Open Access Article
Open Access ArticleQuantifying tensile forces at cell–cell junctions with a DNA-based fluorescent probe†
Bin
Zhao
 a,
Ningwei
Li
b,
Tianfa
Xie
a,
Ningwei
Li
b,
Tianfa
Xie
 b,
Yousef
Bagheri
b,
Yousef
Bagheri
 a,
Chungwen
Liang
c,
Puspam
Keshri
a,
Yubing
Sun
a,
Chungwen
Liang
c,
Puspam
Keshri
a,
Yubing
Sun
 *b and
Mingxu
You
*b and
Mingxu
You
 *a
*a
aDepartment of Chemistry, University of Massachusetts, Amherst, Massachusetts 01003, USA. E-mail: mingxuyou@umass.edu
bDepartment of Mechanical & Industrial Engineering, University of Massachusetts, Amherst, Massachusetts 01003, USA. E-mail: ybsun@umass.edu
cComputational and Modeling Core, Institute for Applied Life Sciences (IALS), University of Massachusetts, Amherst, Massachusetts 01003, USA
First published on 29th July 2020
Abstract
Cells are physically contacting with each other. Direct and precise quantification of forces at cell–cell junctions is still challenging. Herein, we have developed a DNA-based ratiometric fluorescent probe, termed DNAMeter, to quantify intercellular tensile forces. These lipid-modified DNAMeters can spontaneously anchor onto live cell membranes. The DNAMeter consists of two self-assembled DNA hairpins of different force tolerance. Once the intercellular tension exceeds the force tolerance to unfold a DNA hairpin, a specific fluorescence signal will be activated, which enables the real-time imaging and quantification of tensile forces. Using E-cadherin-modified DNAMeter as an example, we have demonstrated an approach to quantify, at the molecular level, the magnitude and distribution of E-cadherin tension among epithelial cells. Compatible with readily accessible fluorescence microscopes, these easy-to-use DNA tension probes can be broadly used to quantify mechanotransduction in collective cell behaviors.
Introduction
Intercellular mechanical forces, especially tensile forces, play important roles in development, wound healing and cancer invasion.1–3 These tensile forces at cell–cell junctions actively reshape the tissues during morphogenesis in embryos and in quiescent adult tissues,4–6 such as epithelial and endothelial monolayers.7,8 For example, cadherins constitute a superfamily of cell–cell adhesion molecules that are expressed in various types of cells.9,10 It is known that cadherins can sense and mediate tensile forces at cell–cell junctions,11 which are required for regulating cellular homeostasis and collective migration during embryo development, wound healing, and pulmonary system homeostasis.6,12–17 Elucidating the mechanisms of cadherin-mediated force sensing and transduction is therefore critical for revealing the fundamental principles in the collective organizations and motions of cell populations.18,19 However, mapping the spatiotemporal dynamics of intercellular forces is still challenging. Tools for the precise, quantitative, and real-time measurement of tensile forces at cell–cell junctions are still largely missing.20–22Generally, two strategies are currently available to estimate intercellular forces. Monolayer stress microscopy utilizes cell–matrix traction force data to deduce mechanical forces at cell–cell junctions, with the assumption that total forces experienced by each cell remain zero.23 Traction force microscopy has been used to elucidate the relationships between the total cellular forces on extracellular matrix and the endogenous intercellular forces.24 However, these methods can only be applied to a monolayer of cells. It requires extensive image analysis and data processing. Moreover, the force deduced is not specific for certain junctional molecules. Similarly, intercellular forces between a pair of cells have been measured by microfabricated cantilever pillars.13 However, in addition to the above-mentioned drawbacks, these cantilever pillars can only measure forces between a pair of cells, one at a time, and require advanced microfabrication facilities.
In another strategy, genetically encoded protein-based tension probes have been developed to measure intercellular forces mediated by cadherins or platelet endothelial cell adhesion molecule.8,25–27 However, the routine use of these sensors is limited due to their labor-intensive design and validation. The functions of many junctional proteins will be disrupted after insertion of a large protein sensor (∼500 amino acids). The small force measurement range (1–12 pN) and low sensitivity of fluorescence signals of these probes (∼10-fold lower than sensors using common organic dyes)28 further hinders the widespread applications of these genetically encoded probes.
We have recently developed a new DNA-based probe to visualize intercellular tensile forces at cell–cell junctions.29 In this system, a pair of cholesterol anchors was used to insert a DNA hairpin probe onto cell membranes.30,31 Once the intercellular tensile force exceeds the threshold value to unfold the DNA hairpin, the separation of a fluorophore–quencher pair results in the activation of fluorescence signals. These DNA probes are well suited for intercellular force measurement. First, the probes function simply by incubation with target cells. There is no need for cloning or transfection. Secondly, different mechanosensitive ligands or receptors can be directly conjugated within these DNA probes, which allow the facile study of specific junctional molecule-mediated force transduction. Thirdly, by tuning the sequence and duplex length of the DNA hairpin, the force tolerance of the probe can be rationally adjusted in a large range.32–34 Moreover, a broad choice of organic fluorophores and quenchers allows highly sensitive imaging of tensile forces.
However, this “first-generation” DNA tension probe has several limitations. First, it cannot be used to quantify the intercellular forces due to the heterogeneous membrane distribution of the probes. Secondly, each DNA hairpin unfolds in a narrow threshold force range (±2 pN),35 it is challenging to use a single probe to measure a large range of intercellular forces. In natural mechanotransduction process, indeed, different levels of tensile forces could exist simultaneously. In addition, many collective cell behavior studies require a long-term measurement of intercellular forces.17,36–38 However, current lipid-modified DNA probes have a limited anchoring persistence on the cell membranes (∼2 to 4 h). To overcome all these limitations, in this study we have developed the “second-generation” DNA-based Membrane Tension Ratiometric Probe, termed “DNAMeter”.
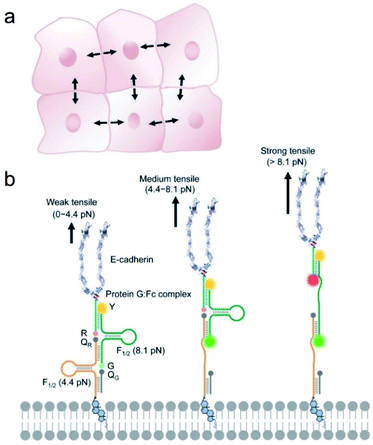
The DNAMeter was designed to be highly adaptable, consisting of two self-assembled DNA hairpins with different threshold forces and a lipid tail to anchor onto live cell membranes (Fig. 1). To quantify the intercellular tension based on the fluorescence signal, an internal reference fluorophore was introduced to normalize the membrane distribution of the DNAMeter. In addition, two orthogonal fluorophore–quencher pairs were conjugated at the end of each DNA hairpin to report different magnitudes of forces. By measuring each reporter-to-reference fluorescence intensity ratio, molecular scale intercellular force distributions can be quantified at cell–cell junctions. Using intercellular E-cadherin tension measurement as an example, we demonstrated here a quantitative and general approach to map spatiotemporal distributions of tensile forces at the molecular level during long-term collective cell behaviors.
Results and discussion
Design and characterization of the DNAMeter
The DNAMeter is designed based on the self-assembly of four oligonucleotide strands (Fig. 1b and Table S1†). Two of the strands contain a 25-nucleotide-long DNA hairpin with 22% and 66% G/C base pairs to detect weak and strong tensile force, respectively. As an internal reference, a TAMRA dye (λex/λem: 557/579 nm, denoted as Y) was modified at one end of the 66%GC DNA hairpin strand. To detect the folding/unfolding switch of the 66%GC DNA hairpin, a Cy5-QSY®21 fluorophore–quencher pair (λex/λem: 640/659 nm, denoted as R-QR) was conjugated next to the end of this hairpin. Similarly, a FAM-Dabcyl fluorophore–quencher pair (λex/λem: 488/519 nm, denoted as G-QG) was used to measure the folding/unfolding of the 22%GC DNA hairpin. After calculating the unfolding free energy of hairpin at zero force and the free energy of stretching the corresponding single stranded DNA,33,35 based on a worm-like chain model,39,40 the tensile force threshold (F1/2) of the 22%GC and 66%GC DNA hairpin was determined to be 4.4 pN and 8.1 pN, respectively (Materials and methods and Table S2†). Here, F1/2 is defined as the force at which the DNA hairpin has 50% probability of being unfolded.When experiencing a weak tensile force (<4.4 pN), both FAM-Dabcyl and Cy5-QSY®21 pairs remain at close proximity, resulting in low fluorescence level in both reporter channels (denoted as G−/R−). In contrast, a strong tensile force (>8.1 pN) results in the stretching out of both 22%GC and 66%GC hairpins. Both fluorophores will separate from the corresponding quencher, leading to an increase in both FAM and Cy5 fluorescence signal (denoted as G+/R+). In another case, a medium tensile force (4.4–8.1 pN) opens up the 22%GC hairpin, but not the 66%GC hairpin, so only the FAM signal will be activated (denoted as G+/R−). As a result, we can image different ranges of tensile forces based on the two reporter channels.
To test the efficiency of this probe design, we prepared an E-cadherin-modified DNAMeter (termed EC-DNAMeter) to quantify E-cadherin-mediated intercellular tensile forces at the Madin–Darby canine kidney (MDCK) epithelial cell–cell junctions. E-cadherin is a transmembrane protein, which cytoplasmic domain can bind with beta-catenin, alpha-catenin, and then experience forces generated by the actomyosin cytoskeleton.10,25 The EC-DNAMeter was prepared using a Protein G linker to couple the IgG/Fc-fused extracellular domain of E-cadherin with the DNAMeter (Materials and methods). Compared with direct chemical conjugation, the Protein G linker helps to avoid the loss of E-cadherin activities.29,41 In addition, to allow the probe to insert onto MDCK cell membranes, a cholesterol anchor was conjugated at the other end of the DNAMeter.
After demonstrating the formation of the DNAMeter in a gel mobility shift assay (Fig. S1†), the cell membrane insertion efficiency of the DNAMeter was studied. Here, we prepared a non-quenched DNAMeter (nqDNAMeter) by using DNA strands that are not modified with Dabcyl and QSY®21 quenchers (Scheme S1†). The fluorescence of the nqDNAMeter is always on and is independent of intercellular forces. As a result, the cell membrane fluorescence intensity can be used to indicate the concentration of the immobilized probes. Indeed, obvious fluorescence signal on MDCK cell membranes was shown shortly after adding these nqDNAMeter probes (Fig. S2†).
We have further studied the membrane anchoring efficiency of the DNAMeter containing one or two cholesterol tail. Interestingly, one cholesterol-modified nqDNAMeter (1Chol-nqDNAMeter) exhibited higher insertion efficiency (2.1-fold) on MDCK cell membranes than the more hydrophobic two cholesterol-modified one (2Chol-nqDNAMeter) (Fig. S2†). This might be due to the relatively larger critical micelle concentration value of the 1Chol-nqDNAMeter as compared to 2Chol-nqDNAMeter.42 As a result, more monomeric nqDNAMeter could exist in the solution when one cholesterol was anchored. Indeed, our recent data indicated that the cell membrane anchoring of the lipid–DNA conjugates stems mainly from the monomeric form, instead of the aggregation form.43 Previous studies have suggested that ∼100 pN tensile force is required to extract a cholesterol from lipid bilayers.44 As a result, the membrane insertion of the cholesterol should be quite stable during the unfolding of DNA hairpins (4.4 pN and 8.1 pN). Unless specifically indicated, one cholesterol-based construct was used for the following studies.
We next asked if the DNAMeter probe may be activated by the cis receptor–ligand interactions on the same cell membrane, rather than between neighboring cells. To study if the DNA probes prefer to “stand” (favoring trans interactions) or “lie down” (favoring cis interactions) on membrane surfaces, we performed atomistic molecular dynamics simulation in a DNAMeter/lipid bilayer membrane system (see Materials and methods). Our simulation results indicated that the tilting angle (θ) of the DNAMeter with respect to the membrane surface is always within 30° (see Fig. S3†). Indeed, these membrane-anchored DNA probes prefer to “stand” (θ < 30°) on the membrane surface and favor the sensing of trans interactions between cells. The reason for DNA probes to maintain such orientation is likely attributed to the electrostatic repulsion between DNA strands and cell membranes, which are both negatively charged. It is also worth mentioning that trans interactions between two E-cadherins are much stronger than cis.45 As a result, the EC-DNAMeter will be mostly activated by trans interactions between neighboring cells.
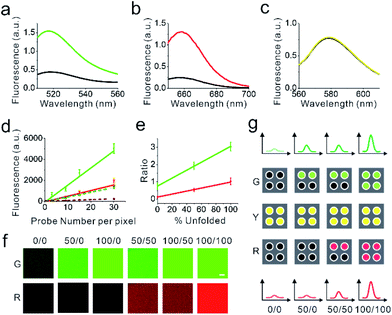
We next asked if we could distinguish the unfolding and the folding state of DNA hairpins based on their fluorescence intensities. To determine the fluorescence of the unfolded DNAMeter, we prepared a de-quenched probe (dqDNAMeter) by incubating the DNAMeter with DNA strands that are complementary to the 22%GC and 66%GC hairpins, respectively (Scheme S1, Fig. S4†). Based on the fluorescence intensity ratio between the DNAMeter and dqDNAMeter, in the absence of external forces, the quenching efficiency for FAM and Cy5 in the DNAMeter was measured to be 70% and 81%, respectively (Fig. 2a and b). Meanwhile, the DNAMeter and dqDNAMeter exhibited the same TAMRA fluorescence intensity, which can act as a standard reference to normalize probe concentrations (Fig. 2c). In addition, after incubating 1 μM DNAMeter or dqDNAMeter with MDCK cells for 30 min, similarly, almost the same TAMRA fluorescence was observed. In contrast, 6.7-fold and 3.1-fold activation of Cy5 and FAM fluorescence exhibited after the unfolding of DNA hairpins (Fig. S5†). These results indicate that the folding and unfolding of 22%GC and 66%GC hairpins indeed can be visualized based on changes in the fluorescence intensity.
Calibration of the DNAMeter on supported lipid monolayers
We next asked if we could further quantify the percentage of unfolded DNA hairpins based on the fluorescence intensities. For this purpose, we prepared a supported lipid monolayer system using soybean polar extract.46 Cholesterol-modified DNAMeter can anchor into this monolayer and diffuse freely.30 By mixing the soybean polar extract with different amount of DNA probes, we can precisely control the membrane density of the DNAMeter on lipid monolayers. After preparing a series of monolayers with different probe densities, we measured the corresponding membrane fluorescence intensity with a spinning disk confocal fluorescence microscope. The same setup and parameters of the microscope was used for the following cellular measurements as well.The obtained fluorescence intensities were then plotted as a function of probe densities for the calibration. A linear correlation between the fluorescence intensity and the DNAMeter concentration was observed for all the fluorophores, including FAM (G), Cy5 (R), and TAMRA (Y) (Fig. 2d and e). Similarly, a linear correlation was observed with all these fluorophores in the dqDNAMeter as well (Fig. 2d and e). After subtracting the background fluorescence for each channel, the fluorescence intensity ratio of both FAM/TAMRA (G/Y) and Cy5/TAMRA (R/Y) is independent on the probe concentration due to the linear correlation between the probe density and fluorescence (Fig. 2d). Such concentration-independent G/Y and R/Y ratio was observed with both the DNAMeter and dqDNAMeter, while the dqDNAMeter exhibited a 4.0-fold and 8.3-fold higher intensity ratio. The G/Y and R/Y ratio can thus be used to quantify the membrane dqDNAMeter-to-DNAMeter probe density ratio, as well as the percentage of unfolded 22%GC and 66%GC DNA hairpins, respectively.
Our next goal is to validate if the G/Y and R/Y ratio can be used to quantify the percentage of unfolded DNA hairpins. We prepared mixtures of dqDNAMeter and DNAMeter, with a ratio of 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 0.5
1, 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5, and 1
0.5, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0. Indeed, both G/Y and R/Y ratio are linearly correlated with the percentage of the unfolded dqDNAMeter (Fig. 2e). We have also tested if the G/Y and R/Y ratio can orthogonally report the unfolding of 22%GC and 66%GC DNA hairpins, respectively. By adding only a complementary DNA strand to either 22%GC or 66%GC DNA hairpin, we prepared de-quenched DNAMeter with only one hairpin being unfolded. After mixing different ratios of these two partially unfolded DNAMeter, indeed, the FAM and Cy5 signal can be used to quantify the amount of unfolded 22%GC or 66%GC DNA hairpin, without interfering with each other (Fig. 2f). All these results indicated that we could quantify the unfolding of DNA hairpins in the DNAMeter by measuring the G/Y and R/Y ratio. Based on the standard calibration curve (Fig. 2d), we can also use the TAMRA fluorescence to quantify the number of probes per individual pixel of images. As a result, we can quantitatively determine not only the percentage, but also the number of unfolded DNA hairpins from the images (Fig. 2g).
0. Indeed, both G/Y and R/Y ratio are linearly correlated with the percentage of the unfolded dqDNAMeter (Fig. 2e). We have also tested if the G/Y and R/Y ratio can orthogonally report the unfolding of 22%GC and 66%GC DNA hairpins, respectively. By adding only a complementary DNA strand to either 22%GC or 66%GC DNA hairpin, we prepared de-quenched DNAMeter with only one hairpin being unfolded. After mixing different ratios of these two partially unfolded DNAMeter, indeed, the FAM and Cy5 signal can be used to quantify the amount of unfolded 22%GC or 66%GC DNA hairpin, without interfering with each other (Fig. 2f). All these results indicated that we could quantify the unfolding of DNA hairpins in the DNAMeter by measuring the G/Y and R/Y ratio. Based on the standard calibration curve (Fig. 2d), we can also use the TAMRA fluorescence to quantify the number of probes per individual pixel of images. As a result, we can quantitatively determine not only the percentage, but also the number of unfolded DNA hairpins from the images (Fig. 2g).
Imaging and quantification of E-cadherin-mediated tensile forces
Before imaging intercellular forces, we wondered if the addition of DNAMeter would impair the adhesion and mechanical function of cell–cell junctions. We first studied the effect of membrane-anchored EC-DNAMeter on the force-dependent recruitment of vinculin and β-catenin to the adherens junctions.47,48 Immunofluorescence staining was used to image the cellular locations of vinculin or β-catenin in MDCK cells before and after adding the DNAMeter. MDCK cells have been widely used as a model cell line to study E-cadherin-mediated tensile forces.49 No significant difference in the junction vinculin or β-catenin fluorescence was observed (Fig. S6†). We have also used Western blot to study the effect of DNAMeter on the membrane expression of another critical cell–cell adhesion protein, β-catenin.50,51 Again, the amount of β-catenin in MDCK cell membranes was quite similar in the presence or absence of EC-DNAMeter anchoring (Fig. S6†). As a result, the addition of DNAMeter will not influence the mechanotransduction at cell–cell junctions.We also studied the effect of DNAMeter on the cell viability. For this purpose, propidium iodide staining was conducted to assess the viability of MDCK cells with or without EC-DNAMeter probe treatment (Fig. S6†). Minimal cell death was observed in both cases. These results indicated that the modification of the DNAMeter probe or lipid–DNA conjugate would not affect MDCK cell viability.
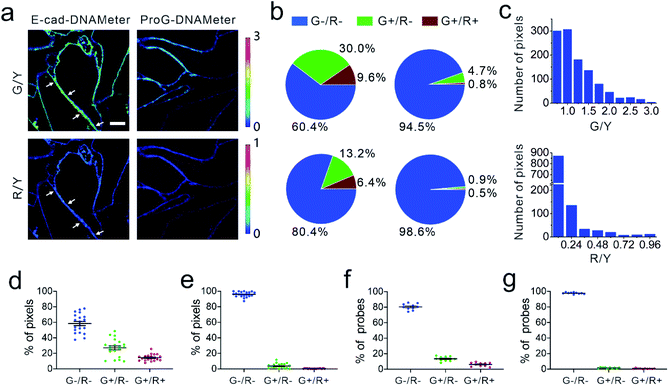
We next applied the EC-DNAMeter to image E-cadherin-mediated intercellular tensile forces at MDCK cell–cell junctions. After incubating the pre-assembled EC-DNAMeter with MDCK cells for 1 h, the cell membrane fluorescence signal of FAM (λex/λem: 488/530 nm), TAMRA (λex/λem: 561/590 nm), and Cy5 (λex/λem: 640/675 nm) were imaged with a spinning disk confocal microscope (Fig. 3a). Here, we denoted the fluorescence of FAM, TAMRA, and Cy5 as G, Y, and R, respectively. For a given DNAMeter-modified cell membrane (Y+), the weak (<4.4 pN), medium (4.4–8.1 pN), and strong (>8.1 pN) E-cadherin-mediated intercellular tensile forces can be visualized based on the fluorescence distribution of G−/R−, G+/R−, and G+/R+, respectively. A large number of G+/R− and some G+/R+ pixels were clearly observed at MDCK cell–cell junctions (Fig. 3a). To test if the fluorescence activation is indeed mediated by E-cadherin interactions, we prepared a control DNAMeter without the modification of E-cadherin, denoted as ProG-DNAMeter. As expected, limited FAM and Cy5 fluorescence was observed, while the TAMRA fluorescence was similar as that of the EC-DNAMeter. These results indicated that we could visualize E-cadherin-mediated tensile forces using the EC-DNAMeter.
We next asked if we could quantify the distribution of different magnitudes of tensile forces at cell–cell junctions. Here, we quantified the force distribution by either the number of bright pixels or the number of unfolded DNA probes. To calculate the number or percentage of pixels considering as G+ or R+, we first measured the membrane statistical fluorescence distribution of the negative control, the ProG-DNAMeter (Fig. 2e). A threshold value of G/Y > 1.0 and R/Y > 0.24 was determined to distinguish the pixels experiencing tensile forces above 4.4 pN and 8.1 pN, respectively. After counting the total number of probe-immobilized pixels (Y+) at cell–cell junctions, we quantified the percentage of junction pixels experiencing the tensile forces (Fig. 3b). For example, at a representative cell–cell junction (Fig. 3a), the weak (<4.4 pN), medium (4.4–8.1 pN), and strong (>8.1 pN) tension was present in ∼60.4%, 30.0%, and 9.6% membrane areas, respectively. We have further calculated these distributions at another 20 cell–cell junctions. On average, under the studied condition when MDCK cells were stably adhered to each other, the fraction of pixels experiencing the weak, medium, and strong tension was 58.5 ± 12.3%, 27.1 ± 11.5% and 14.4 ± 4.2%, respectively (Fig. 3d and Table S3†).
The second approach to quantify the force distributions is based on the number and percentage of the unfolded EC-DNAMeter. As mentioned above, the percentage of unfolded 22%GC and 66%GC DNA hairpin in each pixel can be calculated by measuring the G/Y and R/Y ratio (Fig. 2g). The number of unfolded probes can then be quantified based on the TAMRA fluorescence and the standard calibration curve (Fig. 2f). Our data indicated that 13.2 ± 3.1% and 6.4 ± 2.4% EC-DNAMeter probe was unfolded by 4.4–8.1 pN and >8.1 pN tension, respectively, while 80.4 ± 3.9% probe remained folded (Fig. 3b and c, Materials and methods). As a control, unfolding of the ProG-DNAMeter was negligible (Fig. 3e and g).
When comparing the obtained data from two approaches, we found that by measuring the percentage of unfolded DNA probes, force distributions at different cell–cell junctions were more homogeneous (Fig. 3d and f). Considering these MDCK cells were experiencing similar physical environment and cell–cell adhesions, it may be more accurate to determine force distributions by measuring the percentage of unfolded probes rather than that of bright pixels. Even though the percentage of bright pixels is easier to be quantified, the accuracy of this approach is influenced by the choice of threshold values, e.g., G/Y > 1.0 and R/Y > 0.24 in this case. Meanwhile, even in pixels that are brighter than the threshold value, many DNAMeters can be still in the folded form. In contrast, the percentage of unfolded DNA hairpins can more accurately report, at the molecular level, the force distributions experienced by the DNAMeter.
Dynamics of E-cadherin-mediated tension
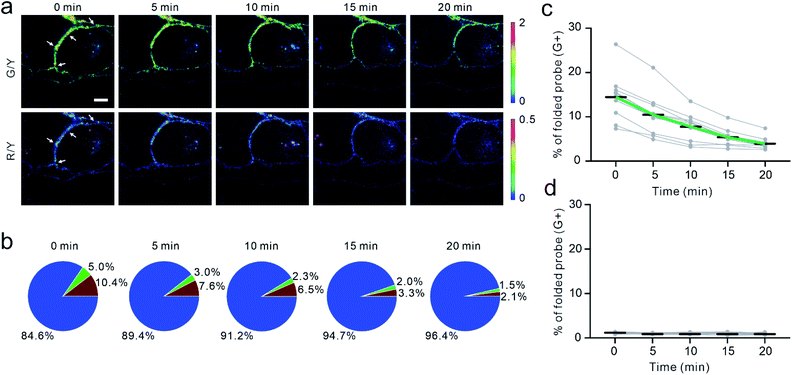
To validate if the EC-DNAMeter indeed measured E-cadherin-mediated tensile forces, we have further studied the effect of ethylene glycol-bis (β-aminoethyl ether)-N,N,N′,N'-tetraacetic acid (EGTA) treatment on MDCK cell adhesions. E-cadherin interactions are gated by extracellular Ca2+ ions that can rigidify the extracellular domains of cadherins and promote cadherin–cadherin junctional interactions.52 As a selective Ca2+ chelating agent, EGTA can disrupt E-cadherin interactions at cell–cell junctions.53 Indeed, after the insertion of the EC-DNAMeter onto MDCK cell membranes, the addition of EGTA triggered a rapid and substantial loss of the fluorescence signal, accompanied with cell dissociations (Fig. 4a and S7†).We next asked if the EC-DNAMeter could be used to monitor the dynamic variations of E-cadherin-mediated intercellular tension after the EGTA treatment. Indeed, at a representative cell–cell junction, within 20 min after adding 10 mM EGTA, the percentage of medium tension (4.4–8.1 pN) gradually decreased from 5.0% to 1.5%, and meanwhile large tension (>8.1 pN) dropped from 10.4% to 2.1% (Fig. 4a and b). Further quantification of more cell–cell junctions confirmed that the EC-DNAMeter could be used to measure the dynamics of intercellular E-cadherin tension. Interestingly, a linear decrease in the number of membrane probes experiencing medium or large tensile forces (>4.4 pN) was observed after adding EGTA, with a rate constant ∼53 μm−2 min−1 (Fig. 4c). In a control experiment, a constant unfolding percentage of the ProG-DNAMeter (∼1.1%) was shown before and after adding EGTA (Fig. 4d, S8, and S9†).
As another validation, we applied the EC-DNAMeter to monitor the ML-7-induced changes in the E-cadherin tension. ML-7 can inhibit the activity of myosin light chain kinase and impair the ability of cells to concentrate E-cadherin at cell–cell junctions.54 As expected, the treatment of ML-7 induced a gradual decrease in the G/Y and R/Y ratio at MDCK cell–cell junctions (Fig. S10†). For example, at a representative junction, after adding 100 μM ML-7, the percentage of large tension (>8.1 pN) gradually decreased within 20 min from 6.3% to 0.5%, and meanwhile medium tension (4.4–8.1 pN) dropped from 10.2% to 0.5% (Fig. S10†). The statistical analysis of more cell–cell junctions further confirmed this observation (Fig. S11†). In contrast, the control probe, ProG-DNAMeter, displayed a constant unfolding percentage at ∼0.8% (Fig. S10 and S11†). Indeed, the EC-DNAMeter can be used to study the dynamic E-cadherin tensions at cell–cell junctions.
Force mapping during collective cell migration
Cooperative intercellular forces drive cellular motions and play vital roles in collective cell migration.23,55 We asked if the DNAMeter could be used to quantify intercellular tensions during collective migration of an epithelial monolayer. Epithelial migration occurs on a time scale of hours-to-days. We first wondered if the DNAMeter allows long-term force measurement. For the sake of simplicity, we prepared a non-quenched EC-DNAMeter containing only a 22%GC DNA hairpin (nqEC22-DNAMeter). After incubating this nqEC22-DNAMeter with a confluent MDCK cell monolayer for 1 h, the probe persistence on the cell membrane was studied. Our results indicated that in a complete growth medium, the cell membrane fluorescence would completely disappear within 3 h (Fig. S12†). Since the growth medium is needed for collective epithelial migrations, the DNAMeter cannot be directly used for the force mapping.To achieve a long-term force measurement, we asked if the cell membrane probe density could be recovered by simply adding fresh DNAMeter. To test that, we first anchored 0.2 μM nqEC22-DNAMeter onto MDCK cell membrane in HEPES-buffered saline, followed by replacing with complete growth medium. After 3 h incubation, almost no fluorescence was observed on the cell membrane. By further replacing the growth medium with buffer containing 0.2 μM of fresh nqEC22-DNAMeter, again, strong fluorescence and a similar level of membrane probe density (95.6 ± 4.5%) was observed at cell–cell junctions (Fig. S13†). Moreover, this loss-and-regain of cell membrane probes can be repeated for at least 5 cycles without significant reduction in the efficiency (Fig. S13 and S14†). In addition, considering the relatively weak E-cadherin binding affinity,45 intercellular E-cadherin interactions are highly dynamic. Newly added EC-DNAMeter probes can quickly engage in these natural cadherin interactions. As a result, the DNAMeter can now be used to study long-term cellular events.
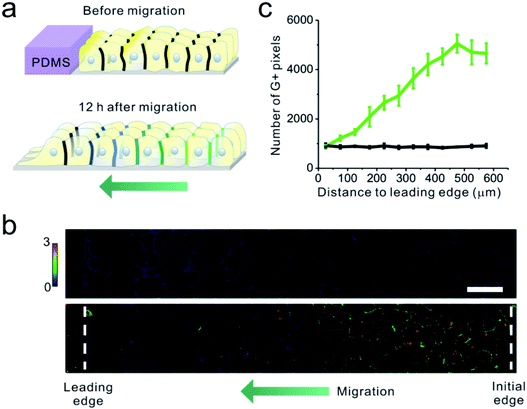
Finally, we applied the EC22-DNAMeter to measure intercellular E-cadherin tensions during collective epithelial migrations. A slab of polydimethylsiloxane (PDMS) was pre-attached onto a substrate, and then a confluent MDCK cell monolayer was formed adjacent to the PDMS slab.37 The interface between the monolayer and PDMS was defined as the initial edge. After the removal of the PDMS slab, the exposed free space triggers the migration of the cell sheet, emulating the wound healing process. After the initial force mapping with the EC22-DNAMeter, we replaced the HEPES-buffered saline with the complete cell growth medium. Following another 12 h of cell growth and migration, fresh EC22-DNAMeter was added to measure intercellular E-cadherin tensions (Fig. 5a and S15†). Before removing the PDMS slab, intercellular forces mediated by E-cadherin were rarely observed within the cell sheet (Fig. 5b). After allowing the cell sheet to migrate for 12 h, interestingly, junctional pixels of high G/Y ratio were clearly observed in the regions ∼15 cell lengths from the leading edge of migration (Fig. 5b). We have further quantified the correlation between the number of pixels experiencing >4.4 pN forces and their distance to the leading edge (Fig. S16† and 5c, Materials and methods). As the distance increased, the percentage of E-cadherins undergoing intercellular tensions also linearly increased. In comparison, negligible forces were observed throughout the imaging zone before removing the PDMS (Fig. 5c). Overall, these observations are in good agreement with some previous studies on the global force distributions during this process.17,37,56
Conclusions
In this study, we have developed a DNA-based nanoprobe to quantify, at the molecular level, E-cadherin-mediated tensile forces at cell–cell junctions. The so-called DNAMeter exhibits several unique features. First, the intrinsic modularity and precise self-assembly of the DNA scaffold allows the accurate positioning of specific number of reference fluorophores, reporter fluorophores and quenchers.33,57–59 As a result, a facile ratiometric quantification of tensile forces can be achieved. Predictably, through the rational tuning of the sequence and length of the DNA hairpin, the threshold force can be tailored in a large range to study different intensities of intercellular forces.32,33 By further conjugating two hairpins into one self-assembled “rod”-like DNA structure, a large range of tensile forces can be measured simultaneously. Compared to two separated membrane DNA hairpin probes, the conjugated DNAMeter allows the use of one reference fluorophore to characterize the membrane distributions of both hairpins. Supposedly, more hairpins can be incorporated into the DNAMeter to realize delicate quantification of an even larger range of forces.Compared to the traction force microscopy,23,24 the beauty of the DNAMeter is its capability to distinguish tension mediated by a particular protein from the total forces at cell–cell junctions. Compatible with readily accessible fluorescence microscopes, the DNAMeter is also easy to prepare and use. By simply incubating with the target cells, the DNAMeter can be spontaneously anchored onto cell membrane to report the tensions. Compared to fluorescent protein-based sensors and cantilever pillars,13,25 there is no requirement for the cloning or microfabrication. In addition, the obtained fluorescence signals can be straightforwardly converted into mechanical forces without the need of complicated data processing or analysis. As a natural component in the cell plasma membrane, the cholesterol anchors can freely diffuse along the membrane.30 In addition, the cholesterol–DNA conjugates can also be effectively removed if desired (Fig. S13 and S14†).
We have demonstrated in detail two approaches to quantify the force distributions by either the number of bright pixels or the unfolded DNA probes. Both approaches can be facilely applied for mapping intercellular forces. In addition, the EC-DNAMeter has been used to quantify intercellular E-cadherin tension during the collective migration of cell sheet. In principle, the DNAMeter can also be used to quantify three-dimensional protein-specific intercellular forces in physiologically relevant multi-layer cell assemblies or tissues.60,61 Our study demonstrated the ability of the DNAMeter to quantify and real-time monitor mechanical forces within a colony of cells. With a broad choice of fluorophores and quenchers, the DNAMeter can be further used to simultaneously measure intercellular forces among different receptor–ligand pairs, and to study the correlations between forces and the concentration gradients of morphogens or signaling molecules.62 Further applications of the DNAMeter will allow the construction of more accurate mechanical models to study mechanotransduction during embryogenesis, morphogenesis, and various physiological and pathological processes.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge NIH R35GM133507, a start-up grant from UMass Amherst and IALS M2M seed grant to M. You and NSF CMMI 1662835 to Y. Sun. We are grateful to Dr James Chambers for the assistance in fluorescence imaging, and Dr Tianxi Yang for manuscript preparation. We also thank every other member of the You Lab and Dr Craig Martin for useful discussion.Notes and references
- G. Charras and A. S. Yap, Curr. Biol., 2018, 28, R445–R457 CrossRef CAS PubMed.
- M. A. Wozniak and C. S. Chen, Nat. Rev. Mol. Cell Biol., 2009, 10, 34–43 CrossRef CAS PubMed.
- C. S. Chen, J. Tan and J. Tien, Annu. Rev. Biomed. Eng., 2004, 6, 275–302 CrossRef CAS PubMed.
- E. Levine, C. H. Lee, C. Kintner and B. M. Gumbiner, Development, 1994, 120, 901–909 CAS.
- A. C. Martin, M. Kaschube and E. F. Wieschaus, Nature, 2008, 457, 495 CrossRef PubMed.
- E. Bazellières, V. Conte, A. Elosegui-Artola, X. Serra-Picamal, M. Bintanel-Morcillo, P. Roca-Cusachs, J. J. Muñoz, M. Sales-Pardo, R. Guimerà and X. Trepat, Nat. Cell Biol., 2015, 17, 409–420 CrossRef PubMed.
- B. R. Acharya, S. K. Wu, Z. Z. Lieu, R. G. Parton, S. W. Grill, A. D. Bershadsky, G. A. Gomez and A. S. Yap, Cell Rep., 2017, 18, 2854–2867 CrossRef CAS PubMed.
- D. E. Conway, M. T. Breckenridge, E. Hinde, E. Gratton, C. S. Chen and M. A. Schwartz, Curr. Biol., 2013, 23, 1024–1030 CrossRef CAS PubMed.
- M. Takeichi, Development, 1988, 102, 639–655 CAS.
- F. van Roy and G. Berx, Cell. Mol. Life Sci., 2008, 65, 3756–3788 CrossRef CAS PubMed.
- D. E. Leckband, Q. le Duc, N. Wang and J. de Rooij, Curr. Opin. Cell Biol., 2011, 23, 523–530 CrossRef CAS PubMed.
- C. M. Niessen, D. Leckband and A. S. Yap, Physiol. Rev., 2011, 91, 691–731 CrossRef CAS PubMed.
- Z. Liu, J. L. Tan, D. M. Cohen, M. T. Yang, N. J. Sniadecki, S. A. Ruiz, C. M. Nelson and C. S. Chen, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 9944–9949 CrossRef CAS PubMed.
- J. M. Halbleib and W. J. Nelson, Genes Dev., 2006, 20, 3199–3214 CrossRef CAS PubMed.
- M. P. Stemmler, Mol. BioSyst., 2008, 4, 835–850 RSC.
- A. Brugués, E. Anon, V. Conte, J. H. Veldhuis, M. Gupta, J. Colombelli, J. J. Muñoz, G. W. Brodland, B. Ladoux and X. Trepat, Nat. Phys., 2014, 10, 683–690 Search PubMed.
- X. Trepat, M. R. Wasserman, T. E. Angelini, E. Millet, D. A. Weitz, J. P. Butler and J. J. Fredberg, Nat. Phys., 2009, 5, 426–430 Search PubMed.
- F. van Roy, Nat. Rev. Cancer, 2014, 14, 121–134 CrossRef CAS PubMed.
- D. E. Leckband and J. d. Rooij, Annu. Rev. Cell Dev. Biol., 2014, 30, 291–315 CrossRef CAS PubMed.
- W. J. Polacheck and C. S. Chen, Nat. Methods, 2016, 13, 415–423 CrossRef CAS PubMed.
- P. Roca-Cusachs, V. Conte and X. Trepat, Nat. Cell Biol., 2017, 19, 742–751 CrossRef CAS PubMed.
- M. Eisenstein, Nature, 2017, 544, 255–257 CrossRef CAS PubMed.
- D. T. Tambe, C. Corey Hardin, T. E. Angelini, K. Rajendran, C. Y. Park, X. Serra-Picamal, E. H. Zhou, M. H. Zaman, J. P. Butler, D. A. Weitz, J. J. Fredberg and X. Trepat, Nat. Mater., 2011, 10, 469–475 CrossRef CAS PubMed.
- V. Maruthamuthu, B. Sabass, U. S. Schwarz and M. L. Gardel, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 4708–4713 CrossRef CAS PubMed.
- N. Borghi, M. Sorokina, O. G. Shcherbakova, W. I. Weis, B. L. Pruitt, W. J. Nelson and A. R. Dunn, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 12568–12573 CrossRef CAS PubMed.
- B. D. Hoffman and A. S. Yap, Trends Cell Biol., 2015, 25, 803–814 CrossRef CAS PubMed.
- P. Wang, J. Liang, L. Z. Shi, Y. Wang, P. Zhang, M. Ouyang, D. Preece, Q. Peng, L. Shao, J. Fan, J. Sun, S. S. Li, M. W. Berns, H. Zhao and Y. Wang, ACS Photonics, 2018, 5, 3565–3574 CrossRef CAS PubMed.
- C. Jurchenko and K. S. Salaita, Mol. Cell. Biol., 2015, 35, 2570–2582 CrossRef CAS PubMed.
- B. Zhao, C. O'Brien, A. P. K. K. K. Mudiyanselage, N. Li, Y. Bagheri, R. Wu, Y. Sun and M. You, J. Am. Chem. Soc., 2017, 139, 18182–18185 CrossRef CAS PubMed.
- M. You, Y. Lyu, D. Han, L. Qiu, Q. Liu, T. Chen, C. Wu, L. Peng, L. Zhang, G. Bao and W. Tan, Nat. Nanotechnol., 2017, 12, 453–459 CrossRef CAS PubMed.
- A. Bunge, M. Loew, P. Pescador, A. Arbuzova, N. Brodersen, J. Kang, L. Dähne, J. Liebscher, A. Herrmann, G. Stengel and D. Huster, J. Phys. Chem. B, 2009, 113, 16425–16434 CrossRef CAS PubMed.
- X. Wang and T. Ha, Science, 2013, 340, 991–994 CrossRef CAS PubMed.
- Y. Zhang, C. Ge, C. Zhu and K. Salaita, Nat. Commun., 2014, 5, 5167 CrossRef CAS PubMed.
- B. L. Blakely, C. E. Dumelin, B. Trappmann, L. M. McGregor, C. K. Choi, P. C. Anthony, V. K. Duesterberg, B. M. Baker, S. M. Block, D. R. Liu and C. S. Chen, Nat. Methods, 2014, 11, 1229–1232 CrossRef CAS PubMed.
- M. T. Woodside, W. M. Behnke-Parks, K. Larizadeh, K. Travers, D. Herschlag and S. M. Block, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 6190–6195 CrossRef CAS PubMed.
- N. M. Kronenberg, P. Liehm, A. Steude, J. A. Knipper, J. G. Borger, G. Scarcelli, K. Franze, S. J. Powis and M. C. Gather, Nat. Cell Biol., 2017, 19, 864–872 CrossRef CAS PubMed.
- S. R. K. Vedula, M. C. Leong, T. L. Lai, P. Hersen, A. J. Kabla, C. T. Lim and B. Ladoux, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 12974–12979 CrossRef CAS PubMed.
- L. Li, Y. He, M. Zhao and J. Jiang, Burns Trauma, 2013, 1, 21–26 CrossRef PubMed.
- J. F. Marko and E. D. Siggia, Macromolecules, 1995, 28, 8759–8770 CrossRef CAS.
- S. B. Smith, Y. Cui and C. Bustamante, Science, 1996, 271, 795–799 CrossRef CAS PubMed.
- X. Wang, Z. Rahil, I. T. Li, F. Chowdhury, D. E. Leckband, Y. R. Chemla and T. Ha, Sci. Rep., 2016, 6, 21584 CrossRef CAS PubMed.
- H. Liu, Z. Zhu, H. Kang, Y. Wu, K. Sefan and W. Tan, Chem. –Eur. J., 2010, 16, 3791–3797 CrossRef CAS PubMed.
- Y. Bagheri, S. Chedid, F. Shafiei, B. Zhao and M. You, Chem. Sci., 2019, 10, 11030–11040 RSC.
- F. W. S. Stetter, L. Cwiklik, P. Jungwirth and T. Hugel, Biophys. J., 2014, 107, 1167–1175 CrossRef CAS PubMed.
- P. Katsamba, K. Carroll, G. Ahlsen, F. Bahna, J. Vendome, S. Posy, M. Rajebhosale, S. Price, T. M. Jessell, A. Ben-Shaul, L. Shapiro and B. H. Honig, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 11594–11599 CrossRef CAS PubMed.
- J. K. Hannestad, R. Brune, I. Czolkos, A. Jesorka, A. H. El-Sagheer, T. Brown, B. Albinsson and O. Orwar, ACS Nano, 2013, 7, 308–315 CrossRef CAS PubMed.
- F. Twiss, Q. Le Duc, S. Van Der Horst, H. Tabdili, G. Van Der Krogt, N. Wang, H. Rehmann, S. Huveneers, D. E. Leckband and J. J. B. o. De Rooij, Biol. Open, 2012, 1, 1128–1140 CrossRef CAS PubMed.
- R. Seddiki, G. H. N. S. Narayana, P.-O. Strale, H. E. Balcioglu, G. Peyret, M. Yao, A. P. Le, C. Teck Lim, J. Yan, B. Ladoux and R. M. Mège, Mol. Biol. Cell, 2018, 29, 380–388 CrossRef CAS PubMed.
- I. Muhamed, J. Wu, P. Sehgal, X. Kong, A. Tajik, N. Wang and D. E. Leckband, J. Cell Sci., 2016, 129, 1843–1854 CrossRef CAS PubMed.
- X. Tian, Z. Liu, B. Niu, J. Zhang, T. K. Tan, S. R. Lee, Y. Zhao, D. C. Harris and G. J. B. R. I. Zheng, J. Biomed. Biotechnol., 2011, 2011, 567305 Search PubMed.
- A. Hartsock and W. Nelson, Biochim. Biophys. Acta, 2008, 1778, 660–669 CrossRef CAS PubMed.
- S. Pokutta, K. Herrenknecht, R. Kemler and J. Engel, Eur. J. Biochem., 1994, 223, 1019–1026 CrossRef CAS PubMed.
- B. Rothen-Rutishauser, F. K. Riesen, A. Braun, M. Günthert and H. Wunderli-Allenspach, J. Membr. Biol., 2002, 188, 151–162 CrossRef CAS PubMed.
- A. M. Shewan, M. Maddugoda, A. Kraemer, S. J. Stehbens, S. Verma, E. M. Kovacs and A. Yap, Mol. Biol. Cell, 2005, 16, 4531–4542 CrossRef CAS PubMed.
- P. Pandya, J. L. Orgaz and V. Sanz-Moreno, Curr. Opin. Cell Biol., 2017, 48, 87–96 CrossRef CAS PubMed.
- C. Gayrard, C. Bernaudin, T. Déjardin, C. Seiler and N. Borghi, J. Cell Biol., 2018, 217, 1063–1077 CrossRef CAS PubMed.
- S. Surana, J. M. Bhat, S. P. Koushika and Y. Krishnan, Nat. Commun., 2011, 2, 340 CrossRef PubMed.
- K. Chakraborty, A. T. Veetil, S. R. Jaffrey and Y. Krishnan, Annu. Rev. Biochem., 2016, 85, 349–373 CrossRef CAS PubMed.
- S. Ranallo, A. Porchetta and F. Ricci, Anal. Chem., 2019, 91, 44–59 CrossRef CAS PubMed.
- W. R. Legant, A. Pathak, M. T. Yang, V. S. Deshpande, R. M. McMeeking and C. S. Chen, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 10097–10102 CrossRef CAS PubMed.
- C. M. Nelson, J. Cell Biol., 2017, 216, 29–30 CrossRef CAS PubMed.
- T. Tabata and Y. Takei, Development, 2004, 131, 703–712 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and methods, scheme S1, Fig. S1–S16, and tables S1–S4. See DOI: 10.1039/d0sc01455a |
| This journal is © The Royal Society of Chemistry 2020 |