Open Access Article
Open Access ArticleHigh-efficiency dynamic sensing of biothiols in cancer cells with a fluorescent β-cyclodextrin supramolecular assembly†
Zhixue
Liu
,
Weilei
Zhou
,
Jingjing
Li
,
Haoyang
Zhang
,
Xianyin
Dai
,
Yaohua
Liu
and
Yu
Liu
 *
*
State Key Laboratory of Elemento-Organic Chemistry, College of Chemistry, Nankai University, Tianjin 300071, P. R. China. E-mail: yuliu@nankai.edu.cn
First published on 22nd April 2020
Abstract
A unique fluorescent supramolecular assembly was constructed using coumarin-modified β-cyclodextrin as a reversible ratiometric probe and an adamantane-modified cyclic arginine–glycine–aspartate peptide as a cancer-targeting agent via host–guest inclusion complexation. Importantly, the coumarin-modified β-cyclodextrin not only showed higher sensitivity than the parent coumarin derivatives owing to the presence of numerous hydroxyl groups on the cyclodextrin but also provided a hydrophobic cavity for encapsulation of a cancer-targeting agent. The assembly showed a reversible and fast response to biothiols with a micromolar dissociation constant, as well as outstanding cancer cell permeability, which can be used for high-efficiency real-time monitoring of biothiols in cancer cells. This supramolecular assembly strategy endows the fluorescent probe with superior performance for dynamic sensing of biothiols.
Introduction
Biothiols such as cysteine (Cys), homocysteine (Hcy), and glutathione (GSH) perform vital physiological functions, such as maintaining redox homeostasis, activating gene expression, and mitigating damage caused by free radicals and toxins; abnormal levels of biothiols are thought to be involved in many human diseases, including cancer, Alzheimer's and Parkinson's diseases, and AIDS.1–3 Therefore, the development of methods for real-time monitoring of biothiol concentrations in living cells is imperative for gaining a deeper understanding of biothiol-related pathophysiological processes.4–6Efforts have been made to develop fluorescent probes for this purpose because they are non-invasive and simple for rapid detection.7,8 The sensing mechanisms of these probes can be classified as disulfide exchange,9 cleavage of sulfonamide and sulfonate ester,10–12 Michael addition,13,14 cyclization with aldehydes,15 thiol-halogen nucleophilic substitution,16 metal complex–displace coordination,17etc. However, for real-time observation of concentration changes of biothiols, the Michael addition–elimination mechanism is the optimum one because of its reversibility. To date, a number of researchers have focused on modifying Michael addition acceptors to obtain highly sensitive and accurate reversible probes.18,19 For example, Wang and co-workers reported the first reversible fluorescent probe with a large dissociation constant (Kd = 1.6 mM) to quantify intracellular GSH concentrations.20 Then they introduced the cyano group adjacent to the double bond to improve the sensitivity and accuracy of the probe for quantitative real-time imaging of GSH in the cytoplasm (Kd = 3.7 mM),21 mitochondria (Kd = 1.0 mM) and other organelles.22–24 Furthermore, Urano and co-workers used an improved strategy to devise two novel reversible fluorescent probes (Kd = 3 and 0.6 mM) for live-cell imaging and quantification of rapid changes in GSH concentrations.25 Yoon and co-workers also reported a cyano-modified reversible fluorescent probe for real-time quantitative monitoring of cellular GSH (Kd = 2.59 mM).26 Likewise, similar strategies were introduced for designing reversible fluorescent probes by Jiang,27 Zhang,28 Yin,29 and Guo30etc., separately. Even though the above-described fluorescent probes are revolutionary tools for exploring biothiol concentration dynamics under physiological conditions, their Kd values are in the millimolar range; therefore, an interesting approach is to develop reversible fluorescent probes with higher sensitivity (Kd at the micromolar level).
The most common method for enhancing the biothiol-detection efficiency of a fluorescent probe that acts as a Michael acceptor is to introduce an electron-withdrawing substituent adjacent to the α-carbon of the unsaturated double bond, which renders the β-carbon more susceptible to attack by the nucleophilic thiol, thereby accelerating the addition reaction.31,32 The above-described fluorescent probes were in fact modified in this way; however, methods for increasing the nucleophilicity of the thiol group have not been thoroughly explored, despite the fact that the efficiency of a Michael addition depends on the nature of both the acceptor and the nucleophile. In addition, currently available fluorescent probes could be improved by endowing them with cancer-targeting ability and better bioavailability.33 Therefore, the development of methods for enhancing the biothiol-detection efficiency of fluorescent probes and simultaneously endowing them with cancer-targeting ability is urgently needed.
One possible strategy for accomplishing these tasks is to take advantage of supramolecular macrocyclic compounds such as cyclodextrins (CDs). CDs, which are enzymatically modified forms of starch and are composed of six to eight D-glucose units linked by α-1,4-glycosidic bonds,34 can improve the solubility, stability, and bioavailability of organic and inorganic materials.35,36 Hence, CD-based catalysts,37,38 molecular-recognition systems,39,40 metal–organic frameworks,41,42 room-temperature-phosphorescent materials,43–45 artificial actuators,46,47 drug delivery and controlled-release systems,48–50 and light-harvesting systems51 have proved to be fertile research areas in chemistry and the life sciences. Under appropriate conditions, CDs and their derivatives can act as selective catalysts by means of the formation of hydrogen bonds between the hydroxyl groups of the CD and a guest molecule, which can drive a reaction in the desired direction.52 Sakuraba et al. reported that CD derivatives can accelerate aqueous Michael addition reactions involving thiols by means of host–guest interactions.53,54 In addition, Suresh and Pitchumani reported that β-CD-catalyzed Michael reactions involving thiols can take place readily at room temperature without the need for hazardous external acids and bases and that the catalyst can be reused.55 Hydrogen bonding between thiols and the hydroxyl groups of β-CD can weaken the S–H bond, thus enhancing the nucleophilicity of the sulfur atom and facilitating addition reactions with electron-deficient alkenes.56
Taken together, these previously reported results suggest that covalently linking a CD to a fluorescent probe might be a simple and effective method for improving biothiol-detection sensitivity while simultaneously endowing the probe with a cavity for host–guest complexation of a cancer-targeting agent, thus realizing a method for dynamic, targeted imaging of biothiols in cancer cells. In this study, we constructed a novel cancer-targeting fluorescent supramolecular assembly based on coumarin-modified β-cyclodextrin (rCP-βCD) as a reversible ratiometric probe and adamantane-modified cRGD (cRGD-ADA, where cRGD = cyclic arginine–glycine–aspartate motif) as a cancer-targeting agent (Scheme 1). rCP-βCD exhibited a high affinity for biothiols, with a small dissociation constant (Kd) and a short half-life (t1/2). Formation of hydrogen bonds involving the CD hydroxyl groups played a crucial role in improving the biothiol-detection sensitivity, and the hydrophobic cavity could encapsulate cRGD-ADAvia host–guest inclusion complexation to form a supramolecular conjugate denoted as rCP-βCD@cRGD-ADA. This conjugate exhibited outstanding cancer cell permeability; it showed a faster reverse reaction rate for GSH than rCP-βCD, and its biothiol-detection sensitivity was on par with that of rCP-βCD, making it useful for high-efficiency real-time monitoring of dynamic changes of biothiols in cancer cells.
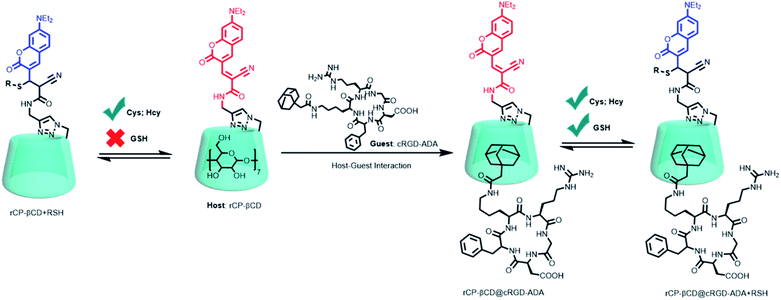 | ||
| Scheme 1 Construction of rCP-βCD@cRGD-ADA and the reversible Michael addition reactions between rCP-βCD@cRGD-ADA and biothiols. | ||
Results and discussion
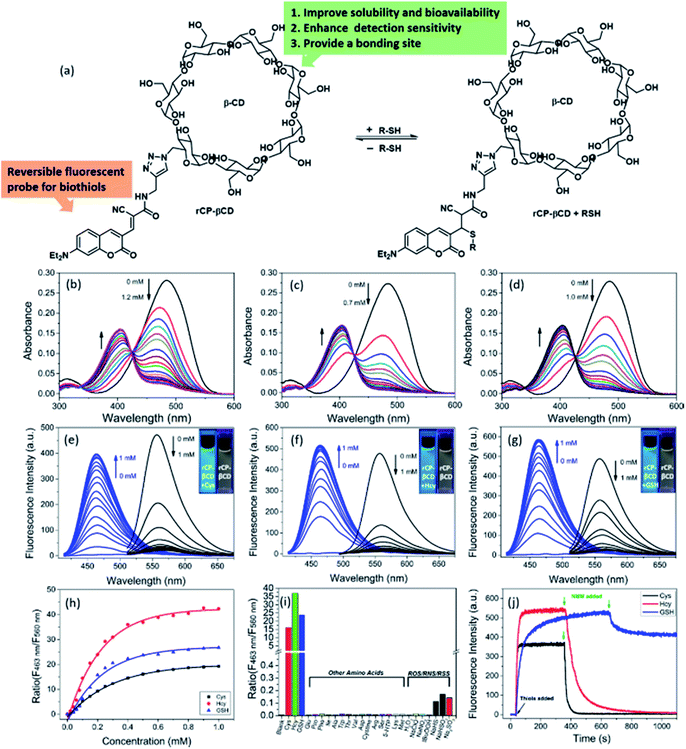
Although coumarin derivatives have been used for dynamic sensing of biothiols, the poor solubility and relatively low cancer targeting detection efficiency of such derivatives have limited their utility for biological applications. To overcome these limits, we used click chemistry to prepare a coumarin-modified β-CD (Fig. 1a). This modification not only improved the solubility and detection sensitivity of the coumarin moiety as a result of the hydrogen bonding interactions with β-CD but also provided a cavity for incorporation of cancer-targeting agents via host–guest inclusion complexation. We used UV-vis absorption and fluorescence spectroscopy to evaluate the performance of rCP-βCD for biothiol detection in pure PBS buffer solution. Upon addition of Cys, Hcy, or GSH to the buffer solution of rCP-βCD, the absorption peak at 485 nm decreased rapidly, and a new peak emerged at 405 nm (Fig. 1b–d). In addition, excellent ratiometric fluorescence responses were observed at excitation wavelengths of 405 and 485 nm, and these responses were accompanied by a large separation (ca. 97 nm) between the wavelengths of the two emission maxima, making this probe suitable for use in laser confocal microscopy (Fig. 1e–g). In the dose–response plots for the reactions between rCP-βCD and the biothiols, we found that the most obvious changes were around 0–0.8 mM (Fig. 1h). Therefore, we deduced that rCP-βCD could be used to monitor intracellular biothiols with high sensitivity instead of distinguishing them. On the basis of a S/N ratio of 3, we determined the limits of detection for Cys, Hcy, and GSH to be 122, 46, and 113 nM, respectively (Fig. S1†). The t1/2 values of rCP-βCD with respect to reactions with Cys, Hcy, and GSH were calculated to be 4.97, 10.39, and 59.70 s, respectively (Fig. S2†). The fast kinetics of the forward reaction enabled us to monitor biothiols at a time resolution in the order of seconds. The fluorescence quantum yields were measured to be 2.05% for rCP-βCD and 13.90%, 14.87%, and 15.03% for rCP-βCD + Cys, rCP-βCD + Hcy, and rCP-βCD + GSH, respectively (Fig. S3†).To assess the stability of rCP-βCD and its selectivity toward biothiols, we measured its UV-vis and fluorescence spectra under physiological conditions. rCP-βCD showed no fluorescence response to pH variations and exhibited excellent responsivity to biothiols at pH 5–9 (Fig. S4 and S5†). In addition, it also showed good selectivity for biothiols with an enhancement of the fluorescence ratio (F463 nm/F560 nm) of at least 2700-fold relative to that of various other intracellular amino acids, as well as a tolerance to various reactive oxygen and nitrogen species, accompanied by negligible interference of reactive sulfur (Fig. 1i, S6 and S7†), suggesting that rCP-βCD was suitable for qualitative detection of biothiols in living cells. To test the reversibility of the reaction of rCP-βCD with biothiols, we performed dilution experiments (Fig. S8†). Partial recovery of rCP-βCD was observed by UV-vis spectroscopy during 10-fold dilution experiments. This result confirms that the reversible reaction between rCP-βCD and biothiols was fast and depended mainly on the concentration of biothiols in the reaction system. The irreversible thiol scavenger N-methylmaleimide (NMM) was also used to explore the reversibility of the reaction. Addition of NMM caused a rapid decrease in the fluorescence at 405 nm (Fig. 1j). However, the response times in the reverse direction were different, especially for GSH, the recovery percentage from rCP-βCD + GSH to rCP-βCD was only 20%, perhaps as a result of differences in the degree of interaction between the biothiols and β-CD, such as hydrogen bonding and complexation.57,58 The result demonstrated that rCP-βCD was suitable for real-time monitoring of intracellular Cys and Hcy, but not of GSH.
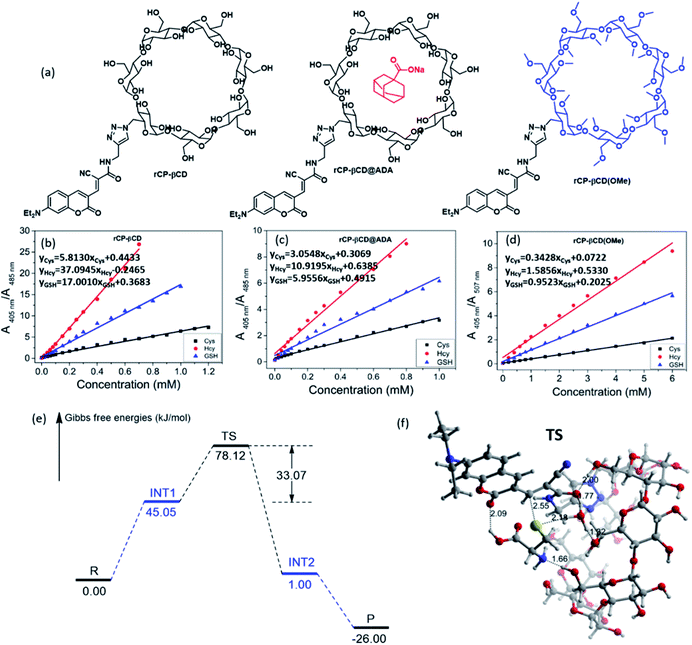
Next, we focused on elucidating the high efficiency of the reaction between rCP-βCD and the biothiols. We began by synthesizing coumarin derivatives bearing an alkyl group (rCP-Al) and a pyridinium moiety (rCP-Py) and then evaluated the efficiency of their respective reactions with the three thiols (Scheme S1†). When the ratio of the two main absorption peaks (A405 nm/A485 nm) was plotted as a function of biothiol concentration, different linear relationships were observed (Fig. S9†). rCP-βCD gave the largest slope among them, which exhibited higher sensitivity toward biothiols relative to the compounds that were not conjugated to β-CD (Fig. 2b). Calculation of the dissociation constants (Kd) by means of the reported procedures20,21,25 revealed that rCP-βCD showed a much lower Kd (micromolar level) than rCP-Al and rCP-Py (millimolar level) (Table S1†). The smaller Kd reflects the stronger affinity between the double bond and mercapto group, indicating that rCP-βCD showed a remarkably high affinity for the biothiols and that the β-CD moiety played an important role in enhancement of the reaction efficiency.
To verify the spatial configuration of coumarin and β-CD, we measured the circular dichroism spectra of rCP-βCD and rCP-βCD-biothiols (Fig. S10†). The spectra of rCP-βCD and rCP-βCD-biothiols displayed no Cotton effects at the testing concentration (10 μM), which suggests that the coumarin was not encapsulated in the hydrophobic cavity of the β-CD. We suggest that self-inclusion was hampered by the relatively low concentration used to measure the spectra and by the relatively long and rigid triazole linker. On the basis of these results, we deduced that hydrogen bonding between β-CD hydroxyl groups and the coumarin moiety improved the efficiency of the reactions with the biothiols.
In order to exclude the influence of inclusion and to confirm the effects of hydrogen bonding, we filled the rCP-βCD cavity with adamantane sodium formate (ADA), and we also generated a permethyl-β-cyclodextrin decorated coumarin rCP-βCD(OMe) to block the effects of the hydroxyl groups (Fig. 2a). Reactions of rCP-βCD@ADA and rCP-βCD(OMe) with the biothiols showed that occupation of the cavity by ADA caused only a slight change in Kd relative to that of rCP-βCD (Fig. 2c, S11a–c and Table S1†), whereas methylation of the β-CD hydroxyl groups resulted in an obvious increase in Kd (Fig. 2d, S11d–f and Table S1†). In addition to confirming that the hydroxyl groups played a decisive role in the enhancement of reaction efficiency, these results also suggested that rCP-βCD could host a cancer-targeting agent without any interference with its ability to react with biothiols. After this, to evaluate the effect of the covalent bond between the probe and β-CD in rCP-βCD, we prepared nonbonded complexes between β-CD and rCP-Al and between β-CD and rCP-Py with β-CD, and evaluated their reactions with the biothiols (Fig. S12†). Under these conditions, no obvious spectral change was observed upon addition of the biothiols in the absence or presence of β-CD, which demonstrated that simply mixing a fluorescent probe with β-CD did not improve the reaction efficiency and that the covalent connection between β-CD and the probe was necessary.
To identify the mechanisms of the reaction between rCP-βCD and biothiols, DFT calculations were performed (Fig. S13, S14 and Table S2†). We found that there are two steps including dehydrogenation of biothiols and nucleophilic attack in the process of C–S bond formation but usually not synchronous addition (Fig. 2e and S13†). Of note, the water molecule complexed β-CD plays an important role in the process. When the mercapto group loses the H to a water molecule, an intermediate cation and anion pair form and the Gibbs free energy increases up to 45.05 kJ mol−1. The Gibbs free energy barrier is 33.07 kJ mol−1 for the formation of the C–S bond to form INT2via a transition state (TS) (Fig. 2f), which means that it is very easy for this step of the reaction to occur. At the same time, cooperative proton-transfer occurs at the hydroxyl of β-CD. After this, the carbanion of INT2 accepts a proton to form the stable product P. On the whole, the changes of the Gibbs free energy are acceptable in the mechanism, so that it is reasonable and confirms the reason for efficient detection of rCP-βCD. Therefore, on the basis of the experimental results and DFT calculation study, we propose a mechanism for the enhanced Michael addition reactions through supramolecular interaction between rCP-βCD and the biothiols. First, the rigid triazole linker and the hydrogen bonding between the hydroxyl group of β-CD and the unsaturated carbonyl group of the conjugated coumarin derivative jointly stabilizes the configuration of rCP-βCD. Second, the biothiols could be fixed by the hydroxyl group of β-CD and ester carbonyl, as well as one H2O which facilitates the nucleophilic addition reaction of thiol groups.
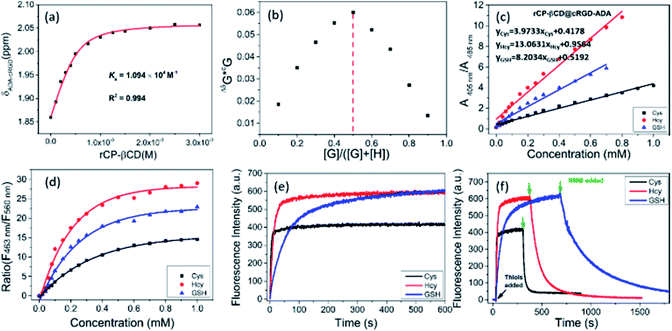
To target cancer cells via ligand–receptor-mediated endocytosis, we chose cRGD because it targets integrin αvβ3, which is overexpressed on tumor cells.59,60 The cRGD was modified with adamantane (cRGD-ADA), which could bind to rCP-βCD by means of adamantane–β-CD interactions and lead to cell-specific internalization. 1H NMR titration was used to investigate the complexation behavior of rCP-βCD and cRGD-ADA (Fig. S15†). When rCP-βCD was added to cRGD-ADA (0.5 mM), the proton signals of cRGD-ADA gradually shifted downfield as the rCP-βCD concentration increased from 0 to 3.0 mM, indicating inclusion of the ADA moiety in the rCP-βCD cavity. By nonlinear least-squares fitting of the titration data, we determined the binding constant (KS) for the complex of rCP-βCD@cRGD-ADA to be 1.1 × 104 M−1 (Fig. 3a). In addition, the stoichiometry of the complex was calculated to be 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 from Job's plot, in which the maximum was observed at a molar fraction of 0.5 (Fig. 3b and S16†). Meanwhile, the fluorescence quantum yields were measured to be 1.81% for rCP-βCD@cRGD-ADA at 560 nm (Fig. S17†).
1 from Job's plot, in which the maximum was observed at a molar fraction of 0.5 (Fig. 3b and S16†). Meanwhile, the fluorescence quantum yields were measured to be 1.81% for rCP-βCD@cRGD-ADA at 560 nm (Fig. S17†).
Next, we confirmed the ability of rCP-βCD@cRGD-ADA to respond to biothiols. Upon addition of biothiols to the complex, it showed a detection performance similar to that of rCP-βCD (Fig. 3c and S18a–c†). In addition, excellent ratiometric fluorescence responses were observed under the same conditions (Fig. S18d–i†). In the dose–response plots for the reactions of rCP-βCD with biothiols, we observed obvious changes in the biothiol concentration range of 0–0.8 mM (Fig. 3d). By plotting absorption peak ratios (A405 nm/A485 nm) as a function of biothiol concentration, we determined Kd(Cys), Kd(Hcy), and Kd(GSH) to be 0.252, 0.077, and 0.122 mM, respectively. In addition, we tested the detection performance of rCP-βCD@cRGD-ADA towards biothiols in FBS and culture medium. As shown in Fig. S19,† the assembly exhibited excellent ratiometric fluorescence responses towards biothiols, which means that the FBS and culture medium shows no interference in the detection. Next, we tracked and tested the stability of the supramolecular assembly by means of fluorescence spectra (Fig. S20†). Within 24 h, no obvious fluorescence quenching occurred, demonstrating that the assembly has a good stability in the complex environment. The above results demonstrated that the rCP-βCD@cRGD-ADA was stable and could be used to sense biothiols in a complex environment, such as FBS and culture medium. Calculation of the rate constants showed that the reactions of rCP-βCD@cRGD-ADA with biothiols were very fast (t1/2(Cys) = 3.02 s, t1/2(Hcy) = 10.49 s, and t1/2(GSH) = 57.55 s; Fig. 3e). Taken together, these results demonstrate that the rCP-βCD@cRGD-ADA supramolecular conjugate detected biothiols with an efficiency on par with that of rCP-βCD.
After this, NMM was added to demonstrate the reversibility of the reactions. The addition of NMM caused a rapid decrease in the fluorescence generated by excitation at 405 nm, and rCP-βCD@cRGD-ADA exhibited a faster reverse response rate for GSH than rCP-βCD which attained full-recovery in 16 min (Fig. 3f and S21†). This difference probably arose from occupation of the β-CD cavity by ADA and the interaction between β-CD and cRGD, which would have reduced the interaction between β-CD and GSH and thus accelerated the depletion of GSH. However, 1H NMR spectroscopy did not capture obvious changes in the chemical shifts of the proton signals of β-CD or the biothiols, possibly owing to the presence of water (Fig. S22†). In addition, the addition of H2O2 also confirmed the reversibility of rCP-βCD@cRGD-ADA towards biothiols with the fluorescence spectra (Fig. S23†). In summary, the t1/2 values for rCP-βCD@cRGD-ADA with respect to reactions with the biothiols were very small, indicating that the reaction kinetics were fast and that this system could be used to reliably monitor the biothiol concentration dynamics.
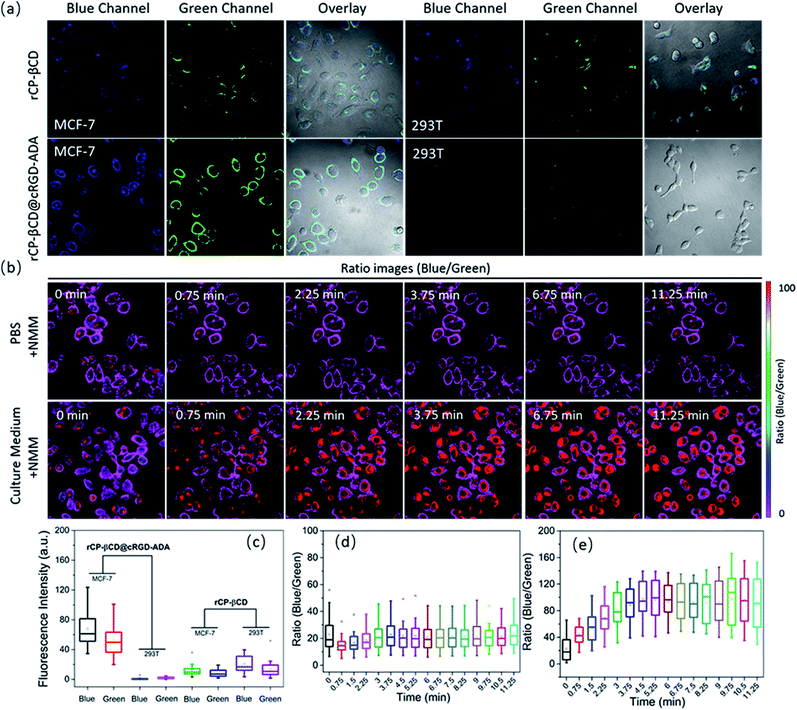
To explore the utility of the rCP-βCD@cRGD-ADA complex for monitoring intracellular biothiol concentration dynamics in living cells, we carried out real-time imaging using live MCF-7 breast cancer cells. First, to clearly compare the internalization of rCP-βCD@cRGD-ADA between normal cells and cancer cells, MCF-7 and 293T cells were incubated separately with rCP-βCD and rCP-βCD@cRGD-ADA for 2 h (Fig. 4a and S24†). Compared with 293T cells, rCP-βCD@cRGD-ADA emitted bright visible fluorescence in MCF-7 cells in both the blue and green channels. Conversely, rCP-βCD exhibited weak fluorescence in both the MCF-7 and 293T cells. Notably rCP-βCD@cRGD-ADA gave brighter blue and green fluorescence than rCP-βCD did, confirming that rCP-βCD@cRGD-ADA was internalized in larger quantities and reacted with intracellular biothiols because of the presence of the cRGD cancer-targeting moiety. These results demonstrated that the cancer cell targeting ability is from the RGD. Statistical analysis of the fluorescence intensity data for the two channels confirmed the results indicated by the images (Fig. 4c). In addition, similar cell uptake experiments with rCP-βCD@cRGD-ADA in A549, KYSE-150 and COS-7 cells were also performed (Fig. S25†). The results demonstrated that rCP-βCD@cRGD-ADA was internalized in larger quantities than in normal cells. To monitor the biothiol concentration dynamics in the cells, we added NMM to perturb the intracellular redox homeostasis. Upon treatment of MCF-7 cells in PBS with a bolus of NMM, ratiometric imaging of the cells showed that the blue/green ratio initially slightly decreased (within 0.75 min) and then increased back to an equilibrium value after 3.75 min (Fig. 4b [upper panels], 4d and S26†). However, when the cells were treated with NMM in culture medium rather than in PBS, a different response was observed; specifically, the ratio initially increased over the course of 3.75 min and then remained elevated (Fig. 4b [lower panels], 4e and S27†). In contrast, there was no obvious change in the blue/green ratio when cells were incubated in NMM-free culture medium for 10.5 min (Fig. S28†). These results demonstrate that the rCP-βCD@cRGD-ADA could be used to monitor various biothiols. Importantly, because rCP-βCD@cRGD-ADA detected the biothiols with high sensitivity, we were still able to capture the rapid increase in the biothiol concentration necessary to maintain intracellular redox homeostasis upon depletion of biothiols by treatment with NMM. Moreover, real-time biothiol imaging with rCP-βCD@cRGD-ADA in A549 and KYSE-150 cells upon NMM treatment was also explored (Fig. S29a and S30a†). Upon treatment with a bolus of NMM, ratiometric imaging of the cells showed the decrease of the blue/green ratio which confirmed the reduction of biothiols. The related statistical analysis of the fluorescence intensity data is displayed in Fig. S29b and S30b.† From representative blue/green ratio images, obvious biothiol concentration variations in A549 and KYSE-150 cells were observed (Fig. S31†). These experiments demonstrate that rCP-βCD@cRGD-ADA could reversibly react with intracellular biothiols and sense intracellular biothiol concentration dynamics.
Conclusions
In summary, we have developed a unique fluorescent supramolecular assembly consisting of rCP-βCD and cRGD-ADA for high-sensitivity dynamic sensing of intracellular biothiols. The rCP-βCD moiety exhibited high affinity for biothiols, with a small Kd at the micromolar level and a fast response rate in the order of seconds. When β-CD was replaced with an alkyl, pyridinium, or permethyl-β-CD moiety or with adamantine-modified rCP-βCD, only rCP-βCD@ADA showed a high biothiol-detection efficiency, on par with that of rCP-βCD. These results indicate that the hydroxyl groups of β-CD played a key role in improving the sensitivity of the probe, in addition to providing a hydrophobic cavity for encapsulation of an agent for targeted cancer-cell imaging. On the other hand, taking Cys as the model, DFT calculations gave a reasonable mechanism for enhanced Michael addition reactions which indicated that β-CD played an important role in this process. When rCP-βCD was combined with cRGD-ADA, the cancer-targeting agent was encapsulated in the β-CD cavity by means of host–guest inclusion complexation. The resulting supramolecular assembly rCP-βCD@cRGD-ADA exhibited a biothiol-detection efficiency similar to that of rCP-βCD and a faster reverse reaction rate, as well as outstanding cancer cell permeability. These properties allowed us to use it for real-time monitoring of biothiol concentration dynamics in living cells. In addition, a direct comparison between QG-1 (ref. 26) and rCP-βCD@cRGD-ADA was performed. Structurally, due to the existence of β-cyclodextrin, the dynamic detection sensitivity to biothiols could be significantly enhanced by the hydrogen bonding among β-CD hydroxyl groups, coumarin moiety and biothiols. Meanwhile, the solubility and biocompatibility of the β-cyclodextrin modified probe were also greatly improved. On the other hand, the encapsulated cRGD-ADA was able to improve the uptake of rCP-βCD@cRGD-ADA by cancer cells, endowing it with cancer-targeting ability. In the present research, we confirmed that this supramolecular assembly strategy could provide a novel approach for high-efficiency dynamic real-time monitoring of biothiols in living cells.Experimental
All chemical reagents and solvents for synthesis were purchased from commercial sources (Aladdin Industrial Corporation, Tokyo Chemical Industry and Sigma-Aldrich Chemical) and were used without further purification. Adamantane modified cRGD was purchased from GL Biochem (Shanghai) Ltd All moisture-sensitive reactions were carried out under an atmosphere of nitrogen. Ultrapure water was used after passing through a water ultra-purification system. 1H NMR and 13C NMR spectra were recorded on an Ascend 400 MHz (BRUKER) at room temperature. High-resolution mass spectra (HRMS) were measured on a 6520 Q-TOF LC/MS (Agilent). Absorption spectra were recorded on a UV-vis spectrophotometer (UV-2700, Shimadzu), and steady-state fluorescence emission spectra were recorded in a conventional quartz cell (10 × 10 × 45 mm) at 25 °C on a Varian Cary Eclipse equipped with a Varin Cary single-cell Peltier accessory to control temperature. Absolute fluorescence quantum yields were recorded on a FLS980 instrument (Edinburgh Instruments Ltd, Livingstone, UK). CD spectra were recorded on a Jasco J 715 CD spectrophotometer. Confocal fluorescence and bright-field imaging were recorded with an FV1000 (Olympus).Solution preparation
rCP-βCD stock solution was made by dissolving it in water. rCP-Al, rCP-Py and rCP-βCD(OMe) were dissolved in DMSO. rCP-βCD@ADA was obtained by dissolving rCP-βCD and amantadine sodium formate together in water. rCP-βCD@cRDG-ADA was obtained by dissolving rCP-βCD and cRGD-ADA together in water. All the above stock solutions were prepared with a final concentration of 1 mM and kept at −20 °C before use. GSH, Cys, Hcy and other analytes were dissolved in water, and NEM was dissolved in DMSO. In addition, all testing solutions were prepared by diluting stock solutions with PBS. Mixing was usually done by adding the analyte solution (for example, Cys solution) into the probe solution. Cell staining solution was made by diluting 1 mM rCP-βCD or rCP-βCD@cRDG-ADA in water for a final probe concentration of 10 μM. 10 mM PBS buffers were adjusted to different pHs by using 2 M NaOH solution and 2 M HCl. The UV-vis absorption changes and fluorescence spectra of 10 μM rCP-βCD in buffers in the pH range of 5–9 were recorded with or without biothiols.Determination of Kd
The Kd for biothiols was calculated according to a reported equation.10,15 However, plotting A405nm/A485 nm as a function of GSH concentration can afford a linear relationship, and the reciprocal of the slope can be approximated as Kd.13 The bonding constant Ks and Job's plot of cRGD-ADA (G) and rCP-βCD (H) were obtained by using 1H NMR spectra in D2O. Ks: 1H NMR spectra of cRGD-ADA (0.5 mM) with addition of 0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.75, 1.0, 1.25, 1.5, 2.0, 2.5, and 3.0 mM rCP-βCD in D2O. The Ks of the complex was determined by nonlinear least-squares fitting of (δ2.05 ppm–δ1.85 ppm) and concentration of rCP-βCD. Job's plot: 1H NMR spectra of different ratios (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 8
1, 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 7
2, 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 6
3, 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, 5
4, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, 4
5, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6, 3
6, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7, 2
7, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8, and 1
8, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) between rCP-βCD and cRGD-ADA in D2O ([cRGD-ADA] + [rCP-βCD] = 2 mM). The stoichiometry of the complex was calculated by plotting the chemical shift value (δ2.05 ppm–δ1.85 ppm) and different ratios.
9) between rCP-βCD and cRGD-ADA in D2O ([cRGD-ADA] + [rCP-βCD] = 2 mM). The stoichiometry of the complex was calculated by plotting the chemical shift value (δ2.05 ppm–δ1.85 ppm) and different ratios.
Theoretical calculation
All quantum chemical calculations were carried out using the DFT method with the M06-2X functional61,62 with the Gaussian 16 program, and the solution-phase environment was treated with the universal solvation SMD model63 in a water solvent. Geometry optimizations were performed at the M06-2X-D3/6-31G(d,p)//SMDwater level.64,65 Harmonic vibrational frequency calculations were performed at the same level for thermal corrections of Gibbs free energies making sure that the local minimum points had no imaginary frequencies, while the saddle points had only one imaginary frequency. Single-point energy calculations were carried out at the M06-2X-D3/6-311 + g(df,pd)//SMDwater level based on these optimized structures. Intrinsic reaction coordinates66,67 (IRCs) were calculated to confirm that the transition state structure is correct on the potential energy surface for the mechanisms. Computed structures are displayed with CYLVIEW.Cell culture and fluorescence imaging experiment. MCF-7, 293T, A549, KYSE-150 and COS-7 cells were grown in 1640 and F12 media containing 10% FBS, 1% penicillin, and 1% streptomycin at 37 °C in 5% CO2. Cells were plated on confocal dishes and allowed to adhere for 24 hours. For the confocal fluorescence imaging experiment, cells were washed with PBS and then incubated with rCP-βCD and rCP-βCD@cRGD-ADA in culture medium for another 2 h at 37 °C. The cell staining experiment was performed after washing with PBS 3 times. Experiments to assess reversible variation were performed under different conditions (culture medium or PBS) supplemented with NMM. Emission was collected at 430–510 nm (excited at 405 nm) for the blue channel and at 525–625 nm (excited at 480 nm) for the green channel.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant nos. 21807038, 21432004 and 21772099) and China Postdoctoral Science Foundation (2019M651006).References
- H. Fan, G. Yan, Z. Zhao, X. Hu, W. Zhang, H. Liu, X. Fu, T. Fu, X. B. Zhang and W. Tan, Angew. Chem., Int. Ed., 2016, 55, 5477–5482 CrossRef CAS PubMed.
- Y. Zhang, X. Wang, X. Bai, P. Li, D. Su, W. Zhang, W. Zhang and B. Tang, Anal. Chem., 2019, 91, 8591–8594 CrossRef CAS PubMed.
- C. X. Yin, K. M. Xiong, F. J. Huo, J. C. Salamanca and R. M. Strongin, Angew. Chem., Int. Ed., 2017, 56, 13188–13198 CrossRef CAS PubMed.
- C. S. Lim, G. Masanta, H. J. Kim, J. H. Han, H. M. Kim and B. R. Cho, J. Am. Chem. Soc., 2011, 133, 11132–11135 CrossRef CAS PubMed.
- J. Liu, Y. Q. Sun, Y. Huo, H. Zhang, L. Wang, P. Zhang, D. Song, Y. Shi and W. Guo, J. Am. Chem. Soc., 2014, 136, 574–577 CrossRef CAS PubMed.
- X. Han, X. Song, F. Yu and L. Chen, Chem. Sci., 2017, 8, 6991–7002 RSC.
- X. Chen, Y. Zhou, X. Peng and J. Yoon, Chem. Soc. Rev., 2010, 39, 2120–2135 RSC.
- M. She, Z. Wang, T. Luo, B. Yin, P. Liu, J. Liu, F. Chen, S. Zhang and J. Li, Chem. Sci., 2018, 9, 8065–8070 RSC.
- D. Jung, S. Maiti, J. H. Lee, J. H. Lee and J. S. Kim, Chem. Commun., 2014, 50, 3044–3047 RSC.
- D. Kand, P. K. Mishra, T. Saha, M. Lahiri and P. Talukdar, Analyst, 2012, 137, 3921–3924 RSC.
- J. Shao, H. Sun, H. Guo, S. Ji, J. Zhao, W. Wu, X. Yuan, C. Zhang and T. D. James, Chem. Sci., 2012, 3, 1049–1061 RSC.
- M. Li, X. Wu, Y. Wang, Y. Li, W. Zhu and T. D. James, Chem. Commun., 2014, 50, 1751 RSC.
- C. Yin, F. Huo, J. Zhang, R. Martinez-Manez, Y. Yang, H. Lv and S. Li, Chem. Soc. Rev., 2013, 42, 6032–6059 RSC.
- X. Zhou, X. Jin, G. Sun, D. Li and X. Wu, Chem. Commun., 2012, 48, 8793–8795 RSC.
- A. Barve, M. Lowry, J. O. Escobedo, K. T. Huynh, L. Hakuna and R. M. Strongin, Chem. Commun., 2014, 50, 8219–8222 RSC.
- C. Xu, H. Li and B. Yin, Biosens. Bioelectron., 2015, 72, 275–281 CrossRef CAS PubMed.
- H. S. Jung, X. Chen, J. S. Kim and J. Yoon, Chem. Soc. Rev., 2013, 42, 6019–6031 RSC.
- A. Y. Cho and K. Choi, Chem. Lett., 2012, 41, 1611–1612 CrossRef CAS.
- D. Cao, Z. Liu, P. Verwilst, S. Koo, P. Jangjili, J. S. Kim and W. Lin, Chem. Rev., 2019, 119, 10403–10519 CrossRef CAS PubMed.
- X. Jiang, Y. Yu, J. Chen, M. Zhao, H. Chen, X. Song, A. J. Matzuk, S. L. Carroll, X. Tan, A. Sizovs, N. Cheng, M. C. Wang and J. Wang, ACS Chem. Biol., 2015, 10, 864–874 CrossRef CAS PubMed.
- X. Jiang, J. Chen, A. Bajic, C. Zhang, X. Song, S. L. Carroll, Z. L. Cai, M. Tang, M. Xue, N. Cheng, C. P. Schaaf, F. Li, K. R. MacKenzie, A. C. M. Ferreon, F. Xia, M. C. Wang, M. Maletic-Savatic and J. Wang, Nat. Commun., 2017, 8, 16087 CrossRef CAS PubMed.
- J. Chen, X. Jiang, C. Zhang, K. R. MacKenzie, F. Stossi, T. Palzkill, M. C. Wang and J. Wang, ACS Sens., 2017, 2, 1257–1261 CrossRef CAS PubMed.
- J. Chen, X. Jiang, S. Carroll, J. Huang and J. Wang, Org. Lett., 2015, 17, 5978–5981 CrossRef CAS PubMed.
- X. Jiang, C. Zhang, J. Chen, S. Choi, Y. Zhou, M. Zhao, X. Song, X. Chen, M. Maletic-Savatic, T. Palzkill, D. Moore, M. C. Wang and J. Wang, Antioxid. Redox Signaling, 2019, 30, 1900–1910 CrossRef CAS PubMed.
- K. Umezawa, M. Yoshida, M. Kamiya, T. Yamasoba and Y. Urano, Nat. Chem., 2017, 9, 279–286 CrossRef CAS PubMed.
- Z. Liu, X. Zhou, Y. Miao, Y. Hu, N. Kwon, X. Wu and J. Yoon, Angew. Chem., Int. Ed., 2017, 56, 5812–5816 CrossRef CAS PubMed.
- M. Tian, M. Yang, Y. Liu and F.-L. Jiang, ACS Appl. Bio Mater., 2019, 2, 4503–4514 CrossRef CAS.
- L. Nie, C. Gao, T. Shen, J. Jing, S. Zhang and X. Zhang, Anal. Chem., 2019, 91, 4451–4456 CrossRef CAS PubMed.
- Y. Yue, F. Huo, P. Ning, Y. Zhang, J. Chao, X. Meng and C. Yin, J. Am. Chem. Soc., 2017, 139, 3181–3185 CrossRef CAS PubMed.
- M. Ren, L. Wang, X. Lv, Y. Sun, H. Chen, K. Zhang, Q. Wu, Y. Bai and W. Guo, Analyst, 2019, 144, 7457–7462 RSC.
- I. M. Serafimova, M. A. Pufall, S. Krishnan, K. Duda, M. S. Cohen, R. L. Maglathlin, J. M. McFarland, R. M. Miller, M. Frodin and J. Taunton, Nat. Chem. Biol., 2012, 8, 471–476 CrossRef CAS PubMed.
- C. U. Lee and T. N. Grossmann, Angew. Chem., Int. Ed., 2012, 51, 8699–8700 CrossRef CAS PubMed.
- K. Z. Gu, W. H. Zhu and X. J. Peng, Sci. China: Chem., 2019, 62, 189–198 CrossRef CAS.
- X. Ma and Y. Zhao, Chem. Rev., 2015, 115, 7794–7839 CrossRef CAS PubMed.
- H. B. Cheng, Y. M. Zhang, Y. Liu and J. Y. Yoon, Chem, 2019, 5, 553–574 CAS.
- D. Prochowicz, A. Kornowicz and J. Lewinski, Chem. Rev., 2017, 117, 13461–13501 CrossRef CAS PubMed.
- K. Surendra, N. S. Krishnaveni, Y. V. Nageswar and K. R. Rao, J. Org. Chem., 2003, 68, 4994–4995 CrossRef CAS PubMed.
- N. S. Krishnaveni, K. Surendra, M. A. Reddy, Y. V. Nageswar and K. R. Rao, J. Org. Chem., 2003, 68, 2018–2019 CrossRef CAS PubMed.
- Z. Xu, S. Jia, W. Wang, Z. Yuan, B. Jan Ravoo and D. S. Guo, Nat. Chem., 2019, 11, 86–93 CrossRef CAS PubMed.
- J. Shi, Q. Deng, Y. Li, Z. Chai, C. Wan, H. Shangguan, L. Li and B. Tang, Chem.–Asian J., 2019, 14, 847–852 CrossRef CAS PubMed.
- Y. Chen, B. Yu, Y. Cui, S. Xu and J. Gong, Chem. Mater., 2019, 31, 1289–1295 CrossRef CAS.
- D. Shen, G. Wang, Z. Liu, P. Li, K. Cai, C. Cheng, Y. Shi, J. M. Han, C. W. Kung, X. Gong, Q. H. Guo, H. Chen, A. C. Sue, Y. Y. Botros, A. Facchetti, O. K. Farha, T. J. Marks and J. F. Stoddart, J. Am. Chem. Soc., 2018, 140, 11402–11407 CrossRef CAS PubMed.
- X. Yu, W. Liang, Q. Huang, W. Wu, J. J. Chruma and C. Yang, Chem. Commun., 2019, 55, 3156–3159 RSC.
- C. Zhao, Y. Jin, J. Wang, X. Cao, X. Ma and H. Tian, Chem. Commun., 2019, 55, 5355–5358 RSC.
- D. Li, F. Lu, J. Wang, W. Hu, X. M. Cao, X. Ma and H. Tian, J. Am. Chem. Soc., 2018, 140, 1916–1923 CrossRef CAS PubMed.
- X. Ji, M. Ahmed, L. Long, N. M. Khashab, F. Huang and J. L. Sessler, Chem. Soc. Rev., 2019, 48, 2682–2697 RSC.
- S. Ikejiri, Y. Takashima, M. Osaki, H. Yamaguchi and A. Harada, J. Am. Chem. Soc., 2018, 140, 17308–17315 CrossRef CAS PubMed.
- G. Yu, X. Zhao, J. Zhou, Z. Mao, X. Huang, Z. Wang, B. Hua, Y. Liu, F. Zhang, Z. He, O. Jacobson, C. Gao, W. Wang, C. Yu, X. Zhu, F. Huang and X. Chen, J. Am. Chem. Soc., 2018, 140, 8005–8019 CrossRef CAS PubMed.
- H. Jin, L. Yang, M. J. R. Ahonen and M. H. Schoenfisch, J. Am. Chem. Soc., 2018, 140, 14178–14184 CrossRef CAS PubMed.
- Y. M. Zhang, Y. H. Liu and Y. Liu, Adv. Mater., 2020, 32, e1806158 CrossRef PubMed.
- J. J. Li, Y. Chen, J. Yu, N. Cheng and Y. Liu, Adv. Mater., 2017, 29, e1701905 CrossRef PubMed.
- K. Takahashi, Chem. Rev., 1998, 98, 2013–2034 CrossRef CAS PubMed.
- H. Sakuraba, Y. Tananaka and F. Toda, J. Inclusion Phenom. Mol. Recognit. Chem., 1991, 11, 195–204 CrossRef CAS.
- K. Harano, H. Kiyonaga and T. Hisano, Tetrahedron Lett., 1991, 32, 7557–7558 CrossRef CAS.
- P. Suresh and K. Pitchumani, Tetrahedron: Asymmetry, 2008, 19, 2037–2044 CrossRef CAS.
- N. S. Krishnaveni, K. Surendra and K. R. Rao, Chem. Commun., 2005, 669–671 RSC.
- M. Garcia-Fuentes, A. Trapani and M. J. Alonso, Eur. J. Pharm. Biopharm., 2006, 64, 146–153 CrossRef CAS PubMed.
- W. Sun, J. Liu, M. Cui, F. Song and S. Liu, Rapid Commun. Mass Spectrom., 1999, 13, 950–954 CrossRef CAS.
- M. H. Lee, J. Y. Kim, J. H. Han, S. Bhuniya, J. L. Sessler, C. Kang and J. S. Kim, J. Am. Chem. Soc., 2012, 134, 12668–12674 CrossRef CAS PubMed.
- J. Chen, H. Ni, Z. Meng, J. Wang, X. Huang, Y. Dong, C. Sun, Y. Zhang, L. Cui, J. Li, X. Jia, Q. Meng and C. Li, Nat. Commun., 2019, 10, 3546 CrossRef PubMed.
- Y. Zhao and D. G. Truhlar, Theor. Chem. Acc., 2008, 120, 215–241 Search PubMed.
- Y. Zhao and D. G. Truhlar, Acc. Chem. Res., 2008, 41, 157–167 CrossRef CAS PubMed.
- A. V. Marenich, C. J. Cramer and D. G. Truhlar, J. Phys. Chem. B, 2009, 113, 6378–6396 CrossRef CAS PubMed.
- S. Grimme, J. Antony, S. Ehrlich and H. Krieg, J. Chem. Phys., 2010, 132, 154104–154118 CrossRef PubMed.
- P. C. Hariharan and J. A. Pople, Theor. Chim. Acta, 1973, 28, 213–222 CrossRef CAS.
- C. Gonzalez and H. B. Schlegel, J. Chem. Phys., 1989, 90, 2154–2161 CrossRef CAS.
- C. Gonzalez and H. B. Schlegel, J. Phys. Chem., 1990, 94, 5523–5527 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures and characterization. See DOI: 10.1039/d0sc00414f |
| This journal is © The Royal Society of Chemistry 2020 |