaffold. Pioneering work led by Chang et al. developed peroxyresorufin-1 (PR1) for H2O2 detection whereby resorufin is masked with boronic esters.22,23 Boronic esters have been identified as a relevant sensing group for both H2O2 and ONOO− detection. However, in an environment with both species present, boronic esters preferentially react with ONOO− due to the inherent faster reactivity of ONOO− in comparison to H2O2.24 Previously, we have demonstrated PR1's ability to preferentially detect ONOO− over H2O2in vitro.25 Consequently, we decided to investigate functionalized synthetic derivatives of PR1.26 This led to the development of Pinkment-OH for the design of dual analyte AND-logic probes, Pinkment-OTBS (ONOO− “AND” fluoride) and Pinkment-OAc (H2O2 “AND” esterase) using Pinkment-OH (Fig. 1) as a synthetic starting point. These results revealed the potential of Pinkment-OH to be used as a synthetic platform for the development of ONOO− selective fluorescence probes with additional sensing, targeting or drug units. Here, we have serendipitously discovered that the benzyl unit of our ROS Pinkment fluorescent probe can be functionalized with a functional unit of choice without compromising ROS selectivity. As a result, Pinkment-OH was successfully shown as a synthetic platform to develop ONOO− selective fluorescent probes with additional functional units (Fig. 1).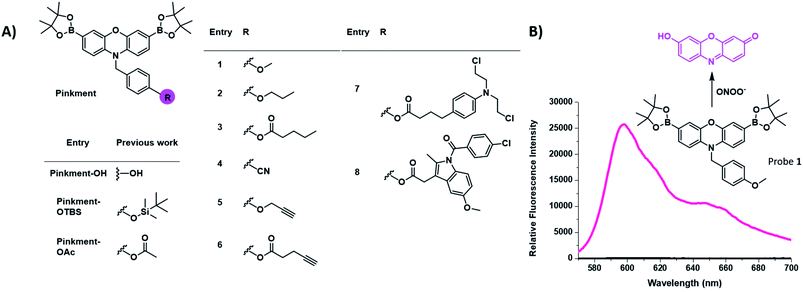 | ||
| Fig. 1 (A) Chemical structure of the resorufin-based probes for the sensing of ONOO−, including the previously reported Pinkment-OH “plug and play” scaffold and dual analyte probes Pinkment-OTBS and Pinkment-OAc.10 (B) Unexpected fluorescence turn on response of 1 in the presence of ONOO−. | ||
Results and discussion
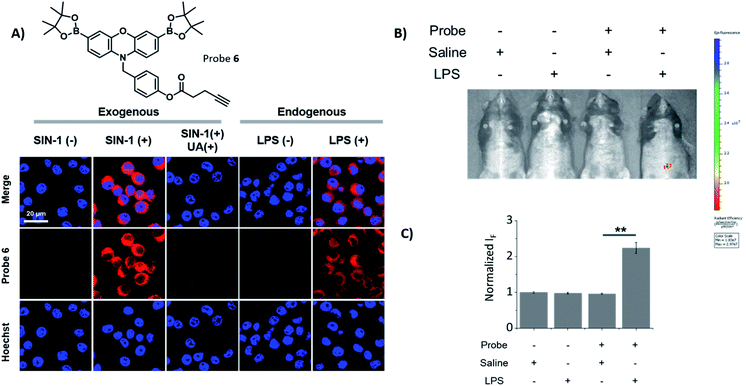
Initially, our focus was on continuing the development of “AND”-based logic-gates for biological application.1,17,26 This led to the elaboration of probe 1, which was accessed in a simple three step synthesis (Scheme S1†). Unexpectedly, we discovered that 1 “turned on” in the sole presence of ONOO− (Fig. 1B and S1†). This led to the development of Pinkment probes 2 and 3 to further confirm this observation. These probes were accessible from the synthetic platform Pinkment-OH, whose 6-step synthesis has been previously reported by our group.26 Nucleophilic substitution by Pinkment-OH using 1-bromopropane and pentanoyl chloride respectively gave 2 and 3 in moderate yields: 50% and 51% respectively (Scheme S2 and S3†). Probes 2 and 3 showed good selectivity towards ONOO− over other ROS species (Fig. S2–S8†). Surprisingly, the probes demonstrated a high sensitivity towards ONOO− requiring concentrations in the low micromolar range. Both 2 and 3 displayed increased solubility in comparison to 1. We decided to further explore this unexpected result by introducing a terminal nitrile group. Probe 4 was accessible in a facile three-step synthesis (Scheme S4†) in the same manner as 1. Again, good selectivity and sensitivity for ONOO− was observed (Fig. S2–S8†). From these results, we realized that the Pinkment benzyl unit can be functionalized with any unit of choice without compromising the ROS selectivity. Thus, we rationalized that Pinkment-OH offers a unique platform for the design of ONOO− selective fluorescence based probes that can be tailored towards a range of applications.27–29 This led to the development of alkyne-based Pinkment probes 5 and 6 that have potential to be used in “click” chemistry.29 These probes were accessed from Pinkment-OH and prepared in moderate yields: 48% and 47% for 5, and 6 respectively (Scheme S5 and S6†). Fluorescence studies of 5 and 6 established good sensitivity and selectivity towards ONOO− over other ROS (Fig. S2–S8†).We then turned our attention to assessing the imaging capacity of probes 2 and 3 and the potential “click” based probes 5 and 6 in cells and live animals. To demonstrate their suitability as imaging tools, all four probes were evaluated for cellular toxicity in murine RAW 264.7 macrophages using a MTS cell proliferation assay. Probes 2, 3, 5 and 6 were incubated at different concentrations ranging from 5 to 40 μM for 24 h (Fig. S9†). Probes 2, 5 and 6 were found to be non-toxic. In contrast, probe 3 decreased the cell viability of RAW 264.7 macrophages by 40% at a concentration of 40 μM compared to control conditions. As a result, 3 was not taken forward for further cell studies since high concentrations of the probe are required for in vivo studies.
Probes 2, 5 and 6 were shown to be non-toxic, and were evaluated with exogenous ONOO−, using SIN-1 (500 μM) in RAW 264.7 macrophages (Fig. 2A and S10†). Each probe alone demonstrated minimal fluorescence in cells, the addition of SIN-1 led to a significant enhancement in intracellular fluorescence at a wavelength corresponding to the dye, resorufin, therefore, suggesting the intracellular reaction of the probe with ONOO− and their suitability for use as fluorescence-based probes. The SIN-1 generated fluorescence signal was then evaluated with the ONOO− scavenger, uric acid.30 As expected, uric acid attenuated the fluorescent increase that was induced by SIN-1 for all probes, thus confirming the ONOO− mediated increase in fluorescence intensity. Next, we evaluated the capability of 2, 5 and 6 to detect endogenous ONOO− in LPS primed RAW 264.7 macrophages. All three probes were shown to detect endogenous ONOO− in LPS primed RAW 264.7 macrophages (Fig. 2A and S11†), confirming their promise for the imaging of LPS-induced inflammatory responses. In addition, HeLa and A549 cell lines treated with or without SIN-1 were used to illustrate the versatility of the Pinkment probes (Fig. S12 and S13†).
Encouraged by these cell imaging results, we used a known LPS-induced inflammation mouse model31 for the in vivo detection of ONOO− (Fig. 2B). The injection of LPS (2 mg mL−1 in saline) to the abdominal region of mice followed by the injection of 6 (200 μM) led to its fluorescence activation. The quantified fluorescence intensity in the probe(+)/LPS(+) group was significantly larger than that in the probe(+)/LPS(−) group (Fig. 1C), demonstrating the potential of using 6 for the monitoring of ONOO−in situ during acute inflammation.
In order to follow our current interest in theranostics,32 we then turned our attention towards the potential of Pinkment-OH for the design of fluorescence-based drug releasing probes. Therefore, we used the drugs chlorambucil and indomethacin to afford two distinct theranostic probes 7 and 8, respectively (Fig. 1A). Chlorambucil is used to treat chronic lymphatic leukemia33 and indomethacin is used as a non-steroidal anti-inflammatory drug (NSAID).34,35 Both 7 and 8 were easily accessible from Pinkment-OH (Scheme S7 and S8†).
Mass spectrometry confirmed and validated the simultaneous release of each drug and fluorescent resorufin dye (Fig. S14 and S15†). Therefore, the enhancement in the fluorescence intensity over time indicates the release of each drug. As such, time-dependent fluorescence experiments with 7 and 8 in the presence of ONOO− were performed to illustrate the time dependence of the drug release. These experiments revealed a maximum fluorescence response after ∼10 min (Fig. S16†).
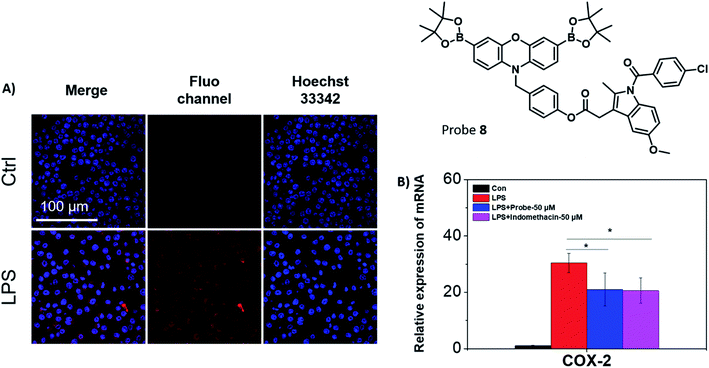
Fluorescence studies were carried out including ROS selectivity, H2O2 titration and ONOO− titration studies (Fig. S17–S21†) and demonstrated high sensitivity towards these inflammation-based biomarkers. Following these initial studies, we evaluated both 7 and 8 in RAW 264.7 macrophages towards exogenous ONOO− detection (Fig. S22†). The presence of SIN-1 significantly enhanced the intracellular fluorescence of 7 and 8, confirming the applicability of the probes in vitro. Despite 7 displaying significant promise, the creation of an appropriate model system to differentiate between cancerous and healthy cells would require a significant amount of development and as such was beyond the scope of this current research. Therefore, only 8 was further evaluated, since its cellular behaviour was easier to monitor. Endogenous ONOO− was also detected by 8 in RAW 264.7 macrophages (Fig. 3A). Indomethacin, a NSAID, is an effective and non-selective inhibitor of cyclooxygenase-1 (COX-1) and cyclooxygenase-2 (COX-2), of which COX-2 is mainly responsible for the inflammatory response.36 The therapeutic effects on the LPS-induced inflammatory responses in RAW 264.7 macrophages were further investigated using 8. RAW 264.7 macrophages were treated with LPS and the expression of the pro-inflammatory gene (COX-2) was investigated using qRT-PCR in the presence or absence of 8 (Fig. 3B).37 The mRNA level of COX-2 decreased in the presence of 8 (50 μM) in comparison to the LPS-induced group. A similar effect to the LPS-induced group was observed with indomethacin alone. This suggests that 8 can monitor ONOO− production in acute inflammation, and in addition, reduce the inflammatory response by releasing indomethacin.
Conclusions
The ability of the Pinkment scaffold to be functionalised with any unit of choice without compromising the overall ROS selectivity, opens up new possibilities for the design of highly specific ONOO− probes that can be used in a variety of applications. In this work, we have successfully illustrated the applicability of Pinkment-based probes for diagnostic and theranostics applications. Our probes displayed good selectivity and sensitivity towards ONOO− over a range of other ROS. Cellular studies with the Pinkment probes led to the identification of alkyne-functionalised Pinkment probe 6 as a suitable candidate for in vivo studies using an inflammatory mouse model. These promising results led us to design potential theranostic probes 7 and 8 with candidate 8 displaying promising properties in vitro. We believe this work demonstrates Pinkment-OH as a useful synthetic platform to enable the rapid development of a ONOO− fluorescent probe that can be tailored to the needs of the chemical biologist. In particular, the alkyne Pinkment probes offer the possibility of attaching any desired unit via click chemistry. Therefore, we anticipate that the Pinkment scaffold can be further elaborated for the development of dual analyte, organelle targeting and theranostic probes for a range of diagnostic and theranostic applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
All animal experiments were carried out under the Guidelines for Care and Use of Laboratory Animals of Shanghai Institute of Materia Medica (SIMM), Chinese Academy of Sciences and approved by the Institutional Animal Care and Use Committee (IACUC) of SIMM (Shanghai, China). M. W. would like to thank the EPSRC for funding (i) EP/L016354/1 and the CDT in Sustainable Chemical Technologies. M. L. O. and A. C. S. would like to thank the University of Bath and EPSRC for PhD studentships. T. D. J. thanks the Royal Society for a Wolfson Research Merit Award. X. P. H. thanks the National Natural Science Foundation of China (No. 21788102, 91853201, 21722801 and 21776078), the National Key Sci-Tech Special Projects of Infection Diseases of China (2018ZX10732202), the Shanghai Municipal Science and Technology Major Project (No. 2018SHZDZX03), the International Cooperation Program of Shanghai Science and Technology Committee (No. 17520750100) and the Fundamental Research Funds for the Central Universities (222201717003) and the Programme of Introducing Talents of Discipline to Universities (B16017) for financial support. Characterization facilities were provided through the Material and Chemical Characterization Facility (MC2) at the University of Bath (http://go.bath.ac.uk/mc2). In particular, we would like to acknowledge Shaun Reeksting for helping with the more advanced MS studies of the theranostic probes. All data supporting this study are provided in the ESI† accompanying this paper.Notes and references
- X. L. Hu, N. Kwon, K. C. Yan, A. C. Sedgwick, G. R. Chen, X. P. He, T. D. James and J. Yoon, Adv. Funct. Mater., 2020, 30, 1907906 CrossRef CAS.
- D. Wu, A. C. Sedgwick, T. Gunnlaugsson, E. U. Akkaya, J. Yoon and T. D. James, Chem. Soc. Rev., 2017, 46, 7105–7123 RSC.
- S. Erbas-Cakmak, S. Kolemen, A. C. Sedgwick, T. Gunnlaugsson, T. D. James, J. Yoon and E. U. Akkaya, Chem. Soc. Rev., 2018, 47, 2228–2248 RSC.
- A. C. Sedgwick, L. L. Wu, H. H. Han, S. D. Bull, X. P. He, T. D. James, J. L. Sessler, B. Z. Tang, H. Tian and J. Yoon, Chem. Soc. Rev., 2018, 47, 8842–8880 RSC.
- J. Chan, S. C. Dodani and C. J. Chang, Nat. Chem., 2012, 4, 973–984 CrossRef CAS PubMed.
- K. C. Yon, A. C. Sedgwick, Y. Zang, G. R. Chen, X. P. He, J. Li, J. Yoon and T. D. James, Small Methods, 2019, 3, 1900013 CrossRef.
- A. Weidinger and A. V. Kozlov, Biomolecules, 2015, 5, 472–484 CrossRef CAS PubMed.
- B. Ghesquiere, B. W. Wong, A. Kuchnio and P. Carmeliet, Nature, 2014, 511, 167–176 CrossRef CAS PubMed.
- A. Kumar, S. H. Chen, M. B. Kadiiska, J. S. Hong, J. Zielonka, B. Kalyanaraman and R. P. Mason, Free Radical Biol. Med., 2014, 73, 51–59 CrossRef CAS PubMed.
- B. Halliwell, Annu. Rev. Nutr., 1996, 16, 33–50 CrossRef CAS PubMed.
- H. Wiseman and B. Halliwell, Biochem. J., 1996, 313, 17–29 CrossRef CAS PubMed.
- J. S. Beckman and W. H. Koppenol, Am. J. Physiol.: Cell Physiol., 1996, 271, C1424–C1437 CrossRef CAS PubMed.
- M. Valko, D. Leibfritz, J. Moncol, M. T. D. Cronin, M. Mazur and J. Telser, Int. J. Biochem. Cell Biol., 2007, 39, 44–84 CrossRef CAS PubMed.
- H. Y. Tan, N. Wang, S. Li, M. Hong, X. B. Wang and Y. B. Feng, Oxid. Med. Cell. Longevity, 2016, 2016, 2795090 Search PubMed.
- A. C. Sedgwick, R. S. L. Chapman, J. E. Gardiner, L. R. Peacock, G. Kim, J. Yoon, S. D. Bull and T. D. James, Chem. Commun., 2017, 53, 10441–10443 RSC.
- A. C. Sedgwick, H. H. Han, J. E. Gardiner, S. D. Bull, X. P. He and T. D. James, Chem. Commun., 2017, 53, 12822–12825 RSC.
- A. C. Sedgwick, H. H. Han, J. E. Gardiner, S. D. Bull, X. P. He and T. D. James, Chem. Sci., 2018, 9, 3672–3676 RSC.
- A. C. Sedgwick, W. T. Dou, J. B. Jiao, L. L. Wu, G. T. Williams, A. T. A. Jenkins, S. D. Bull, J. L. Sessler, X. P. He and T. D. James, J. Am. Chem. Soc., 2018, 140, 14267–14271 CrossRef CAS PubMed.
- L. L. Wu, H. H. Han, L. Y. Liu, J. E. Gardiner, A. C. Sedgwick, C. S. Huang, S. D. Bull, X. P. He and T. D. James, Chem. Commun., 2018, 54, 11336–11339 RSC.
- A. C. Sedgwick, J. E. Gardiner, G. Kim, M. Yevglevskis, M. D. Lloyd, A. T. A. Jenkins, S. D. Bull, J. Yoon and T. D. James, Chem. Commun., 2018, 54, 4786–4789 RSC.
- L. L. Wu, A. C. Sedgwick, X. L. Sun, S. D. Bull, X. P. He and T. D. James, Acc. Chem. Res., 2019, 52, 2582–2597 CrossRef CAS PubMed.
- B. C. Dickinson, C. Huynh and C. J. Chang, J. Am. Chem. Soc., 2010, 132, 5906–5915 CrossRef CAS PubMed.
- E. W. Miller, A. E. Albers, A. Pralle, E. Y. Isacoff and C. J. Chang, J. Am. Chem. Soc., 2005, 127, 16652–16659 CrossRef CAS PubMed.
- A. Sikora, J. Zielonka, M. Lopez, J. Joseph and B. Kalyanaraman, Free Radical Biol. Med., 2009, 47, 1401–1407 CrossRef CAS PubMed.
- M. Weber, A. B. Mackenzie, S. D. Bull and T. D. James, Anal. Chem., 2018, 90, 10621–10627 CrossRef CAS PubMed.
- M. L. Odyniec, A. C. Sedgwick, A. H. Swan, M. Weber, T. M. S. Tang, J. E. Gardiner, M. Zhang, Y. B. Jiang, G. Kociok-Kohn, R. B. P. Elmes, S. D. Bull, X. P. He and T. D. James, Chem. Commun., 2018, 54, 8466–8469 RSC.
- A. Marrocchi, A. Facchetti, D. Lanari, S. Santoro and L. Vaccaro, Chem. Sci., 2016, 7, 6298–6308 RSC.
- Y. P. Chen, Y. L. Xianyu, J. Wu, B. F. Yin and X. Y. Jiang, Theranostics, 2016, 6, 969–985 CrossRef CAS PubMed.
- H. C. Kolb, M. G. Finn and K. B. Sharpless, Angew. Chem., Int. Ed., 2001, 40, 2004–2021 CrossRef CAS PubMed.
- D. C. Hooper, S. Spitsin, R. B. Kean, J. M. Champion, G. M. Dickson, I. Chaudhry and H. Koprowski, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 675–680 CrossRef CAS PubMed.
- D. Lee, S. Khaja, J. C. Velasquez-Castano, M. Dasari, C. Sun, J. Petros, W. R. Taylor and N. Murthy, Nat. Mater., 2007, 6, 765–769 CrossRef CAS PubMed.
- M. L. Odyniec, H. H. Han, J. E. Gardiner, A. C. Sedgwick, X. P. He, S. D. Bull and T. D. James, Front. Chem., 2019, 7, 775 CrossRef CAS PubMed.
- M. J. Reese, D. W. Knapp, K. M. Anderson, J. A. Mund, J. Case, D. R. Jones and R. A. Packer, PLoS One, 2018, 13, e0203517 CrossRef PubMed.
- K. Seibert, Y. Zhang, K. Leahy, S. Hauser, J. Masferrer and P. Isakson, in Eicosanoids and Other Bioactive Lipids in Cancer, Inflammation, and Radiation Injury 2, Pts A and B, ed. K. V. Honn, S. Nigam and L. J. Marnett, 1997, vol. 400, pp. 167–170 Search PubMed.
- A. A. Onischuk, T. G. Tolstikova, I. V. Sorokina, N. A. Zhukova, A. M. Baklanov, V. V. Karasev, G. G. Dultseva, V. V. Boldyrev and V. M. Fomin, J. Aerosol Med. Pulm. Drug Delivery, 2008, 21, 231–243 CrossRef CAS PubMed.
- C. Chen, Nat. Chem. Biol., 2010, 6, 401–402 CrossRef CAS PubMed.
- M. Kawashima, N. Ogura, M. Akutsu, K. Ito and T. Kondoh, J. Oral Pathol. Med., 2013, 42, 499–506 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Synthetic procedures, 1H, 13C NMR, and mass spectra, UV-Vis and fluorescence spectroscopic analyses, and other materials. See DOI: 10.1039/d0sc02438d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |