Open Access Article
Open Access ArticleApoptosis-inducing activity of a fluorescent barrel-rosette M+/Cl− channel†
Javid Ahmad
Malla
 a,
Rintu M.
Umesh
b,
Amal
Vijay
a,
Arnab
Mukherjee
a,
Rintu M.
Umesh
b,
Amal
Vijay
a,
Arnab
Mukherjee
 a,
Mayurika
Lahiri
a,
Mayurika
Lahiri
 b and
Pinaki
Talukdar
b and
Pinaki
Talukdar
 *a
*a
aDepartment of Chemistry, Indian Institute of Science Education and Research Pune, Dr Homi Bhabha Road, Pashan, Pune 411008, Maharashtra, India. E-mail: ptalukdar@iiserpune.ac.in
bDepartment of Biology, Indian Institute of Science Education and Research Pune, Dr Homi Bhabha Road, Pashan, Pune 411008, Maharashtra, India
First published on 29th January 2020
Abstract
Synthetic transmembrane ion transport systems are emerging as new tools for anticancer therapy. Here, a series of 2-hydroxy-N1,N3-diarylisophthalamide-based fluorescent ion channel-forming compounds are reported. Ion transport studies across large unilamellar vesicles confirmed that the compound with two 3,5-bis(trifluoromethyl)phenyl arms is the most efficient transporter among the series and it facilitates M+/Cl− symport. The compound formed supramolecular ion channels with a single-channel conductance of 100 ± 2 pS, a diameter of 5.06 ± 0.16 Å and a permeability ratio, PCl−/PK+, of 8.29 ± 1. The molecular dynamics simulations of the proposed M2.11 channel (i.e. 11 coaxial layers of a dimeric rosette) with K+ and Cl− in the preequilibrated POPC lipid bilayer with water molecules illustrated various aspects of channel formation and ion permeation. Cell viability assay with the designed compounds indicated that cell death is being induced by the individual compounds which follow the order of their ion transport activity and chloride and cations play roles in cell death. The inherent fluorescence of the most active transporter was helpful to monitor its permeation in cells by confocal microscopy. The apoptosis-inducing activity upon perturbation of intracellular ionic homeostasis was established by monitoring mitochondrial membrane depolarization, generation of reactive oxygen species, cytochrome c release, activation of the caspase 9 pathway, and finally the uptake of the propidium iodide dye in the treated MCF7 cells.
Introduction
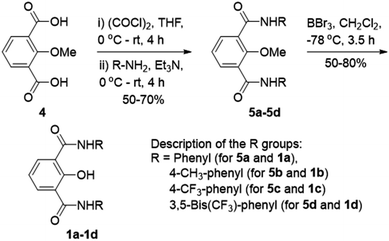
Ion transport across the cell membrane is a very crucial physiological process that ensures the healthy functions of cells.1–8 The cell membrane creates a thermodynamic barrier for polar molecules and ions to pass through its hydrophobic middle core. To ensure the regulated membrane transport of these species, nature has endowed the cells with very special molecular machineries in the form of ion transport systems and other pore forming protein molecules.5,9,10 The ion transport systems involve ion channels which can span the membrane and provide a hydrophilic pathway for ions to pass through.11–13 On the other hand, there are small molecular systems, called ion carriers, which can pick up ions from one side of the membrane and flip to the other side for releasing these ions.14 Any defect or damage in these complex transport systems (channelopathies) can have drastic effects on the physiological functions of the cell.15 The typical examples of channelopathies include cystic fibrosis which is caused by the mutation in the CFTR channel.16 Similarly Barter's syndrome, Dent's disease, Long QT syndrome, osteoporosis, epilepsy, Parkinson's disease etc. are other disorders which are caused by channelopathies.15,17,18 To treat these channelopathies, the identification of their causes becomes an important concern to the scientific community. In this regard, various studies have come out and the process continues. However, to solve these kinds of problems chemists have come up with an alternative approach to replace the defective channels with structurally simple organic motifs, which can restore the ion transport in defective cells by functioning as ion channels or ion carriers.19–22 So synthetic ion transport systems may open new opportunities for the treatment of these channelopathies. Along with these findings, the renewed interest in the field of synthetic ion transport systems has emerged after anticancer activity was observed in prodigiosin,23–25 a naturally occurring ionophore, which acts as a chloride and HCl carrier. Afterward, various synthetic chloride ion carriers26–30 and chloride channels9 were evaluated for anticancer activity and were found to induce apoptosis in cancer cells through the caspase-dependent intrinsic pathway. In the field of symporters, the first synthetic KCl symporter was reported by Smith and coworkers, using a ditopic receptor for NaCl and KCl transport.31 This was followed by Kyu-Sung Jeong and coworkers, who used an azacrown as a K+ binding motif and urea as a chloride binding motif.32 Gale and coworkers developed their calix[4]pyrrole as CsCl and RbCl transporters where Cl− binding was through H-bonding and Cs+ was tightly bound to pyrrole rings.33 Matile and coworkers reported ditopic transport systems operating via halogen bonding.34 These two types of transport systems work using cation activated anion transport.34 The latest example of a metal halide symport was presented by Madhavan and coworkers, with a peptide connected to an aminopicolinic acid unit, having more preference for cations over anions.35 However, none of these symporters have been evaluated for biological impacts.Herein, we report a series of fluorescent molecules that form self-assembled barrel-rosette channels to facilitate the symport of M+ and Cl− across liposomes. These channels allow the transport of Cl− and K+ as well as Na+ across the cell membrane to induce apoptotic cell death. The design included the central core of 2-hydroxy-N1,N3-diarylisophthalamide connected to two identical aromatic side arms such as phenyl, 4-methylphenyl, 4-trifluoromethylphenyl or 3,5-bis(trifluoromethyl)phenyl to get molecules 1a–1d (Fig. 1A). It was envisaged that the intramolecular C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯H–O and C–O⋯H–N interactions would provide a preorganized geometry of individual monomers. The intermolecular π–π stacking interactions among aromatic rings and hydrogen bonding interactions among amide moieties would lead to the aggregation of individual monomers in the membrane to form an active channel (Fig. 1B).3 Theoretical estimations, based on the calculator plugins of the MarvinSketch program,36 indicated that the variation of terminal aromatic groups from phenyl to 4-methylphenyl, 4-trifluoromethylphenyl and 3,5-bis(trifluoromethyl)phenyl would decrease the pKa of both amide protons (Table S1†). Therefore, in the resulting supramolecular channel, the amide protons of 1d would contribute to the maximum extent in forming intra- and intermolecular (with neighboring monomers and water molecules present in the cavity and anions) hydrogen bonds. Estimated pKa = 6.42 for the 2-hydroxyl group of each compound indicated that in the supramolecular channel, one or more hydroxyl groups may get deprotonated at physiological pH providing cation binding sites. The recognition sites for cations can also be provided by either a C–F or a C
O⋯H–O and C–O⋯H–N interactions would provide a preorganized geometry of individual monomers. The intermolecular π–π stacking interactions among aromatic rings and hydrogen bonding interactions among amide moieties would lead to the aggregation of individual monomers in the membrane to form an active channel (Fig. 1B).3 Theoretical estimations, based on the calculator plugins of the MarvinSketch program,36 indicated that the variation of terminal aromatic groups from phenyl to 4-methylphenyl, 4-trifluoromethylphenyl and 3,5-bis(trifluoromethyl)phenyl would decrease the pKa of both amide protons (Table S1†). Therefore, in the resulting supramolecular channel, the amide protons of 1d would contribute to the maximum extent in forming intra- and intermolecular (with neighboring monomers and water molecules present in the cavity and anions) hydrogen bonds. Estimated pKa = 6.42 for the 2-hydroxyl group of each compound indicated that in the supramolecular channel, one or more hydroxyl groups may get deprotonated at physiological pH providing cation binding sites. The recognition sites for cations can also be provided by either a C–F or a C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O moiety via cation–dipole interactions. We envisaged that the 2-hydroxyl group would also be useful in generating inherent fluorescence of these molecules due to the push–pull effect. The MarvinSketch program36 also predicted that the lipophilicity (logP)37,38 of the compounds would increase as 1a < 1b < 1c < 1d contributing to the better membrane permeability of the channel formed by 1d.
O moiety via cation–dipole interactions. We envisaged that the 2-hydroxyl group would also be useful in generating inherent fluorescence of these molecules due to the push–pull effect. The MarvinSketch program36 also predicted that the lipophilicity (logP)37,38 of the compounds would increase as 1a < 1b < 1c < 1d contributing to the better membrane permeability of the channel formed by 1d.
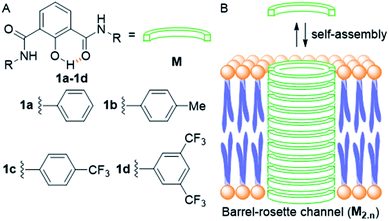 | ||
| Fig. 1 Structure of designed channel-forming compounds 1a–1d (A); self-assembly of individual monomers M into rosette channel M2.n in the lipid bilayer membrane (B). | ||
Results and discussion
Compounds 1a–1d were synthesized from protected acid 4 by converting it to the acid chloride using oxalyl chloride,39 which was then coupled with the respective aromatic amine to get the corresponding amide 5a–5d (Scheme 1). The product was subjected to O-methyl deprotection using 1 M BBr3 in dichloromethane40 to get the corresponding final compound 1a–1d with a free hydroxyl group. All newly synthesized compounds were purified by column chromatography, and characterized by 1H NMR, 13C NMR, HRMS, IR, and melting point. The single-crystal X-ray diffraction study of 1c provided the molecular geometry containing the intramolecular C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯H–O and C–O⋯H–N hydrogen bonding interactions (Fig. S11†).
O⋯H–O and C–O⋯H–N hydrogen bonding interactions (Fig. S11†).
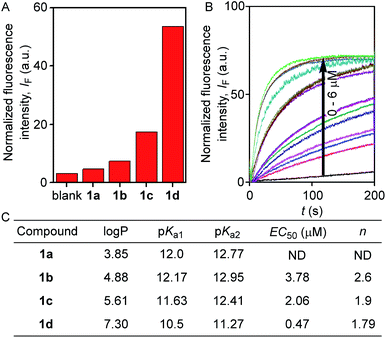
The ion transport activity of 2-hydroxy-N1,N3-diarylisophthalamide derivatives 1a–1d was checked by different fluorometric methods across egg yolk phosphatidylcholine unilamellar vesicles (EYPC-LUVs).41–45 At first, large EYPC-LUVs, entrapping the pH-sensitive dye 8-hydroxypyrene-1,3,6-trisulfonate (HPTS, pKa = 7.2), were prepared, and NaOH was added in the extravesicular buffer to create a pH gradient (i.e., ΔpH = 0.8). The rate of change in the fluorescence intensity was monitored after the addition of 1a–1d, separately, and finally, Triton X-100 was added to lyse the vesicles for the complete leakage of the HPTS dye.46 The comparison of activities showed the activity sequence 1a < 1b < 1c ≪ 1d, inferring 1d as the most efficient transporter of ions (Fig. 2A). The activity sequence of 1a–1d suggests that the pKa values of the amide protons have major control over the transport rate as compared to logP values. The EC50 value, i.e., the concentration required to reach half of the maximum activity, was calculated to be 0.48 μM (compound to lipid ratio = 0.58 mol%), and the Hill coefficient, n, was calculated to be almost equal to 2 (Fig. 2C). These data suggest that a noncovalent dimer of the compound is the active structure for the supramolecular nanochannel assembly.47 The Hill analysis could not be performed with the dose-dependent activity data of 1a due to its precipitation at higher concentrations.
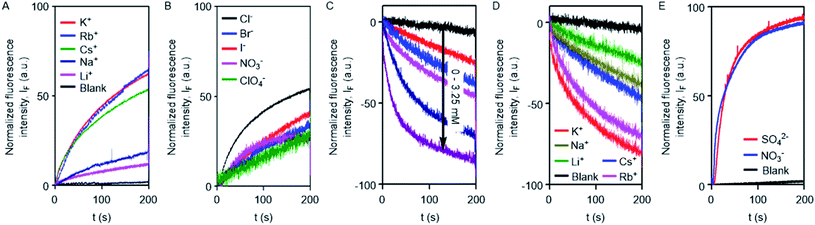
The excellent ion transport activity of 1d inspired us to investigate the ion selectivity of this channel forming molecule. First, the ion transport activity across EYPC-LUVs⊃HPTS with intravesicular NaCl and an isoosmolar extravesicular MCl (where, M+ = Li+, Na+, K+, Rb+, and Cs+) was used to determine the selectivity among the different cations.48–50 The data provided the selectivity sequence as K+ ≈ Rb+ > Cs+ > Na+ > Li+ (Fig. 3A). To see if there is any role of anions involved in the transport process, the anion selectivity study was performed by varying the intravesicular and extravesicular anions of NaX salts (X− = Cl−, Br−, I−, NO3−, and ClO4−), and changes in the ion transport rate were checked upon applying a pH gradient of 0.8 (pHin = 7.0 and pHout = 7.8).48,51,52 Interestingly, the data confirmed a considerable selectivity of the channel towards Cl− compared to the other anions (Fig. 3B). The selectivity studies suggested that both inorganic cations and anions are involved in the transport across the channel formed by 1d.
To further confirm the Cl− ion transport activity of 1d, the EYPC-LUVs⊃lucigenin was prepared by entrapping the lucigenin dye and NaNO3 salt and investigated in detail by monitoring the fluorescence intensity of the intravesicular lucigenin dye at λem = 535 nm (λex = 450 nm).53,54 Subsequently, a Cl−/NO3− gradient was applied by the addition of a concentrated solution of 2 M NaCl in the extravesicular buffer. Concentration-dependent quenching of lucigenin fluorescence was observed upon addition of channel forming compound 1d (Fig. 3C). These data show that the supramolecule formed by the compound can allow the influx of Cl− across liposomes. The variation of extravesicular cations corroborated with cation selectivity obtained by the HPTS assay (Fig. 3D). So, to get further support for the symport mechanism of ion transport, the valinomycin coupled assay with 1d was carried out.29 KCl was added to the extravesicular buffer to create the Cl−/NO3− gradient. Then valinomycin was added along with 1d to monitor any cooperative effect. There was no enhancement of the transport rate, which suggests that there is no cooperative transport by valinomycin and 1d (Fig. S5†). So the operating transport mechanism should be the symport mechanism. To further confirm the symport mechanism, the EYPC liposomes were prepared by entrapping NaCl (200 mM) and 1.0 mM of the lucigenin dye. Then, isoosmolar Na2SO4 was added to the extravesicular solution and Cl− efflux was monitored. A similar experiment was also performed with extravesicular isoosmolar NaNO3. The results indicate that there is no difference in the transport rate of two ions by 1d (Fig. 3E), suggesting that the symport mechanism of ion transport is operative.28
To investigate the integrity of the membrane after addition of 1d or the formation of supramolecular pores in the membrane, the leakage of carboxyfluorescein (CF) was monitored across EYPC-LUVs⊃CF.55 It is well known that a high concentration of CF molecules inside the liposomes leads to the fluorescence quenching of the dye due to collisions among the molecules. However, efflux of these molecules from the liposomes into the external buffer restores their fluorescence (λem = 517 nm, λex = 492 nm). When compound 1d was added to the liposomes, there was no significant enhancement in the fluorescence intensity of CF (Fig. S9†), indicating that neither the membrane is getting damaged, nor the larger pores are being formed by 1d.
To confirm the formation of the transmembrane ion channel by compound 1d, the ionic conductance across the planar lipid bilayer membrane was measured.56 In this experiment, two compartments (cis and trans chambers) containing KCl solution (1.0 M) were separated by a planar lipid bilayer membrane composed of diphytanoyl phosphatidylcholine (diPhyPC) lipid.57,58 The addition of 1d (2.0 μM) to the system led to the distinct channel openings and closing at different holding potentials, confirming the formation of ion channels (Fig. 4A, B). The single-channel conductance was found to be 100 ± 2 pS, and the channel diameter = 5.06 ± 0.16 Å was calculated by the Hille equation (eqn (1))
| 1/g = (l + πd/4) × (4ρ/ πd2) | (1) |
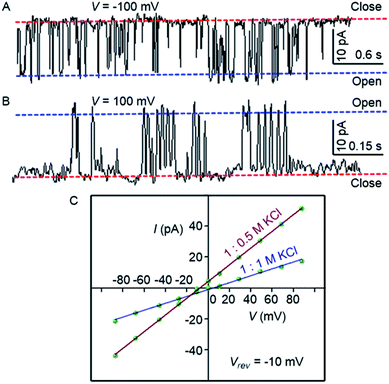 | ||
| Fig. 4 Single-channel conductance of 1d (2.0 μM) recorded at −100 mV (A) and at 100 mV (B) in symmetrical KCl solutions. I–V plots of 1d in symmetrical and unsymmetrical concentrations of KCl (C). | ||
The variation of current with voltage (I–V plot) in the presence of the symmetric solution of KCl (1.0 M each) showed a linear variation, i.e., ohmic behavior, which confirms the non-dipole nature of the channel. When the ion selectivity of the channel was checked using unsymmetric solutions of KCl in two chambers (i.e., 1.0 M in cis and 0.5 M KCl in trans), it was observed that the rate of Cl− transport is higher than the rate of K+ transport with a permeability ratio PCl−/PK+ = 8.29 ± 1 (Fig. 4C). These data suggest that the electrogenic transport of Cl− gives the current signals during the recordings.
To get further support for channel formation, the temperature dependent ion transport in DPPC-LUVs⊃HPTS was studied. Compound 1d showed some decrease in the activity at 30 °C compared to that at 45 °C (Fig. S10†). This behavior is normal for various biological and synthetic ion channels due to hindered partition at lower temperature.41
Based on the crystal structure of 1c (Fig. S12†), it is evident that each 2-hydroxy-N1,N3-diarylisophthalamide molecule exists predominantly in an intramolecular C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯H–O and C–O⋯H–N hydrogen bonded preorganized geometry. The conformational study of 1d using the Conflex 8 program59,60 also indicated similar hydrogen bonded conformations of the di-amide core (Fig. S12†). At first, eleven molecules of 1d in the Conf-1a were placed on top of another to form one side of the channel. Subsequently, the other side of the channel was constructed in the same way and the two halves of the channel were arranged face-to-face to form the barrel-rosette channel. The constructed channel was further optimized using MOPAC2012 (ref. 61) software with the PM6-DH+ (ref. 62) method to get the final channel (Fig. S13A†). This optimized channel was then placed in the POPC lipid bilayer membrane, and molecular dynamics (MD) simulations were carried out (see ESI† for methods). The equilibrated channel is shown in Fig. 5A. Fig. S13B† shows the densities of various constituents of the channel indicating the proper formation of the channel. The top view of the channel with a pore diameter is shown in Fig. S13C.† Fig. S13D† shows that the variation in the pore diameter is minimal around an average of 5.3 Å during the simulations, in excellent agreement with the experiment.
O⋯H–O and C–O⋯H–N hydrogen bonded preorganized geometry. The conformational study of 1d using the Conflex 8 program59,60 also indicated similar hydrogen bonded conformations of the di-amide core (Fig. S12†). At first, eleven molecules of 1d in the Conf-1a were placed on top of another to form one side of the channel. Subsequently, the other side of the channel was constructed in the same way and the two halves of the channel were arranged face-to-face to form the barrel-rosette channel. The constructed channel was further optimized using MOPAC2012 (ref. 61) software with the PM6-DH+ (ref. 62) method to get the final channel (Fig. S13A†). This optimized channel was then placed in the POPC lipid bilayer membrane, and molecular dynamics (MD) simulations were carried out (see ESI† for methods). The equilibrated channel is shown in Fig. 5A. Fig. S13B† shows the densities of various constituents of the channel indicating the proper formation of the channel. The top view of the channel with a pore diameter is shown in Fig. S13C.† Fig. S13D† shows that the variation in the pore diameter is minimal around an average of 5.3 Å during the simulations, in excellent agreement with the experiment.
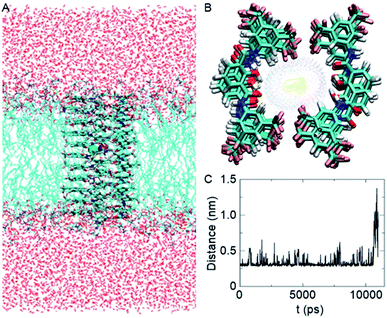 | ||
| Fig. 5 Equilibrated channel-POPC/water system (A), top view of the zoomed channel without lipid membrane (B), and variation of the distance between K+ and Cl− ions during the production run (C). | ||
In the simulated channel, upon inspection at various time frames (Fig. S14A–D†), multiple noncovalent interactions were found to stabilize the channel structure and helps in ion recognition in the channel lumen. The stacking interactions among aromatic rings (among successive central phenyl rings and among successive aromatic arms) and the hydrophobic interactions among CF3 groups were found to be present. Moreover, intermolecular C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯H–O(phenolic) and C–O⋯H–N interactions were also evident in the channel. The water molecules inside the channel formed a continuous array with multiple intermolecular H–O⋯H–O interactions with neighbouring water molecules. The water molecules were also involved in hydrogen bonding with C–F and C
O⋯H–O(phenolic) and C–O⋯H–N interactions were also evident in the channel. The water molecules inside the channel formed a continuous array with multiple intermolecular H–O⋯H–O interactions with neighbouring water molecules. The water molecules were also involved in hydrogen bonding with C–F and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups of the channel forming molecules (Fig. S14E†). The K+ ion was involved in cation–dipole interactions with neighbouring F-centres of the channel forming molecules and with O-centres of the neighbouring water molecules (Table S2†). The Cl− ion was involved in C–O–H⋯Cl− interactions with channel forming molecules and O–H⋯Cl− interactions with neighbouring water molecules (Table S3†). Interestingly, the permeation of K+ and Cl− in the channel took place with an interionic distance within 5 Å, confirming the presence of electrostatic interactions (Fig. 5C, S16†). In the bulk water, the ions got separated due to hydration.
O groups of the channel forming molecules (Fig. S14E†). The K+ ion was involved in cation–dipole interactions with neighbouring F-centres of the channel forming molecules and with O-centres of the neighbouring water molecules (Table S2†). The Cl− ion was involved in C–O–H⋯Cl− interactions with channel forming molecules and O–H⋯Cl− interactions with neighbouring water molecules (Table S3†). Interestingly, the permeation of K+ and Cl− in the channel took place with an interionic distance within 5 Å, confirming the presence of electrostatic interactions (Fig. 5C, S16†). In the bulk water, the ions got separated due to hydration.
The inherent fluorescence of 1d encouraged us to use it for confocal live cell imaging to check whether the molecules of the compound stay only in the membrane or can permeate inside the cell as well. The live cell imaging of 1d in the human epithelial breast cancer cell line, MCF7, indicated that the compound does not remain confined to the cell membrane only, but permeates into the cytosolic part as well (Fig. 6).20 Next, we monitored the time of insertion of the compound in the cell using real-time analysis. The results indicated that the compound enters the cells within a few seconds of incubation (see uploaded video†). Interestingly, the real-time analysis also indicated a change in the cell morphology within a few minutes and a decrease in the cell volume was observed. Such a decrease in the cell volume is indicative of the apoptotic volume decrease (AVD), which is a well-known initial event of apoptosis.63–65
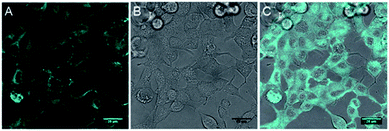 | ||
| Fig. 6 Live cell imaging of 1d at 10 μM in MCF7 cell line; fluorescence (A), DIC image (B) and overlay image (C). Cyan color was given for better contrast. Scale bar 80 μm. | ||
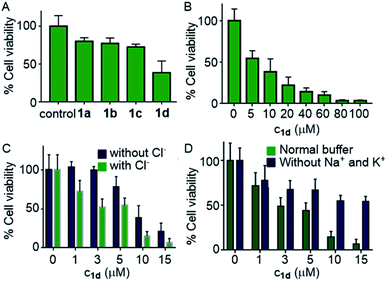
Encouraged by this observation, we evaluated the viability of cells in the presence of compounds 1a–1d. A single-point screening of the cell viability was done by the MTT assay after incubation of 1a–1d in the MCF7 cell line for 24 hours.9 As expected, compound 1d displayed maximum cell killing activity (Fig. 7A). Next, the dose-dependent effects of 1d on MCF7 cells were studied which showed a decrease in the cell viability upon increasing the concentration of the compound giving the IC50 value of approximately 5.0 μM (Fig. 7B).
Next, the chloride mediated cell death was studied by comparing the cell viability both in the presence and absence of chloride ions in the culture medium. The chloride mediated apoptosis is well studied in the literature.9,26,66 So two different types of HBSS (Hanks balanced salt solution) were prepared, with and without Cl− ions for using as the extracellular media. The MCF7 cells were suspended in both the media and incubated with compound 1d at different concentrations around the IC50 value. As expected, with chloride ions in the extracellular buffer the compound showed more cell death compared to that without chloride ions (Fig. 7C). This experiment clearly demonstrates that the chloride-mediated cell death takes place. As the compound is a M+/Cl− symporter, we decided to evaluate the effect of K+ ions and Na+ ions on the cell death. Since the concentration of Na+ ions is very high outside the cell compared to K+ ions, we expected a better involvement of Na+ ions, if the symport process is taking place. So to evaluate the effect of cations, we again prepared two different HBSS buffers, with and without K+ and Na+ ions and used as the extracellular media. Compound 1d showed higher cell viability which was observed for 1d without both K+ and Na+ ions in the extracellular media in comparison to that when studied in the presence of normal HBSS buffer (containing Na+, K+, and Cl− ions) (Fig. 7D). These results confirm that the cell death is mediated by Cl− and cations.
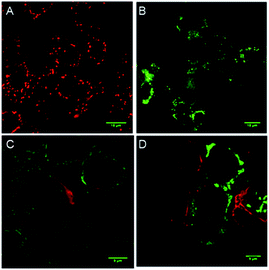
To evaluate whether cell death is being mediated by necrosis or apoptosis, the events involved in the apoptotic pathway were evaluated in detail. The observed AVD is already an indication of apoptosis. Apoptosis is also evidenced by the disruption of mitochondrial membrane potential (MMP),67,68 which subsequently results in cytochrome c release. The release of cytochrome c in the cytoplasm switches on the apoptotic signaling cascade.69–71 Firstly, we monitored the change in mitochondrial membrane potential by using the MMP sensitive JC-1 dye. This dye exhibits red fluorescence emission due to the formation of J-aggregates in the healthy mitochondrial membrane, while depolarization of the mitochondrial membrane leads to dispersion of the dye in the cytosol, resulting in green fluorescence emission. The MCF7 cells were incubated with compound 1d (10 μM), followed by the treatment with the JC-1 dye. The cells were analysed under a confocal microscope to monitor the changes in the red and green fluorescence in the treated cells. The study showed a prominent decrease in the red fluorescence and a concomitant increase in the green emission (Fig. 8).70,72 The quantification of the pixel intensity ratio (green/red = 2.57)9 in treated cells compared to the control (green/red = 0.263) confirmed the enhancement in the depolarization of MMP due to the change in ionic homeostasis of cells.
This change in ionic homeostasis of cells leads to a disturbance in the electron transport chain in the mitochondrial respiratory cycles, which results in reactive oxygen species (ROS) production.73–76 Thus, 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA),68,77 a ROS probe, was used to monitor ROS generation in cells. The probe remains in the non-fluorescent state in the diacetate protected form. However, upon cellular internalization, the hydrolysis of the ester groups by the esterases followed by the ROS-mediated oxidation generates a green fluorescent 2′,7′-dichlorofluorescein (DCF). The MCF7 cells were treated with 1d in a time-dependent manner and then stained with H2DCFDA, and the cells were analysed under a confocal microscope. A significant enhancement in the green fluorescence was observed, which clearly demonstrates the generation of ROS as a result of ion transport by 1d (Fig. S19†).
The elevated ROS levels are known to cause the opening of the mitochondrial permeability transition pores (PTPs),78,79 which are formed by natural ion channels present in the mitochondrial membrane. This process results in the disruption of the mitochondrial outer membrane and a subsequent release of cytochrome c from the mitochondrial membrane into cytosol.80,81 Therefore, to monitor the release of cytochrome c into the cytosol, immunostaining with a specific antibody was used. The MCF7 cells were treated with 1d and then processed for immunofluorescence to see if any release of cytochrome c is taking place. A considerable enhancement in the fluorescence intensity and dispersion of the fluorescence signal over cytosol (Fig. 8) indicates the release of cytochrome c.
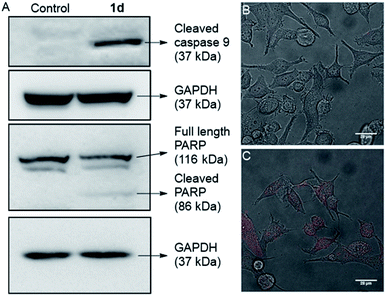
In the intrinsic apoptotic pathway, the released cytochrome c binds to the Apaf-1 to form an apoptosome. Then, the cytochrome c/Apaf-1 complex activates the caspase 9 pathway, which then activates the caspase 3 pathway.69,82–84 Therefore, we analysed the caspase 9 pathway by immunoblot analysis of the cleaved caspase 9 level in MCF7 cells upon addition of 1d. The incubation of the cells with 1d led to a significant increase in the expression of cleaved caspase 9 (Fig. 9). The increase in the levels of caspase 9 was quantified with respect to the GAPDH (loading control). So, the expression of cleaved caspase 9 confirms the activation of the intrinsic pathway of apoptosis. To further confirm the apoptotic pathway, the expression of cleaved poly(ADP-ribose) polymerase (PARP) was checked. PARP cleavage by endogenous caspases is a very well-known phenomenon.85–87 PARP-1 cleavage prevents DNA repair to facilitate apoptosis. A significant amount of degradation of full-length PARP-1 (116 kDa) with a concomitant increase of cleaved PARP-1 (86 kDa) was observed in the immunoblot analysis of MCF7 cells incubated with 10 μM concentration of 1d (Fig. 9A).
For a final validation of the overall process, the propidium iodide (PI) assay was used. The dye binds to the nuclear constituents of the cell, typically DNA and RNA, however, it is impermeable to the healthy cells.88 The dye can easily cross the cell membrane of damaged cells and bind to the nuclear constituents. So the MCF7 cells were treated with 1d for 24 h and then stained with PI. The live cell imaging of the cells clearly showed that the PI uptake is very prominent in the case of treated cells, compared to control cells (Fig. 9B, C, and S20†). So these data finally validate that the cells have been damaged by treatment with 1d.
Conclusions
In summary, we have synthesized fluorescent 2-hydroxy-N1,N3-diarylisophthalamides which form efficient ion channels in the lipid bilayer membranes. The compound with two 3,5-bis(trifluoromethyl)phenyl arms was the most active among the entire series when ion transport activity was measured across large unilamellar vesicles. The study also confirmed the symport of M+ and Cl−. The channel formation was also validated by planar bilayer conductance measurement studies, which gave the channel diameter = 5.06 ± 0.16 Å. The permeability ratio, PCl−/PK+ = 8.29 ± 1, indicates higher selectivity for Cl− compared to K+. We have also constructed an atomistic model of the channel and performed MD simulations to show various interactions of the channel molecules with themselves and with the ions. The simulations show that the pore diameter is maintained at an average value of 5.3 Å. Also, both cations and ions permeate together through the channel, keeping a smaller distance till it reaches the bulk water. Inherent fluorescence of the compound was helpful in the live cell imaging of MCF7 cells, which indicated the presence of the compound in the cell membrane and in the cytoplasmic content. The channel facilitates the transport of chloride and cations across the cellular membrane, and the process resulted in the perturbation of the ionic homeostasis of cells leading to significant cell death. The mechanism of cell death was confirmed by the mitochondrial membrane depolarization and reactive oxygen species generation. These processes resulted in the cellular apoptosis, which was confirmed by cytochrome c release, activation of the caspase 9 pathway, PARP cleavage, and staining of nuclear contents by propidium iodide. To the best of our knowledge, the present study demonstrates the first example of an inherently fluorescent synthetic channel with an ambivalent chemical motif for complexation and transport of both cations and chloride ions simultaneously. Therefore, this system can be considered as a synthetic congener of naturally occurring sodium-potassium-chloride co-transporters (NKCCs). Such design will encourage the future development of synthetic channels for biomedical applications such as cancer treatment, channel replacement therapy etc.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Ms. Aishwarya Venkataravi of IISER Pune for the valuable discussions about microscopy. We also thank Ms. Shivani Sharma for her help in solving the single crystal XRD data. We would like to acknowledge IISER, Pune microscopy facility. This work was supported by SERB, Govt. of India (EMR/2016/001897) and Department of Biotechnology (DBT), Govt. of India (BT/HRD/NBA/36/06/2018).Notes and references
- W. D. Stein, Channels, Carriers, and Pumps: An Introduction to Membrane Transport, Academic Press, San Diego, CA, 1990 Search PubMed.
- P. Agre, Angew. Chem., Int. Ed., 2004, 43, 4278–4290 CrossRef CAS PubMed.
- T. J. Jentsch, C. A. Hubner and J. C. Fuhrmann, Nat. Cell Biol., 2004, 6, 1039–1047 CrossRef CAS.
- L. Song, M. R. Hobaugh, C. Shustak, S. Cheley, H. Bayley and J. E. Gouaux, Science, 1996, 274, 1859–1866 CrossRef CAS PubMed.
- J. T. Davis, O. Okunola and R. Quesada, Chem. Soc. Rev., 2010, 39, 3843–3862 RSC.
- S. B. Hladky, J. Membr. Biol., 1979, 46, 213–237 CrossRef CAS.
- S. Y. Chiu and G. F. Wilson, J. Physiol., 1989, 408, 199–222 CrossRef CAS.
- T. E. DeCoursey, K. G. Chandy, S. Gupta and M. D. Cahalan, Nature, 1984, 307, 465–488 CrossRef CAS.
- T. Saha, A. Gautam, A. Mukherjee, M. Lahiri and P. Talukdar, J. Am. Chem. Soc., 2016, 138, 16443–16451 CrossRef CAS.
- T. W. Allen, O. S. Andersen and B. Roux, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 117–122 CrossRef CAS.
- M. Bayrhuber, T. Meins, M. Habeck, S. Becker, K. Giller, S. Villinger, C. Vonrhein, C. Griesinger, M. Zweckstetter and K. Zeth, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 15370–15375 CrossRef CAS.
- D. A. Doyle, J. M. Cabral, R. A. Pfuetzner, A. Kuo, J. M. Gulbis, S. L. Cohen, B. T. Chait and R. MacKinnon, Science, 1998, 280, 69–77 CrossRef CAS.
- R. MacKinnon, Angew. Chem., Int. Ed., 2004, 43, 4265–4277 CrossRef CAS.
- X. Wu and P. A. Gale, J. Am. Chem. Soc., 2016, 138, 16508–16514 CrossRef CAS.
- B. Dworakowska and K. Dolowy, Acta Biochim. Pol., 2000, 47, 685–703 CrossRef CAS.
- P. A. Gale, J. T. Davis and R. Quesada, Chem. Soc. Rev., 2017, 46, 2497–2519 RSC.
- O. K. Steinlein, Chem. Rev., 2012, 112, 6334–6352 CrossRef CAS.
- J. J. Kasianowicz, Chem. Rev., 2012, 112, 6215–6217 CrossRef CAS PubMed.
- H. Li, H. Valkenier, L. W. Judd, P. R. Brotherhood, S. Hussain, J. A. Cooper, O. Jurček, H. A. Sparkes, D. N. Sheppard and A. P. Davis, Nat. Chem., 2015, 8, 24 CrossRef.
- S. N. Berry, V. Soto-Cerrato, E. N. W. Howe, H. J. Clarke, I. Mistry, A. Tavassoli, Y. T. Chang, R. Pérez-Tomás and P. A. Gale, Chem. Sci., 2016, 7, 5069–5077 RSC.
- N. L. Mora, A. Bahreman, H. Valkenier, H. Li, T. H. Sharp, D. N. Sheppard, A. P. Davis and A. Kros, Chem. Sci., 2016, 7, 1768–1772 RSC.
- B. Shen, X. Li, F. Wang, X. Yao and D. A. Yang, PLoS One, 2012, 7, e34694 CrossRef CAS PubMed.
- P. A. Gale, R. Pérez-Tomás and R. Quesada, Acc. Chem. Res., 2013, 46, 2801–2813 CrossRef CAS.
- A. Furstner, Angew. Chem., Int. Ed., 2003, 42, 3582–3603 CrossRef PubMed.
- R. Perez-Tomas and M. Vinas, Curr. Med. Chem., 2010, 17, 2222–2231 CrossRef CAS PubMed.
- S. K. Ko, S. K. Kim, A. Share, V. M. Lynch, J. Park, W. Namkung, W. Van Rossom, N. Busschaert, P. A. Gale, J. L. Sessler and I. Shin, Nat. Chem., 2014, 6, 885–892 CrossRef CAS PubMed.
- V. Soto-Cerrato, P. Manuel-Manresa, E. Hernando, S. Calabuig-Fariñas, A. Martínez-Romero, V. Fernández-Dueñas, T. Sahlholm, K. Knöpfel, M. García-Valverde, A. M. Rodilla, E. Jantus-Lewintre, R. Farràs, F. Ciruela, R. Pérez-Tomás and R. Quesada, J. Am. Chem. Soc., 2015, 137, 15892–15898 CrossRef CAS PubMed.
- N. Busschaert, M. Wenzel, M. E. Light, P. Iglesias-Hernández, R. Pérez-Tomás and P. A. Gale, J. Am. Chem. Soc., 2011, 133, 14136–14148 CrossRef CAS PubMed.
- T. Saha, M. S. Hossain, D. Saha, M. Lahiri and P. Talukdar, J. Am. Chem. Soc., 2016, 138, 7558–7567 CrossRef CAS PubMed.
- L. A. Jowett, E. N. W. Howe, V. Soto-Cerrato, W. Van Rossom, R. Pérez-Tomás and P. A. Gale, Sci. Rep., 2017, 7, 9397 CrossRef PubMed.
- A. V. Koulov, J. M. Mahoney and B. D. Smith, Org. Biomol. Chem., 2003, 1, 27–29 RSC.
- J. H. Lee, Y. R. Choi, P. Kang, M. G. Choi and K. S. Jeong, J. Org. Chem., 2014, 79, 6403–6409 CrossRef CAS.
- C. C. Tong, R. Quesada, J. L. Sessler and P. A. Gale, Chem. Commun., 2008, 47, 6321–6323 RSC.
- A. Vargas Jentzsch, D. Emery, J. Mareda, P. Metrangolo, G. Resnati and S. Matile, Angew. Chem., Int. Ed., 2011, 50, 11675–11678 CrossRef CAS.
- D. Basak, S. Sridhar, A. K. Bera and N. Madhavan, Org. Biomol. Chem., 2016, 14, 4712–4717 RSC.
- Marvin 5.8.0, ChemAxon, 2012, http://www.chemaxon.com Search PubMed.
- V. Saggiomo, S. Otto, I. Marques, V. Félix, T. Torroba and R. Quesada, Chem. Commun., 2012, 48, 5274–5276 RSC.
- C. A. Lipinski, F. Lombardo, B. W. Dominy and P. J. Feeney, Adv. Drug Delivery Rev., 1997, 23, 3–25 CrossRef CAS.
- A. L. Trifonov, V. V. Levin, M. I. Struchkova and A. D. Dilman, Org. Lett., 2017, 19, 5304–5307 CrossRef CAS PubMed.
- T. Routasalo, J. Helaja, J. Kavakka and A. M. P. Koskinen, Eur. J. Org. Chem., 2008, 2008, 3190–3199 CrossRef.
- A. L. Sisson, M. R. Shah, S. Bhosale and S. Matile, Chem. Soc. Rev., 2006, 35, 1269–1286 RSC.
- P. Talukdar, G. Bollot, J. Mareda, N. Sakai and S. Matile, J. Am. Chem. Soc., 2005, 127, 6528–6529 CrossRef CAS PubMed.
- S. V. Shinde and P. Talukdar, Chem. Commun., 2018, 54, 10351–10354 RSC.
- A. Roy, A. Gautam, J. A. Malla, S. Sarkar, A. Mukherjee and P. Talukdar, Chem. Commun., 2018, 54, 2024–2027 RSC.
- T. Saha, S. Dasari, D. Tewari, A. Prathap, K. M. Sureshan, A. K. Bera, A. Mukherjee and P. Talukdar, J. Am. Chem. Soc., 2014, 136, 14128–14135 CrossRef CAS.
- T. Saha, A. Roy, M. L. Gening, D. V. Titov, A. G. Gerbst, Y. E. Tsvetkov, N. E. Nifantiev and P. Talukdar, Chem. Commun., 2014, 50, 5514–5516 RSC.
- S. Litvinchuk, G. Bollot, J. Mareda, A. Som, D. Ronan, M. R. Shah, P. Perrottet, N. Sakai and S. Matile, J. Am. Chem. Soc., 2004, 126, 10067–10075 CrossRef CAS.
- J. A. Malla, A. Roy and P. Talukdar, Org. Lett., 2018, 20, 5991–5994 CrossRef CAS.
- A. Gilles and M. Barboiu, J. Am. Chem. Soc., 2016, 138, 426–432 CrossRef CAS.
- C. Ren, J. Shen and H. Zeng, J. Am. Chem. Soc., 2017, 139, 12338–12341 CrossRef CAS PubMed.
- C. Ren, X. Ding, A. Roy, J. Shen, S. Zhou, F. Chen, S. F. Yau Li, H. Ren, Y. Y. Yang and H. Zeng, Chem. Sci., 2018, 9, 4044–4051 RSC.
- C. Ren, F. Zeng, J. Shen, F. Chen, A. Roy, S. Zhou, H. Ren and H. Zeng, J. Am. Chem. Soc., 2018, 140, 8817–8826 CrossRef CAS PubMed.
- B. P. Benke, P. Aich, Y. Kim, K. L. Kim, M. R. Rohman, S. Hong, I. C. Hwang, E. H. Lee, J. H. Roh and K. Kim, J. Am. Chem. Soc., 2017, 139, 7432–7435 CrossRef CAS.
- C. R. Yamnitz, S. Negin, I. A. Carasel, R. K. Winter and G. W. Gokel, Chem. Commun., 2010, 46, 2838–2840 RSC.
- M. J. Langton, L. M. Scriven, N. H. Williams and C. A. Hunter, J. Am. Chem. Soc., 2017, 139, 15768–15773 CrossRef CAS PubMed.
- M. Jung, H. Kim, K. Baek and K. Kim, Angew. Chem., Int. Ed., 2008, 47, 5755–5757 CrossRef CAS PubMed.
- N. Sakai and S. Matile, Langmuir, 2013, 29, 9031–9040 CrossRef CAS PubMed.
- A. V. Jentzsch, D. Emery, J. Mareda, S. K. Nayak, P. Metrangolo, G. Resnati, N. Sakai and S. Matile, Nat. Commun., 2012, 3, 905 CrossRef PubMed.
- H. Goto, S. Obata, N. Nakayama and K. Ohta, CONFLEX 8, CONFLEX Corporation, Tokyo, Japan, 2012 Search PubMed.
- H. Goto and E. Osawa, J. Am. Chem. Soc., 1989, 111, 8950–8951 CrossRef CAS.
- J. J. P. Stewart, MOPAC2012, Stewart Computational Chemistry: Colorado Springs, CO, USA, 2012 Search PubMed.
- P. Májek and R. Elber, J. Chem. Theory Comput., 2010, 6, 1805–1817 CrossRef.
- C. D. Bortner and J. A. Cidlowski, Cell Death Differ., 2002, 9, 1307 CrossRef CAS PubMed.
- R. Núñez, S. M. Sancho-Martínez, J. M. L. Novoa and F. J. López-Hernández, Cell Death Differ., 2010, 17, 1665 CrossRef PubMed.
- M. A. Model, Am. J. Physiol.: Cell Physiol., 2014, 306, C417–C424 CrossRef CAS PubMed.
- N. Busschaert, S. H. Park, K. H. Baek, Y. P. Choi, J. Park, E. N. W. Howe, J. R. Hiscock, L. E. Karagiannidis, I. Marques, V. Félix, W. Namkung, J. L. Sessler, P. A. Gale and I. Shin, Nat. Chem., 2017, 9, 667 CrossRef CAS PubMed.
- S. Sakamuru, M. S. Attene-Ramos and M. Xia, Methods Mol. Biol., 2016, 1473, 17–22 CrossRef PubMed.
- S. Patil, M. M. Kuman, S. Palvai, P. Sengupta and S. Basu, ACS Omega, 2018, 3, 1470–1481 CrossRef CAS PubMed.
- A. Ashkenazi, Nat. Rev. Drug Discovery, 2008, 7, 1001–1012 CrossRef CAS PubMed.
- S. T. Smiley, M. Reers, C. Mottola-Hartshorn, M. Lin, A. Chen, T. W. Smith, G. D. Steele and L. B. Chen, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 3671–3675 CrossRef CAS PubMed.
- N. Zamzami and G. Kroemer, Curr. Biol., 2003, 13, R71–R73 CrossRef CAS PubMed.
- A. Cossarizza, M. Baccarani-Contri, G. Kalashnikova and C. A. Franceschi, Biochem. Biophys. Res. Commun., 1993, 197, 40–45 CrossRef CAS PubMed.
- S. S. Sabharwal and P. T. Schumacker, Nat. Rev. Cancer, 2014, 14, 709–721 CrossRef CAS PubMed.
- G. Y. Liou and P. Storz, Free Radical Res., 2010, 44, 479–496 CrossRef CAS PubMed.
- L. A. Sena and N. S. Chandel, Mol. Cell, 2012, 48, 158–167 CrossRef CAS PubMed.
- S. Galadari, A. Rahman, S. Pallichankandy and F. Thayyullathil, Free Radicals Biol. Med., 2017, 104, 144–164 CrossRef CAS PubMed.
- D. Wu and P. Yotnda, J. Visualized Exp., 2011, 57, 3357 Search PubMed.
- A. P. Halestrap, J. Mol. Cell. Cardiol., 2009, 46, 821–831 CrossRef CAS PubMed.
- M. Crompton, Biochem. J., 1999, 341, 233–249 CrossRef CAS PubMed.
- M. Madesh and G. Hajnoczky, J. Cell Biol., 2001, 155, 1003–1015 CrossRef CAS PubMed.
- V. P. Skulachev, Apoptosis, 2006, 11, 473–485 CrossRef CAS.
- S. P. Cullen and S. J. Martin, Cell Death Differ., 2009, 16, 935–938 CrossRef CAS.
- J. Wu, T. Liu, J. Xie, F. Xin and L. Guo, Cell. Mol. Life Sci., 2006, 63, 949–957 CrossRef CAS.
- Y. Shi, Mol. Cell, 2002, 9, 459–470 CrossRef CAS PubMed.
- S. H. Park, Y. P. Choi, J. Park, A. Share, O. Francesconi, C. Nativi, W. Namkung, J. L. Sessler, S. Roelens and I. Shin, Chem. Sci., 2015, 6, 7284–7292 RSC.
- G. V. Chaitanya, A. J. Steven and P. P. Babu, J. Cell Commun. Signal., 2010, 8, 31 CrossRef CAS.
- A. H. Boulares, A. G. Yakovlev, V. Ivanova, B. A. Stoica, G. Wang, S. Iyer and M. Smulson, J. Biol. Chem., 1999, 274, 22932–22940 CrossRef CAS.
- T. Suzuki, K. Fujikura, T. Higashiyama and K. Takata, J. Histochem. Cytochem., 1997, 45, 49–53 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 1904396. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9sc06520b |
| This journal is © The Royal Society of Chemistry 2020 |