Open Access Article
Open Access ArticleRapid differentiation between bacterial infections and cancer using a near-infrared fluorogenic probe†
Ling-Ling
Wu
 ,
Qinghua
Wang
,
Yali
Wang
,
Na
Zhang
,
Qingyang
Zhang
,
Qinghua
Wang
,
Yali
Wang
,
Na
Zhang
,
Qingyang
Zhang
 and
Hai-Yu
Hu
and
Hai-Yu
Hu
 *
*
State Key Laboratory of Bioactive Substances and Function of Natural Medicine, Beijing Key Laboratory of Active Substances Discovery and Drugability Evaluation, Institute of Materia Medica, Chinese Academy of Medical Sciences, Peking Union Medical College, Beijing, 100050, China. E-mail: haiyu.hu@imm.ac.cn
First published on 26th February 2020
Abstract
The reliable differentiation between bacterial infections and other pathologies is crucial for both diagnostics and therapeutic approaches. To accommodate such needs, we herein report the development of an activatable near-infrared fluorescent probe 1 that could be applied in the ultrafast, ultrasensitive and specific detection of nitroreductase (NTR) activity in bacterial pathogens both in vitro and in vivo. Upon reaction with NTR, the nitro-group of the para-nitro phenyl sulfonic moiety present in probe 1 was reduced to an amino-group, resulting in a near-infrared fluorescence turn-on of the latent cyanine 7 fluorophore. Probe 1 was capable of rapid and real-time quantitative detection of 0–150 ng mL−1 NTR with a limit of detection as low as 0.67 ng mL−1in vitro. In addition, probe 1 exhibited an outstanding performance of ultrafast measurements and suitable selectivity toward NTR to accurately sense intracellular basal NTR in ESKAPE bacterial pathogens. Most remarkably, probe 1 was capable of noninvasively identifying bacterial infection sites without showing any significantly increased signal of tumour sites in the same animal within 30 min.
Introduction
The reliable differentiation between bacterial infections and other pathological manifestations such as sterile inflammation and cancer remains an important clinical goal.1,2 There is often significant overlap in the diagnostic features among bacterial infection, inflammation, and cancer both clinically and using conventional imaging techniques, which has resulted in increased urgency to develop novel tools for accurate and swift diagnosis of bacterial infections.3–5 Owing to the development of computed tomography (CT), magnetic resonance imaging (MRI), positron emission tomography (PET), single-photon emission computed tomography (SPECT) and optical imaging techniques,6,7 molecular imaging with bacteria-specific agents may significantly improve patient outcomes by rapidly identifying an infection and monitoring the treatment response. In this regard, using molecular probes for selective sensing and detection of enzyme activity produced by a pathogen represents an attractive approach for identifying bacteria.8 Among such techniques, various activatable near-infrared (NIR) fluorescent tracers have been developed that may allow for the fast, specific and non-invasive localization of bacterial infections in vivo.9We were in particular interested in investigating activatable molecular probes for the detection of nitroreductases (NTRs) in bacteria.10–15 NTRs represent a family of flavin-containing enzymes that are able to effectively metabolize nitro-substituted groups into hydroxylamines or amines in the presence of reduced nicotinamide adenine dinucleotide (NADH) or nicotinamide adenine dinucleotide phosphate (NADPH).16 NTRs are widely expressed in most Gram-positive and Gram-negative bacterial strains, but also in hypoxic tumour cells and tumorous tissues of eukaryotes. This makes this enzyme family an attractive diagnostic target to detect the hypoxic status of cancer cells and potential bacterial infections.17,18 Even though the detection of NTR activity within hypoxic tumours in vitro and in vivo using fluorogenic methods has already been well described, chemical tools for the exploitation of NTRs as diagnostic markers for localizing bacterial infections are still scarce.19 Moreover, NTR-activated fluorogenic probes that can specifically image live bacteria in vivo and do not accumulate at tumour sites have not been investigated thus far.
Antibiotic-modified fluorophores exhibit an enormous potential for imaging infection sites due to their high selectivity for bacterial cell structures compared with mammalian cells.20 Neomycin, an aminoglycoside antibiotic drug, exhibits preferential activity against broad spectrum pathogenic bacteria. Neomycin specifically binds to the A-site on the 16S ribosomal RNA of the 30S ribosome in bacterial cells.21 Recently, we developed various optical/MRI probes by conjugating neomycin with fluorescent/MRI reporters. These probes could be used to selectively recognize and label pathogenic bacteria over macrophage-like cells.12,22 In the present study, we show that NIR optical imaging with a bacteria-specific neomycin-based NTR-activated fluorogenic probe (Scheme 1) could be achieved. The probe used throughout these studies proved to be (i) water-soluble and stable, (ii) having an outstanding performance of ultrafast measurements and excellent selectivity toward NTR to accurately sense intracellular basal NTR activity in live bacteria both in vitro and in vivo, (iii) capable of noninvasively identifying bacterial infection sites without showing a significantly increased signal in the same animal suffering from cancer.
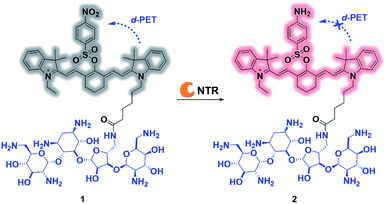 | ||
| Scheme 1 Proposed mechanism for ultrafast detection of NTR activity using the photoactivatable probe 1. NTR = nitroreductase. | ||
Results and discussion
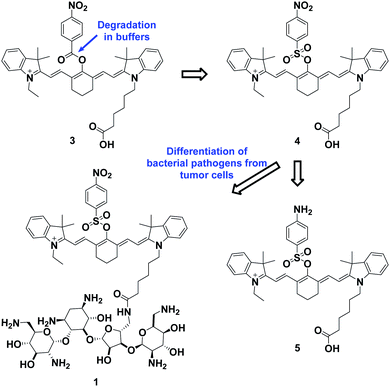
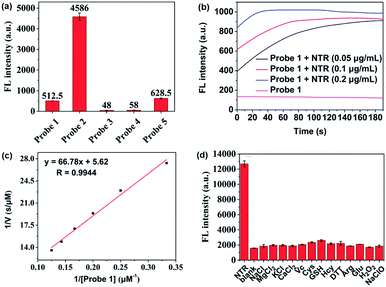
The overall rationale for the design of NTR-activated fluorogenic probes that can specifically image live bacteria in vivo was to conjugate a hydrophobic NTR-responsive NIR fluorogenic probe with neomycin. Previously, Li et al. reported a para-nitro aromatic group decorated cyanine 7 (Cy 7) probe linked via a carboxylic ester bond that displayed a fast response, high sensitivity, and excellent selectivity to NTR.23 Inspired by this structural scaffold, we initially designed and synthesized compound 3 with a carboxylic acid tag for conjugation with 5′′-NH2 neomycin (Fig. 1). However, compound 3 proved to be unstable and the carboxylic ester link proved to be easily hydrolysed in buffered aqueous media (detailed information on the stability of compound 3 can be found in the ESI section, Fig. S2†). Therefore, a more stable linker, sulfonate ester, was selected to link Cy 7 with the para-nitro phenyl group. In doing so, compound 4 was synthesized. We first studied the photochemistry of compound 4 before and after conversion of the nitro group in Tris-buffered saline (TBS, 50 mM Tris/HCl, containing 1.5% DMSO as a co-solvent, pH 7.4). For this purpose, the amino-substituted species 5 was synthesized. Both compounds 4 and 5 showed extended absorption spectra from the visible to NIR regions, with both absorption maxima at approximately 780 nm (Fig. S1†). The emission of compound 5 at 801 nm exhibited an 11-fold increase compared with that of compound 4, indicating that the presence of a sulfonate ester linked electron-withdrawing nitro group could efficiently induce fluorescence quenching via photoinduced electron transfer (PET). Compound 4 featured a near-zero fluorescence background signal (Fig. 2a), which made it a promising NTR-activated fluorogen for subsequent conjugation with neomycin to form a caged fluorescent probe with bacteria-specific in vivo targeting characteristics. Detailed synthetic procedures, full characterization data and the 16S rRNA binding property of probe 1 can be found in the ESI† section.The response function of probe 1 to purified NTR from E. coli was investigated afterwards (Fig. S3†). Fig. 2 showed the fluorescence emission responses of probe 1 in the presence of NADH in Tris-buffered saline. In the absence of NTR, weak fluorescence could be observed, but upon addition of 0.25 μg mL−1 NTR, the fluorescence intensity at 801 nm exhibited an 8-fold signal increase. Moreover, in the presence of dicoumarin, a known NTR inhibitor, a concentration dependent gradual emission reduction could be observed (Fig. S4†). A linear correlation could be determined between the emission intensity at 801 nm and the concentration of NTR in a range of 0–150 ng mL−1. The detection limit (3σ/k) was calculated to be as low as 0.67 ng mL−1 (Fig. S7†). Additionally, probe 1 reached a maximum fluorescence intensity within 1 min at the NTR concentration of 0.20 μg mL−1, and within 3 min even at the NTR concentration of 0.05 μg mL−1. The kinetic parameters of probe 1 activated by NTR were investigated by plotting reciprocal initial reaction rates (1/v) against the increasing probe concentration according to the Lineweaver–Burk plot. The calculated values of the apparent Michaelis–Menten constant Km and the maximum of the initial reaction rate Vmax were 11.88 μM and 0.178 μM s−1, respectively (Fig. 3c and S8†). More importantly, probe 1 at a concentration of 10 μM displayed an excellent selectivity towards NTR over other biologically relevant molecules and other potentially interfering species (Fig. 2d and S5†). Mass spectrometric analysis further substantiated the underlying activation mechanism of probe 1 by NTR as shown in Scheme 1 (Fig. S6†). These experimental data demonstrated that probe 1 could detect NTR activity in vitro in an ultrafast, ultrasensitive and highly specific fashion.
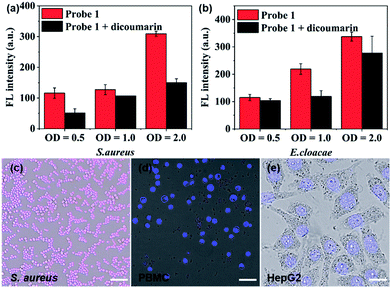
After establishing both excellent selectivity and responsiveness characteristics to NTR in vitro, we sought to evaluate the performance of probe 1 in living cells before carrying out subsequent in vivo studies. The clinically relevant Gram-positive bacterial strain Staphylococcus aureus (S. aureus, ATCC 29213) as well as the Gram-negative bacterial strain Enterobacter cloacae (E. cloacae, ATCC 13047) were selected (Fig. 3a, b and S9†). The cells were incubated at 37 °C in the presence of 10 μM probe 1 with or without the NTR inhibitor (dicoumarin, 1 mM) for 1 h. The emission signal was recorded at 809 nm. As shown in Fig. 3a and b, a bacterial cell concentration-dependent fluorescence intensity increase could be observed, whereas co-incubation with the NTR-inhibitor significantly reduced probe activation. The latter shows that probe 1 proves to be a promising optical imaging candidate for detecting the activity of NTRs in bacterial infections.
Confocal fluorescence imaging was used to study the cellular uptake selectivity and NTR detection properties of probe 1. S. aureus was used as model cells for bacterial infection. The human peripheral blood mononuclear cell line (PBMC) and the human hepatoma cell line (HepG2) were selected to mimic inflammatory tissue and the hypoxic cancer environment, respectively. The in vitro cytotoxicity of probe 1 and 2 to bacterial cells and tumour cells was first investigated using a Minimal Inhibitory Concentration (MIC) or MTS cell-viability assay, respectively (Table S1 and Fig. S10†). The cell viability of bacterial cells and tumour cells remained over 100% at tested concentrations, even up to 50 μM, within 24 h. This result indicated that probes 1 and 2 possessed low cytotoxicity. Subsequently, the response of probe 1 to NTR in living S. aureus, PBMC and HepG2 cells was assessed by confocal fluorescence imaging (Fig. 3c–e and S11†). As shown in Fig. 3c, probe 1 was readily taken up and activated by bacterial cells. However, no NIRF (near-infrared fluorescence) signal could be observed in macrophage cells (Fig. 3d) or hypoxic cancer cells (Fig. 3e). These findings were due to the notion that the neomycin-conjugated probe could not penetrate the cell membrane of mammalian cells. The results further indicate that probe 1 may represent a promising fluorescent bio-probe for cellular imaging with a high fluorescence contrast for NTR visualization in bacterial pathogens.
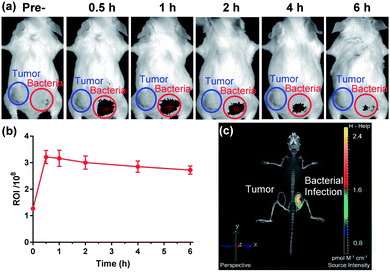
The favourable biological characteristics of probe 1 provided an incentive to the transition from cell assays to animal models. Since NTR represents one of the common biomarkers of both hypoxia and bacterial infection, one critical experiment was to investigate whether probe 1 could exhibit a sufficient imaging specificity between two potentially overlapping diagnostic disease states: bacterial infection and cancer. To test whether the specificity of probe 1 for a bacterial infection was appropriate enough, we designed an experiment to implant both bacterial pathogens and tumour cells into different hind legs of a mouse. CT26 colon cancer cells were injected into the left hind limb (Fig. 4a, blue circle) of each mouse 7 days before in vivo imaging allowing tumours to grow to a size of 100–150 mm3 and live S. aureus (1 × 109 CFU) was injected into the right hind limb (Fig. 4a, red circle) of each mouse 10 min before in vivo imaging. Whole body fluorescence images were obtained using a fluorescence imager (IVIS Spectrum imaging system) after intravenous injection of probe 1 into the tail vein. As shown in Fig. 4, no significant NIRF signal in the CT26 tumour region could be detected compared to the background, whereas a substantial fluorescence signal could be detected through the skin corresponding to a bacterial infection 30 min post-systemic injection (Fig. 4a and S13†). This signal gradually decreased thereafter (Fig. 4b). Afterwards, the images from 3D fluorescence light imaging tomography were combined with images obtained from integrated microCT (FLIT-microCT) to accurately localize the subcutaneous bacterial injection foci in vivo (Fig. 4c, S14 and ESI Movie S1†). 30 min post-injection, the infected tissues exhibited a target-to-background ratio of 2.5 times, background signal-corrected, over the tumour tissue in the contralateral leg. These results revealed that the probe could rapidly and selectively respond to NTRs in the infected region. To the best of our knowledge, this is the first time that a fluorescent probe could be used to selectively image the activity of NTRs in bacterial pathogens both in vitro and in vivo. Additionally, we observed that probe 1 did not accumulate in the metabolically inactive bacteria (neomycin-treated) (Fig. S15†), demonstrating that probe 1 was not binding to the bacteria through non-specific interactions.
Conclusions
It is of concern that there is often a significant and unwanted overlap in the diagnostic features among bacterial infection, inflammation, and cancer. However, the selective recognition of bacterial cells from white blood cells and cancer cells remains an interesting but far less investigated topic. In the study reported herein, we developed a bacteria-specific NIR fluorogenic probe that could be selectively activated in bacteria-infected tissue by enzymatic reduction of an electron-withdrawing nitro-quencher. The probe displayed high sensitivity, excellent selectivity and an ultrafast NTR response. Moreover, it could specifically image live bacterial cells and was not activated or accumulated by either macrophage cells or cancer cells. Most remarkably, the probe was also able to distinguish cancer and bacterial infections in a mouse model, with excellent temporal and spatial resolution. The obtained results may further enhance our understanding of bacterial infections related to molecular events.Ethical statement
Animal studies were conducted following the protocols approved by the Experimental Animal Welfare and Ethics Committee of the Institute of Materia Medica, Chinese Academy of Medical Sciences. This study was performed in strict accordance with the NIH guidelines for the care and use of laboratory animals (NIH Publication no. 85-23 Rev. 1985). The experimental designs were based on the rule of the replacement, refinement, and reduction to reduce suffering of the animals and use the minimum number of animals.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
We would like to thank Xiangchuan Meng for TOC illustration. This work was partially supported by the National Natural Science Foundation of China (NSFC) projects (21778077, 21807116, and 21907108), the Sino-German research project (GZ 1271), the Drug Innovation Major Projects (2018ZX09711-001-005) and the CAMS Innovation Fund for Medical Sciences (CIFMS 2017-I2M-2-004).Notes and references
- G. P. Birch, T. Campbell, M. Bradley and K. Dhaliwal, Frontiers in Oncology, 2019, 9, 1–15 CrossRef PubMed.
- M. van Oosten, M. Hahn, L. M. A. Crane, R. G. Pleijhuis, K. P. Francis, J. M. van Dijl and G. M. van Dam, FEMS Microbiol. Rev., 2015, 39, 892–916 CrossRef CAS PubMed.
- G. G. Zhanel, H. J. Adam, M. R. Baxter, J. Fuller, K. A. Nichol, A. J. Denisuik, A. R. Golden, R. Hink, P. R. S. Lagacé-Wiens, A. Walkty, M. R. Mulvey, F. Schweizer, D. Bay, D. J. Hoban, J. A. Karlowsky, C. A. R. Alliance and CANWARD, J. Antimicrob. Chemother., 2019, 74, iv5–iv21 CrossRef PubMed.
- M. M. Welling, A. W. Hensbergen, A. Bunschoten, A. H. Velders, M. Roestenberg and F. W. B. van Leeuwen, Clinical and Translational Imaging, 2019, 7, 105–124 CrossRef.
- M. Heuker, A. Gomes, J. M. van Dijl, G. M. van Dam, A. W. Friedrich, B. Sinha and M. van Oosten, Clinical and Translational Imaging, 2016, 4, 253–264 CrossRef PubMed.
- J. Wahsner, E. M. Gale, A. Rodríguez-Rodríguez and P. Caravan, Chem. Rev., 2019, 119(2), 957–1057 CrossRef CAS PubMed.
- T. I. Kostelnik and C. Orvig, Chem. Rev., 2019, 119(2), 902–956 CrossRef CAS PubMed.
- B. Mills, M. Bradley and K. Dhaliwal, Clinical and Translational Imaging, 2016, 4, 163–174 CrossRef PubMed.
- H. W. Liu, L. Chen, C. Xu, Z. Li, H. Zhang, X. B. Zhang and W. Tan, Chem. Soc. Rev., 2018, 47, 7140–7180 RSC.
- Y. Ji, Y. Wang, N. Zhang, S. Xu, L. Zhang, Q. Wang, Q. Zhang and H.-Y. Hu, J. Org. Chem., 2019, 84, 1299–1309 CrossRef CAS PubMed.
- Y. Liu, L. Zhang, M. Nazare, Q. Yao and H.-Y. Hu, Acta Pharm. Sin. B, 2018, 8, 401–408 CrossRef PubMed.
- S. Xu, Q. Wang, Q. Zhang, L. Zhang, L. Zuo, J.-D. Jiang and H.-Y. Hu, Chem. Commun., 2017, 53, 11177–11180 RSC.
- K. N. More, T.-H. Lim, J. Kang, H. Yun, S.-T. Yee and D.-J. Chang, Molecules, 2019, 24, 3206 CrossRef CAS PubMed.
- L. Zhou, L. Gong and S. Hu, Spectrochim. Acta, Part A, 2018, 199, 254–259 CrossRef CAS PubMed.
- G. Xu, Y. Tang, Y. Ma, A. Xu and W. Lin, Spectrochim. Acta, Part A, 2018, 188, 197–201 CrossRef CAS PubMed.
- X. Wu, W. Shi, X. Li and H. Ma, Acc. Chem. Res., 2019, 52, 1892–1904 CrossRef CAS PubMed.
- Z. Thiel and P. Rivera-Fuentes, Angew. Chem., Int. Ed., 2019, 58, 11474–11478 CrossRef CAS PubMed.
- A. Chevalier, Y. Zhang, O. M. Khdour, J. B. Kaye and S. M. Hecht, J. Am. Chem. Soc., 2016, 138, 12009–12012 CrossRef CAS PubMed.
- A. G. Vorobyeva, M. Stanton, A. Godinat, K. B. Lund, G. G. Karateev, K. P. Francis, E. Allen, J. G. Gelovani, E. McCormack, M. Tangney and E. A. Dubikovskaya, PLoS One, 2015, 10, e0131037 CrossRef PubMed.
- D. Alsteens, E. Dague, C. Verbelen, G. Andre, V. Dupres and Y. F. Dufrêne, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2009, 1, 168–180 CAS.
- M. Kaul and D. S. Pilch, Biochemistry, 2002, 41, 7695–7706 CrossRef CAS PubMed.
- L. Zhang, Y. Liu, Q. Zhang, T. Li, M. Yang, Q. Yao, X. Xie and H.-Y. Hu, Anal. Chem., 2018, 90, 1934–1940 CrossRef CAS PubMed.
- Y. Li, Y. Sun, J. Li, Q. Su, W. Yuan, Y. Dai, C. Han, Q. Wang, W. Feng and F. Li, J. Am. Chem. Soc., 2015, 137, 6407–6416 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental methods, synthetic procedures, compound characterization, Schemes S1 and S2, Table S1 and Fig. S1–S15 (PDF); Movie S1: Combination of 3D fluorescent light imaging tomography with integrated microCT (FLIT-microCT) (AVI). See DOI: 10.1039/d0sc00508h |
| This journal is © The Royal Society of Chemistry 2020 |