Open Access Article
Open Access ArticleA poly-ADP-ribose polymer-based antibody-drug conjugate†
Xiaojing
Shi‡
a,
Xiao-Nan
Zhang‡
 a,
Jingwen
Chen‡
a,
Qinqin
Cheng
a,
Hua
Pei
b,
Stan G.
Louie
b and
Yong
Zhang
a,
Jingwen
Chen‡
a,
Qinqin
Cheng
a,
Hua
Pei
b,
Stan G.
Louie
b and
Yong
Zhang
 *acde
*acde
aDepartment of Pharmacology and Pharmaceutical Sciences, School of Pharmacy, University of Southern California, Los Angeles, CA 90089, USA
bTitus Family Department of Clinical Pharmacy, School of Pharmacy, University of Southern California, Los Angeles, CA 90089, USA
cDepartment of Chemistry, Dornsife College of Letters, Arts and Sciences, University of Southern California, Los Angeles, CA 90089, USA
dNorris Comprehensive Cancer Center, University of Southern California, Los Angeles, CA 90089, USA
eResearch Center for Liver Diseases, University of Southern California, Los Angeles, CA 90089, USA. E-mail: yongz@usc.edu
First published on 17th August 2020
Abstract
Protein poly-ADP-ribosylation (PARylation) plays vital roles in many aspects of physiology and pathophysiology. This posttranslational modification is catalyzed by poly-ADP-ribose polymerases (PARPs) through additions of ADP-ribose from nicotinamide adenine dinucleotide (NAD+) to protein residues, forming linear or branched poly-ADP-ribose (PAR) polymers. In this study, we explored a new concept of utilizing functionalized PAR polymers for targeted drug delivery. This was achieved by rapid and efficient generation of auto-PARylated PARP1 with 3′-azido ADP-riboses and subsequent conjugations of anti-human epidermal growth factor receptor 2 (HER2) antibodies and monomethyl auristatin F (MMAF) payloads. This designed PARylated PARP1-antibody-MMAF conjugate could potently kill HER2-expressing cancer cells in high specificity. This proof-of-principle work demonstrates the feasibility of production of PAR polymer-based antibody-drug conjugate and its application in targeted delivery. The PAR polymer-based conjugates may lead to new types of therapeutics with potentially improved physicochemical and pharmacological properties.
Introduction
Poly-ADP-ribose polymerases (PARPs) catalyze protein poly-ADP-ribosylation (PARylation). This enzymatic posttranslational modification requires nicotinamide adenine dinucleotide (NAD+) as a donor of ADP-ribose. Upon covalent attachments of ADP-riboses to side chains of various types of amino acid residues, PARPs can continue adding ADP-ribose sequentially at ribosyl 2′-OH positions, resulting in linear or branched poly-ADP-ribose (PAR) polymers with up to 300 ADP-ribose units in length.1,2 As the founding member of the PARP family, PARP1 accounts for 75–95% cellular PARylation activities under genotoxic conditions.3–5 In addition to PARylating many protein substrates, PARP1 undergoes robust auto-PARylation. By adding PAR polymers to itself and other proteins, PARP1-mediated PARylation plays important roles in regulating genome stability, gene expression, protein homeostasis, cell proliferation, differentiation and apoptosis.6–10Antibody-drug conjugates (ADCs) are one of the fast-growing classes of therapeutics that exploit monoclonal antibodies for cell- and tissue-specific drug delivery. The average drug-to-antibody ratios (DARs) of most ADCs currently in clinic are 3.5–4, resulted from a compromise between potency and physicochemical properties.11 Although a higher DAR is often associated with increased in vitro potency, it may unfavorably affect ADC properties. As many payloads used in ADCs are very hydrophobic, an increased DAR could decrease stability, induce aggregation, and paradoxically reduce therapeutic effectiveness.12,13 The choice of drug payloads for ADCs is thus usually limited to highly potent agents.
In comparison, polymer–drug conjugates allow to carry a significant number of payloads. Attachments of small-molecule drugs to synthetic hydrophilic polymers could enhance drug loading capacity and solubility,14 which may translate into improved pharmacological properties. However, due to potential biocompatibility issues, this type of conjugates with unnatural polymer backbones may cause slow drug release and/or incomplete biodegradation, raising concerns on their efficacy and safety profiles.15–17
PAR is a natural form of hydrophilic polymer composed of ADP-ribose units connected in a linear or branched fashion. As the products of PARP-catalyzed reactions, PAR polymers vary significantly in length and branching pattern,18 resulting in high degree of heterogeneity. Auto-PARylation of PARP1 could generate PARs ranging from 6-unit oligomers to heterogeneous polymers with an average size of 200 units of ADP-ribose and approximately five branches per polymer in average.19 Cellular PAR could be efficiently degraded by various enzymes. For example, poly(ADP-ribose) glycohydrolase (PARG) can hydrolyze the glycosidic bond between ADP-ribose units to generate free ADP-ribose.20,21 NUDT9, NUDT16, and ENPP1 can cleave the pyrophosphate bond within the ADP-ribose and release phosphoribosyl-AMP from PAR chains.22
We recently developed a ribose-functionalized NAD+ with nicotinamide riboside (NR) 3′-azido group.23 Its substrate activity for PARP1-catalyzed PARylation is comparable to that of NAD+, making it possible to rapidly incorporate 3′-azido ADP-ribose units into PAR polymers in high efficiency. This result raises the question of whether azido-functionalized PAR polymers could serve as carriers of targeting moieties and small-molecule drugs for targeted delivery. Derived from NAD+-dependent enzymatic PARylation, PAR is a hydrophilic and biocompatible polymer. Covalent attachments of small-molecule drugs to functionalized PAR polymers can increase overall drug loading and solubilization, possibly expanding the spectrum of payloads for targeted delivery. Similar to ADCs with pyrophosphate diester-derived linkers,24,25 PAR polymer–drug conjugates may allow to stably carry payloads in plasma and rapidly release drugs upon internalization into target cells. Collectively, PAR polymer-based ADCs may provide improved physicochemical properties, therapeutic efficacy, and safety profiles. To begin to explore this concept, we generated a PAR polymer-based ADC through clicking anti-human epidermal growth factor receptor 2 (HER2) antibody trastuzumab and monomethyl auristatin F (MMAF) with auto-PARylated PARP1 by the 3′-azido NAD+ (Fig. 1). The resulting PARylated PARP1-Fab-MMAF conjugate revealed excellent and specific cytotoxicity for HER2-expressing breast cancer cells, demonstrating the feasibility of applying functionalized PAR polymers to targeted drug delivery.
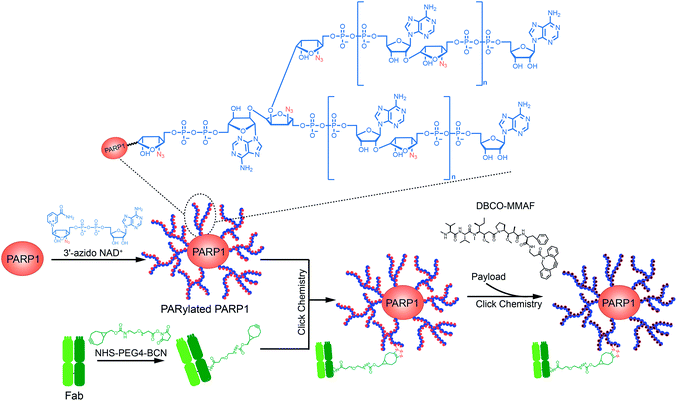 | ||
| Fig. 1 Schematic of the design and generation of a poly-ADP-ribose polymer-based antibody-drug conjugate. | ||
Results and discussion
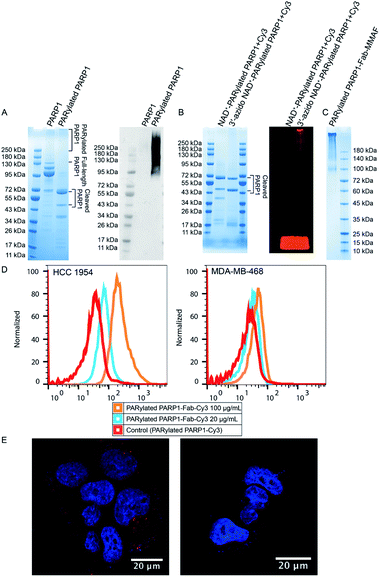
To generate PAR polymers conjugated with targeting moieties and small-molecule drugs, we chose auto-PARylated human PARP1 as model PAR polymers given its strong automodification activity. Anti-HER2 antibody trastuzumab is an approved drug to treat HER2-positive breast cancer.26–29 MMAF is a tubulin inhibitor broadly used in ADCs.30–32 Conjugation of MMAF with several antibodies resulted in ADCs with potent anti-tumor activities.33–36 The free carboxyl group of MMAF also facilitates synthesis of drug-linker conjugates with high potency.37 Trastuzumab and MMAF were selected as a model targeting antibody and cytotoxic payload for synthesizing the PAR polymer-based ADC.First, full-length human PARP1 and the antigen-binding fragment (Fab) of trastuzumab were expressed and purified from Escherichia coli (Fig. S1A†). In addition to the full-length PARP1, two fragments with molecular weights approximately over 95 kDa were co-purified (Fig. 2A and S1A†). Auto-PARylated PARP1 by 3′-azido NAD+ was then prepared (Fig. 1). As revealed by immunoblotting using an anti-PAR antibody, PARP1 could be modified by 3′-azido NAD+-derived PAR polymers (Fig. 2A). SDS-PAGE analysis by Coomassie stain showed cleaved fragments generated during auto-PARylation (Fig. 2A). PARP1 is sensitive to various proteases, which can result in the production of different PARP1 signature fragments.38,39 Weaker signals were also seen for heterogenous auto-PARylated PARP1 as shown in a smearing pattern at upper positions (Fig. 2A). To prepare auto-PARylated PARP1, we tested different temperature, reaction time, and concentrations of PARP1 and 3′-azido NAD+ and chose to incubate 5 μM of PARP1 with 250 μM of 3′-azido NAD+ at 30 °C for 12 hours to ensure efficient auto-PARylation at a relatively high level. To verify the generation of azido-functionalized PAR polymers, dibenzocyclooctyne (DBCO)-Cy3 fluorescent dye was incubated with NAD+- or 3′-azido NAD+-PARylated PARP1. Consistent with the design, in-gel fluorescence imaging revealed fluorescently labeled PAR polymers only for the reaction mixture with PARP1 automodified by 3′-azido NAD+ (Fig. 2B).
To synthesize PARylated PARP1-Fab conjugates, trastuzumab Fab functionalized with bicyclo[6.1.0]nonyne (BCN) was incubated with 3′-azido NAD+-PARylated PARP1, followed by conjugation with DBCO-Cy3 or DBCO-MMAF (Fig. S2†). The generated PARylated PARP1-Fab-Cy3 and PARylated PARP1-Fab-MMAF conjugates were purified by size-exclusion chromatography (Fig. S1B† and 2C). In addition, a DBCO-MMAF conjugate with a disulfide bond in the linker region (denoted by DBCO-S-S-MMAF) was synthesized (Fig. S3†). By replacing DBCO-MMAF with DBCO-S-S-MMAF, PARylated PARP1-Fab-S-S-MMAF conjugate was generated under the same conditions (Fig. S1C†), which facilitated the release of conjugated MMAF payloads by reducing reagents for quantification.
Next, the binding of PARylated PARP1-Fab-Cy3 conjugate to cell-surface HER2 receptor was analyzed by flow cytometry using HER2-positive HCC 1954 and HER2-negative MDA-MB-468 cells (Fig. 2D and S4†). PARylated PARP1-Fab-Cy3 conjugate could bind to HCC 1954 cells in a dose-dependent manner but showed no binding to MDA-MB-468 cells, demonstrating that the conjugated trastuzumab Fab allows to target PAR polymer-based conjugates to HER2-expressing cells. Following flow cytometric analysis, cellular uptake of PARylated PARP1-Fab-Cy3 conjugate was evaluated by confocal microscopy (Fig. 2E). HCC 1954 cells were incubated with PARylated PARP1-Fab-Cy3 conjugate in the absence or presence of trastuzumab Fab at 37 °C. The addition of trastuzumab Fab was to compete with the PARylated PARP1-Fab-Cy3 conjugate for the binding to cell surface HER2 receptors. Confocal imaging revealed red fluorescent puncta indicating internalized PARylated PARP1-Fab-Cy3 conjugates in the absence of trastuzumab Fab (Fig. 2E). The internalization of PARylated PARP1-Fab-Cy3 conjugate was found to be sensitive to binding competition by added trastuzumab Fab. These results suggest that PAR polymer-Fab conjugates could be internalized through HER2 receptor-mediated endocytosis, potentially enabling delivery of conjugated drugs to target cells and tissues.
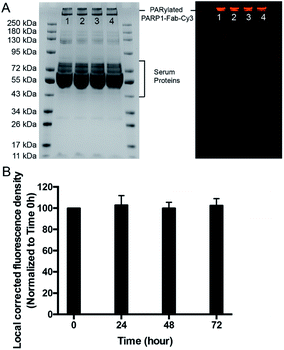
The stability of the generated PARylated PARP1-Fab-Cy3 conjugate in cell culture media was then assessed (Fig. 3). PARylated PARP1-Fab-Cy3 conjugate diluted in RPMI 1640 medium with 10% fetal bovine serum (FBS) was incubated for 0–72 hours at 37 °C. In-gel fluorescence analysis indicated no significant changes in the pattern and fluorescence intensity for the PARylated PARP1-Fab-Cy3 conjugate across various time points. These data suggest that PAR polymer-based conjugates are stable in cell culture media and may allow to stably attach drugs for targeted delivery.
To quantify the total amount of payloads on PARylated PARP1-Fab conjugates, PARylated PARP1-Fab-Cy3 and PARylated PARP1-Fab-S-S-MMAF were treated by phosphodiesterase I (PDE I) and dithiothreitol (DTT), respectively, to release the conjugated Cy3 and MMAF molecules for quantification (Fig. S5†). Based on measured fluorescence intensities of PDE I-treated PARylated PARP1-Fab-Cy3 conjugate, PARylated PARP1-Fab-Cy3 was calculated to carry 45.64 ± 2.45 μmol of Cy3 dye per gram of protein. Through HPLC analysis of released MMAF-SH, each gram of protein for PARylated PARP1-Fab-S-S-MMAF was determined to contain 9.94 ± 0.73 μmol of MMAF molecules, slightly lower than but consistent with amount of conjugated Cy3 dye. Based on the determined number of MMAF payload for PARylated PARP1-Fab-S-S-MMAF, the amounts of conjugated MMAF were calculated for PARylated PARP1-Fab-MMAF conjugate and used for potency determination in comparison with free DBCO-MMAF.
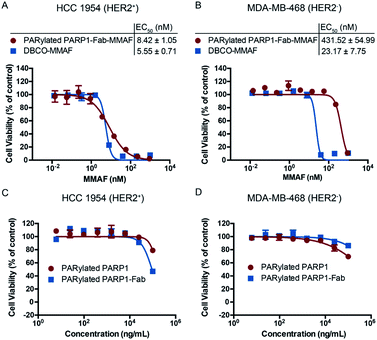
In vitro cytotoxicity assays were then performed using both HCC 1954 and MDA-MB-468 cells (Fig. 4). PARylated PARP1-Fab-MMAF conjugate showed potent cytotoxicity (EC50 = 8.42 ± 1.05 nM) for HER2-expressing HCC 1954 cells, comparable to that of free DBCO-MMAF. In contrast to DBCO-MMAF with an EC50 of 23.17 nM for MDA-MB-468 cells, the PARylated PARP1-Fab-MMAF conjugate revealed significantly decreased cytotoxicity for this HER2-negative cell line (EC50 = 431.52 ± 54.99 nM). As controls, PARylated PARP1 and PARylated PARP1-Fab conjugates have minimal cytotoxicity for both cell lines. Assays with several non-breast cancer cell lines also indicated low cytotoxicity for PARylated PARP1 (Fig. S6†). Additionally, PARylated PARP1-Fab-MMAF conjugates prepared from four different batches display consistent in vitro cytotoxicity for HCC 1954 cells in a range of 504–820 ng mL−1 (Fig. S7†). These results demonstrate the potency and specificity of PARylated PARP1-Fab-MMAF conjugate in killing HER2-expressing breast cancer cells.
Our previous study suggested that payload stably attached to an antibody via an NAD+ analogue-based linker could be rapidly released upon cellular internalization.25 The payload release of the PAR polymer-based ADC in cellular environment will be investigated. In addition to mechanistic studies of the degradation of functionalized PAR polymer and the drug release, future studies include systematic evaluation of pharmacokinetics, therapeutic efficacy, and toxicity of the PARylated PARP1-Fab-MMAF conjugate in comparison to established ADCs and polymer–drug conjugates. Extension of this approach to other disease targets and cytotoxic and non-cytotoxic agents will also be explored. In addition, PAR polymers with two distinct functional groups could be developed to enable orthogonal conjugations of targeting moiety and payload.
Conclusions
In summary, this is the first report of utilizing functionalized PAR polymer–drug conjugates for targeted drug delivery. Using the model anti-HER2 trastuzumab Fab and MMAF payload, PARylated PARP1-Fab-MMAF conjugate could be successfully generated and exhibits potent and specific cytotoxicity toward HER2-expressing cancer cells. In comparison to established systems for targeted drug delivery, the PAR polymer-based ADC represents a novel platform for generating therapeutics with potentially enhanced physicochemical and pharmacological properties.Conflicts of interest
The authors have no conflicts to declare.Acknowledgements
We are grateful to Dr. Peter G. Schultz (The Scripps Research Institute) for providing the pBAD vector encoding trastuzumab Fab. This work was supported in part by University of Southern California School of Pharmacy Start-Up Fund for New Faculty, Sharon L. Cockrell Cancer Research Fund, STOP CANCER Research Career Development Award (to Y. Z.), National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health (NIH) grant R35GM137901 (to Y. Z.), and National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) of the NIH grant P30DK048522 (to USC Research Center for Liver Diseases).References
- R. Alvarez-Gonzalez and M. K. Jacobson, Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo, Biochemistry, 1987, 26(11), 3218–3224 CrossRef CAS PubMed
.
- G. Keith, J. Desgres and G. de Murcia, Use of two-dimensional thin-layer chromatography for the components study of poly(adenosine diphosphate ribose), Anal. Biochem., 1990, 191(2), 309–313 CrossRef CAS PubMed
.
- G. Zarkovic, E. A. Belousova, I. Talhaoui, C. Saint-Pierre, M. M. Kutuzov, B. T. Matkarimov, D. Biard, D. Gasparutto, O. I. Lavrik and A. A. Ishchenko, Characterization of DNA ADP-ribosyltransferase activities of PARP2 and PARP3: new insights into DNA ADP-ribosylation, Nucleic Acids Res., 2018, 46(5), 2417–2431 CrossRef CAS PubMed
.
- W. M. Shieh, J. C. Ame, M. V. Wilson, Z. Q. Wang, D. W. Koh, M. K. Jacobson and E. L. Jacobson, Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers, J. Biol. Chem., 1998, 273(46), 30069–30072 CrossRef CAS PubMed
.
- J. C. Ame, V. Rolli, V. Schreiber, C. Niedergang, F. Apiou, P. Decker, S. Muller, T. Hoger, J. Menissier-de Murcia and G. de Murcia, PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase, J. Biol. Chem., 1999, 274(25), 17860–17868 CrossRef CAS PubMed
.
- B. A. Gibson and W. L. Kraus, New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs, Nat. Rev. Mol. Cell Biol., 2012, 13(7), 411–424 CrossRef CAS PubMed
.
- P. O. Hassa, S. S. Haenni, M. Elser and M. O. Hottiger, Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going?, Microbiol. Mol. Biol. Rev., 2006, 70(3), 789–829 CrossRef CAS PubMed
.
- M. O. Hottiger, Nuclear ADP-Ribosylation and Its Role in Chromatin Plasticity, Cell Differentiation, and Epigenetics, Annu. Rev. Biochem., 2015, 84, 227–263 CrossRef CAS PubMed
.
- K. W. Ryu, D. S. Kim and W. L. Kraus, New Facets in the Regulation of Gene Expression by ADP-Ribosylation and Poly(ADP-ribose) Polymerases, Chem. Rev., 2015, 115(6), 2453–2481 CrossRef CAS PubMed
.
- J. Zheng, H. Xue, H. Zhang, L. Wang, L. Zheng, X. Wang, X. Lin, S. Jin and J. Wu, The influence of therapeutic vaccine candidate against HBeAg pEGFP-N1-C (472-507)-ecdCD40L on dendritic cells, Acta Virol., 2018, 62(2), 157–163 CrossRef CAS PubMed
.
- A. Beck, L. Goetsch, C. Dumontet and N. Corvaia, Strategies and challenges for the next generation of antibody-drug conjugates, Nat. Rev. Drug Discovery, 2017, 16(5), 315–337 CrossRef CAS PubMed
.
- J. Guo, S. Kumar, M. Chipley, O. Marcq, D. Gupta, Z. Jin, D. S. Tomar, C. Swabowski, J. Smith, J. A. Starkey and S. K. Singh, Characterization and Higher-Order Structure Assessment of an Interchain Cysteine-Based ADC: Impact of Drug Loading and Distribution on the Mechanism of Aggregation, Bioconjugate Chem., 2016, 27(3), 604–615 CrossRef CAS PubMed
.
- A. W. Tolcher, Antibody drug conjugates: lessons from 20 years of clinical experience, Annu. Oncol., 2016, 27(12), 2168–2172 CrossRef CAS PubMed
.
- N. Larson and H. Ghandehari, Polymeric conjugates for drug delivery, Chem. Mater., 2012, 24(5), 840–853 CrossRef CAS PubMed
.
- I. Ekladious, Y. L. Colson and M. W. Grinstaff, Polymer-drug conjugate therapeutics: advances, insights and prospects, Nat. Rev. Drug Discovery, 2019, 18(4), 273–294 CrossRef CAS PubMed
.
- K. Knop, R. Hoogenboom, D. Fischer and U. S. Schubert, Poly(ethylene glycol) in drug delivery: pros and cons as well as potential alternatives, Angew. Chem., Int. Ed., 2010, 49(36), 6288–6308 CrossRef CAS PubMed
.
- R. P. Garay, R. El-Gewely, J. K. Armstrong, G. Garratty and P. Richette, Antibodies against polyethylene glycol in healthy subjects and in patients treated with PEG-conjugated agents, Expert Opin. Drug Delivery, 2012, 9(11), 1319–1323 CrossRef CAS PubMed
.
- E. S. Tan, K. A. Krukenberg and T. J. Mitchison, Large-scale preparation and characterization of poly(ADP-ribose) and defined length polymers, Anal. Biochem., 2012, 428(2), 126–136 CrossRef CAS PubMed
.
- C. C. Kiehlbauch, N. Aboul-Ela, E. L. Jacobson, D. P. Ringer and M. K. Jacobson, High resolution fractionation and characterization of ADP-ribose polymers, Anal. Biochem., 1993, 208(1), 26–34 CrossRef CAS PubMed
.
- J. P. Gagne, M. J. Hendzel, A. Droit and G. G. Poirier, The expanding role of poly(ADP-ribose) metabolism: current challenges and new perspectives, Curr. Opin. Cell Biol., 2006, 18(2), 145–151 CrossRef CAS PubMed
.
- V. Schreiber, F. Dantzer, J. C. Ame and G. de Murcia, Poly(ADP-ribose): novel functions for an old molecule, Nat. Rev. Mol. Cell Biol., 2006, 7(7), 517–528 CrossRef CAS PubMed
.
- J. O'Sullivan, M. Tedim Ferreira, J. P. Gagne, A. K. Sharma, M. J. Hendzel, J. Y. Masson and G. G. Poirier, Emerging roles of eraser enzymes in the dynamic control of protein ADP-ribosylation, Nat. Commun., 2019, 10(1), 1182 CrossRef PubMed
.
- X.-N. Zhang, Q. Cheng, J. Chen, A. T. Lam, Y. Lu, Z. Dai, H. Pei, N. M. Evdokimov, S. G. Louie and Y. Zhang, A ribose-functionalized NAD+ with unexpected high activity and selectivity for protein poly-ADP-ribosylation, Nat. Commun., 2019, 10(1), 4196 CrossRef PubMed
.
- J. C. Kern, M. Cancilla, D. Dooney, K. Kwasnjuk, R. Zhang, M. Beaumont, I. Figueroa, S. Hsieh, L. Liang, D. Tomazela, J. Zhang, P. E. Brandish, A. Palmieri, P. Stivers, M. Cheng, G. Feng, P. Geda, S. Shah, A. Beck, D. Bresson, J. Firdos, D. Gately, N. Knudsen, A. Manibusan, P. G. Schultz, Y. Sun and R. M. Garbaccio, Discovery of Pyrophosphate Diesters as Tunable, Soluble, and Bioorthogonal Linkers for Site-Specific Antibody-Drug Conjugates, J. Am. Chem. Soc., 2016, 138(4), 1430–1445 CrossRef CAS PubMed
.
- Z. Dai, X. N. Zhang, F. Nasertorabi, Q. Cheng, J. Li, B. B. Katz, G. Smbatyan, H. Pei, S. G. Louie, H. J. Lenz, R. C. Stevens and Y. Zhang, Synthesis of site-specific antibody-drug conjugates by ADP-ribosyl cyclases, Sci. Adv., 2020, 6(23), eaba6752 CrossRef PubMed
.
- D. Gajria and S. Chandarlapaty, HER2-amplified breast cancer: mechanisms of trastuzumab resistance and novel targeted therapies, Expert Rev. Anticancer Ther., 2011, 11(2), 263–275 CrossRef CAS PubMed
.
- M. A. Bookman, K. M. Darcy, D. Clarke-Pearson, R. A. Boothby and I. R. Horowitz, Evaluation of monoclonal humanized anti-HER2 antibody, trastuzumab, in patients with recurrent or refractory ovarian or primary peritoneal carcinoma with overexpression of HER2: a phase II trial of the Gynecologic Oncology Group, J. Clin. Oncol., 2003, 21(2), 283–290 CrossRef CAS PubMed
.
- J. Baselga, D. Tripathy, J. Mendelsohn, S. Baughman, C. C. Benz, L. Dantis, N. T. Sklarin, A. D. Seidman, C. A. Hudis, J. Moore, P. P. Rosen, T. Twaddell, I. C. Henderson and L. Norton, Phase II study of weekly intravenous trastuzumab (Herceptin) in patients with HER2/neu-overexpressing metastatic breast cancer, Semin. Oncol., 1999, 26(4 Suppl. 12), 78–83 CAS
.
- S. Shak, Overview of the trastuzumab (Herceptin) anti-HER2 monoclonal antibody
clinical program in HER2-overexpressing metastatic breast cancer. Herceptin Multinational Investigator Study Group, Semin. Oncol., 1999, 26(4 Suppl. 12), 71–77 CAS
.
- S. C. Alley, N. M. Okeley and P. D. Senter, Antibody-drug conjugates: targeted drug delivery for cancer, Curr. Opin. Chem. Biol., 2010, 14(4), 529–537 CrossRef CAS PubMed
.
- I. Sassoon and V. Blanc, Antibody-drug conjugate (ADC) clinical pipeline: a review, Methods Mol. Biol., 2013, 1045, 1–27 CrossRef PubMed
.
- S. O. Doronina, T. D. Bovee, D. W. Meyer, J. B. Miyamoto, M. E. Anderson, C. A. Morris-Tilden and P. D. Senter, Novel peptide linkers for highly potent antibody-auristatin conjugate, Bioconjugate Chem., 2008, 19(10), 1960–1963 CrossRef CAS PubMed
.
- D. V. Hingorani, M. K. Doan, M. F. Camargo, J. Aguilera, S. M. Song, D. Pizzo, D. J. Scanderbeg, E. E. W. Cohen, A. M. Lowy, S. R. Adams and S. J. Advani, Precision Chemoradiotherapy for HER2 Tumors Using Antibody Conjugates of an Auristatin Derivative with Reduced Cell Permeability, Mol. Cancer Ther., 2020, 19(1), 157–167 CrossRef CAS PubMed
.
- S. A. Kularatne, V. Deshmukh, J. Ma, V. Tardif, R. K. Lim, H. M. Pugh, Y. Sun, A. Manibusan, A. J. Sellers, R. S. Barnett, S. Srinagesh, J. S. Forsyth, W. Hassenpflug, F. Tian, T. Javahishvili, B. Felding-Habermann, B. R. Lawson, S. A. Kazane and P. G. Schultz, A CXCR4-targeted site-specific antibody-drug conjugate, Angew. Chem., Int. Ed., 2014, 53(44), 11863–11867 CrossRef CAS PubMed
.
- M. S. Sutherland, R. J. Sanderson, K. A. Gordon, J. Andreyka, C. G. Cerveny, C. Yu, T. S. Lewis, D. L. Meyer, R. F. Zabinski, S. O. Doronina, P. D. Senter, C. L. Law and A. F. Wahl, Lysosomal trafficking and cysteine protease metabolism confer target-specific cytotoxicity by peptide-linked anti-CD30-auristatin conjugates, J. Biol. Chem., 2006, 281(15), 10540–10547 CrossRef CAS PubMed
.
- D. Jackson, J. Gooya, S. Mao, K. Kinneer, L. Xu, M. Camara, C. Fazenbaker, R. Fleming, S. Swamynathan, D. Meyer, P. D. Senter, C. Gao, H. Wu, M. Kinch, S. Coats, P. A. Kiener and D. A. Tice, A human antibody-drug conjugate targeting EphA2 inhibits tumor growth in vivo, Cancer Res., 2008, 68(22), 9367–9374 CrossRef CAS PubMed
.
-
S. O. Doronina and P. D. Senter, CHAPTER 4 Auristatin Payloads for Antibody–Drug Conjugates (ADCs) in Cytotoxic Payloads for Antibody–Drug Conjugates, The Royal Society of Chemistry, 2019, pp. 73–99 Search PubMed
.
- G. V. Chaitanya, A. J. Steven and P. P. Babu, PARP-1 cleavage fragments: signatures of cell-death proteases in neurodegeneration, Cell Commun Signal, 2010, 8, 31 CrossRef CAS PubMed
.
- S. Gobeil, C. C. Boucher, D. Nadeau and G. G. Poirier, Characterization of the necrotic cleavage of poly(ADP-ribose) polymerase (PARP-1): implication of lysosomal proteases, Cell Death Differ., 2001, 8(6), 588–594 CrossRef CAS PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc01795g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |