 Open Access Article
Open Access ArticleTackling the chemical diversity of microbial nonulosonic acids – a universal large-scale survey approach†‡
Hugo B. C.
Kleikamp
a,
Yue Mei
Lin
a,
Duncan G. G.
McMillan
 a,
Jeanine S.
Geelhoed
a,
Jeanine S.
Geelhoed
 b,
Suzanne N. H.
Naus-Wiezer
c,
Peter
van Baarlen
b,
Suzanne N. H.
Naus-Wiezer
c,
Peter
van Baarlen
 d,
Chinmoy
Saha
d,
Chinmoy
Saha
 e,
Rogier
Louwen
e,
Dimitry Y.
Sorokin
af,
Mark C. M.
van Loosdrecht
e,
Rogier
Louwen
e,
Dimitry Y.
Sorokin
af,
Mark C. M.
van Loosdrecht
 a and
Martin
Pabst
a and
Martin
Pabst
 *a
*a
aDepartment of Biotechnology, Delft University of Technology, 2629 HZ Delft, The Netherlands. E-mail: m.pabst@tudelft.nl
bDepartment of Biology, University of Antwerp, 2610 Wilrijk, Belgium
cDepartment of Aquatic Ecology, Netherlands Institute of Ecology, 6708 PB Wageningen, The Netherlands
dHost-Microbe Interactomics Group, Animal Sciences, Wageningen University and Research, Wageningen, 6708 WD, The Netherlands
eErasmus MC, University Medical Center Rotterdam, Department of Medical Microbiology and Infectious Diseases, Rotterdam, 3015 CE, The Netherlands
fWinogradsky Institute of Microbiology, Research Centre of Biotechnology, Russian Academy of Sciences, Moscow, Russia
First published on 24th February 2020
Abstract
Nonulosonic acids, commonly referred to as sialic acids, are a highly important group of nine-carbon sugars common to all domains of life. They all share biosynthetic and structural features, but otherwise display a remarkable chemical diversity. In humans, sialic acids cover all cells which makes them important for processes such as cellular protection, immunity and brain development. On the other hand, sialic acids and other nonulosonic acids have been associated with pathological processes including cancer and viral infections. In prokaryotes, nonulosonic acids are commonly associated with pathogens, which developed through molecular mimicry a strategy to circumvent the host's immune response. However, the remarkably large chemical diversity of prokaryotic nonulosonic acids challenges their discovery, and research on molecular characteristics essential for medical applications are often not feasible. Here, we demonstrate a novel, universal large-scale discovery approach that tackles the unmapped diversity of prokaryotic nonulosonic acids. Thereby, we utilize selective chemical labelling combined with a newly established mass spectrometric all-ion-reaction scanning approach to identify nonulosonic acids and other ulosonic acid-like sugars. In doing so, we provide a first molecular-level comparative study on the frequency and diversity across different phyla. We not only illustrate their surprisingly wide-spread occurrence in non-pathogenic species, but also provide evidence of potential higher carbon variants. Many biomedical studies rely on synthetic routes for sialic acids, which are highly demanding and often of low product yields. Our approach enables large-scale exploration for alternative sources of these highly important compounds.
Introduction
Nonulosonic acids (NulOs, including animal-type sialic acids§) are a class of 9-carbon α-keto acid sugars essential to many cellular processes throughout all domains of life.2–6 The first nonulosonic acid (N-acetyl-neuraminic acid or Neu5Ac) was discovered during the 1940s in salivary mucins (from Greek ‘sialon’), establishing the nowadays commonly used abbreviation ‘sialic acids’.7,8 Since then, the broader group of discovered NulO derivatives expanded to some 100 members.9 Neuraminic acid (Neu) and the unmodified variant keto-deoxy-neuraminic acid (Kdn) are commonly associated with animal tissues. Pseudaminic acid (Pse), its stereoisomers legionaminic (Leg) and acinetaminic acid (Aci) are variants exclusive to prokaryotes.10 Neus are involved in processes such as cell–cell interaction, signalling, adhesion, regulation of protein half-life and mediating an immune response. Similarly, derivatives of Pse/Leg have been found as part of bacterial cell surface structures such as lipopolysaccharides, peptidoglycans or glycoproteins of cell surface layers, adhesins, pili and flagella.11–14 Most uniquely, every type of NulO can undergo diversification at multiple positions, which enables an (potential) enormous chemical diversity.9 Where diversification on animal-type NulOs is mostly limited towards acetylation, glycolylation, methylation, and more rarely, phosphorylation or sulphation, diversifications in prokaryotes have been found to be remarkably innovative. This includes additions of formyl, glyceryl, hydroxybutyryl, lactoyl and glutamyl groups, to just name a few out of many more.9 Moreover, new types of modifications are being discovered continuously. Currently there is no good estimate on the natural boundaries of this process in prokaryotes. Therefore, the driving forces behind the evolution of a certain configuration remain elusive.When only considering a number of some 15 possible modifications—occurring at several diversification points—the number of modified NulO derivatives exceeds several thousands, thereby not considering stereochemistry or linkage isomers (Fig. 1A and B). Furthermore, any changes in the chemical structure are likely to affect physicochemical properties and therefore may interfere with highly important molecular recognition processes.
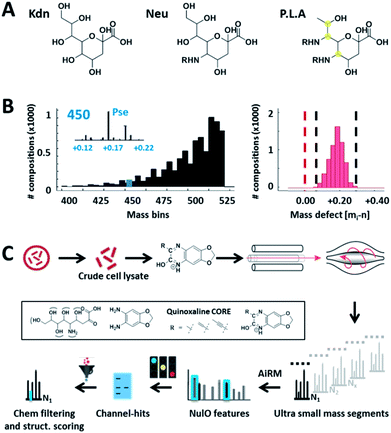 | ||
| Fig. 1 Universal sialic acid large-scale survey approach. (A) outlines the three main NulO core structures known to date: Kdn (deaminoneuraminic acid), Neu (neuraminic acid) and P.L.A = Pse (pseudaminic acid), Leg (legionaminic acid) and Aci (acinetaminic acid). (B) demonstrates the potential diversity generated through (combinations) of (natural) diversifications of the three main NulO core compositions Kdn, Neu and P.L.A (Pse/Leg/Aci). The left graph shows a binned mass histogram (5 Da) of the theoretically possible chemical compositions between 380–520 Da. Already more than 25 different compositions are found within ±0.05 Da of Pse5Ac7Ac. The image on the right shows the mass defect binned for every calculated structure, which all fall within a very narrow window between 0.05–0.25 Da. The mass spectrometric mass defect is defined as the delta of monoisotopic mass and nominal mass. For combinatorial space and abbreviations see ESI-Table, sheet 8.‡ This filters for possible realistic structures during parent NulO annotation. (C) outlines the established large-scale survey pipeline starting from crude cell lysates, α-keto acid specific chemical labelling, small mass channel scanning (2.75 m/z windows, from 380–520 Da), and the automated structural filtering and evidence scoring pipeline to annotate parent NulOs. In summary, channel hits and compositions are discovered based on highly conserved quinoxaline-core fragments together with the diversification-independent carbon chain length and structural features. | ||
Bacterial NulOs have been commonly linked to virulence and pathogenicity, presumed to mimic the host's glycosylation for evading an immune response.15 However, a recent genome level study by Lewis et al. across bacteria and archaea revealed an unexpectedly wide distribution of homologous neuraminic acid biosynthesis (NAB) pathway genes.1 Unfortunately, genome-level studies are only predictive and do not allow for conclusions on active gene products, and the possibility that yet-to-be-discovered NulO pathways have evolved completely independently cannot be ruled out completely. Most importantly however, the above mentioned diversifications processes, which make sialic acids and other NulOs so unique, remain unnoticed because they are not accessible through genome level analysis.16
Furthermore, fully untargeted large-scale molecular level studies on the diversity of natural NulOs is by current state-of-the-art approaches not achievable.
Current detection of sialic acids and other NulOs is achieved by imaging, staining approaches, or by selective fluorescent labelling of the alpha-keto acid group followed by liquid chromatography with or without additional mass-spectrometric detection.6,17,18 Complex lysates, however, produce a large background derived from lower carbon-chain ulosonic acids or common bulk metabolites. Therefore, the classical approach engages pre-fractionation of target conjugates or applies only to samples with low complexity.17,19 Unfortunately, this procedure is laborious and biased, particularly when dealing with uncharacterised (or low abundance) compounds. Alternatively, gas chromatography combined with mass spectrometry has been successfully employed, but this approach also requires pre-fractionation and involves manual data curation of low resolution spectra.20
Over the past decade, new high-resolution mass spectrometers have paved the way to a new era in metabolite analysis. Rapid identification of thousands of compounds in a single experimental measurement is achieved, mostly by using shotgun-type sampling. Due to the stochastic nature of this approach, sub-stoichiometric compounds are overlooked, and frequent co-isolation of closely related metabolite peaks generates hybrid-type and low-informative spectra.
The common route for identifying a given metabolite peak is by matching mass and fragmentation spectra to database library entries.21 More recent advancements include isotope pattern analysis, consider spectral relatedness, and make use of common fragmentation patterns or ultra-high resolution mass spectrometers.21–23 Even so, the virtually large (and yet unmapped) chemical diversification of prokaryotic NulOs, their sub-stoichiometric occurrence, poorly recorded species distribution and difficult to measure nature, hampers the application of shotgun as well as database-matching approaches.
In this study, we describe a nonulosonic acid (NulO) universal screening approach which tackles the (yet unmapped) chemical diversity of prokaryotic nonulosonic acids (NulOs). By doing so, we take advantage of chemical labelling combined with continuous small mass channel mass spectrometric scanning and a systematic matching for NulO unique core features. We applied our approach to a large number of yet unexplored environmental microbes revealing a yet undescribed diversity.
Results and discussions
Given the limitations of current methods, we sought to establish a universal ‘sialo-omics’ approach for untargeted large-scale molecular studies starting from crude cell lysates (Fig. 1C). However, the search for sub-stoichiometric compounds with an unknown chemical composition in complex mixtures compares to the search for a needle in a haystack. To tackle this challenge, we searched for mass spectrometric features which are unique for ulosonic acids but are independent of any diversification. Thereby, we took advantage of the alpha-keto-acid specific labelling reagent 1,2-diamino-4,5-methylene dioxybenzene, which is otherwise utilized for fluorescence detection. However, for our purpose, the probe-of-choice was selected since it significantly shifts the double bond equivalents (+7.5), the mass defect, and since it introduces highly conserved ulosonic acid core fragment features which are different from the bulk cell lysate background. Since lower carbon ulosonic acid molecules are highly common in nature, we aimed to establish additional features which contain structural information. Therefore, we first screened the experimental data acquired in this study for commonly co-occurring fragment compositions using an automated script which confirmed the theoretical fragmentation space (Fig. 2). By doing so, we uncovered universal features which contain the carbon chain length information and which were not influenced by any diversification process (Fig. 2). While the conserved core features are important for identifying ulosonic acids, the carbon chain length features enable differentiation from structurally related eight (KDO) and lower carbon ulosonic acids. However, most importantly, the carbon chain length features provide also information on the degree of saturation and oxidation, and therefore allow differentiation between types of NulOs, such as Neu/Kdn-type and Pse/Leg-type ulosonic acids (Fig. 2A–C). Where this serves as tool to identify derivatives related to known NulOs, the systematic extrapolation of those features makes it universal for the discovery of completely novel types of NulOs. In addition, to maintain high sensitivity for sub-stoichiometric signals, we established a high-resolution mass spectrometric fragmentation of highly small mass segments covering the mass range established by our initial theoretical calculations. The very small mass segments generate hybrid fragmentation spectra of only medium complexity. However, in ‘all-ion-type’ fragmentation techniques, a large number of parent ions contribute to a single spectrum, and the link between parent ions and their product fragment peaks is very difficult to reconstruct.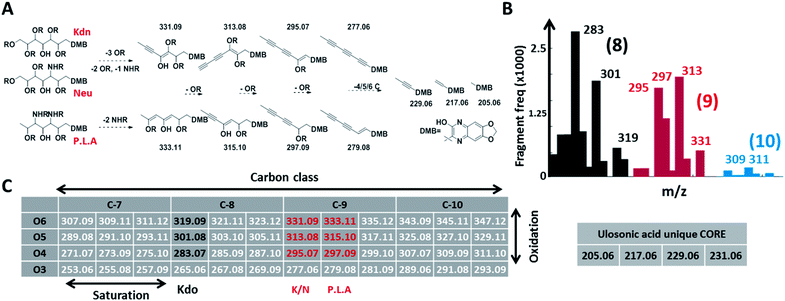 | ||
| Fig. 2 Conserved and diversification independent feature space for nonulosonic acids (NulOs). (A and C) outline the empirical fragmentation space systematically spanning the oxidation and saturation space of ulosonic acids with different carbon chain lengths. More specifically, (A) outlines (from left to right) one universal fragmentation route for the three main sialic acid compositions (Kdn, Neu and Pse/Leg/Aci). (B) shows the frequency (>50 counts, see ESI-Table sheet 4‡) of binned fragments from ulosonic acid related compounds observed in the large-scale study. This confirmed the theoretically established fragment feature space for different ulosonic acid classes. The high frequency for 283 and 301 (8 C, black bars) and 295 and 297 (9 C, red bars) correspond to fragments from KDO, Neu/Kdn and P.L.A respectively. Furthermore, this provided mass spectrometric evidence for potential higher carbon chain ulosonic acids (blue). (C) The alpha-keto acid specific chemical labelling introduced a significantly altered mass defect and fragment features such as the (quinoxaline-based) ulosonic acid unique core fragments and carbon chain length features. Deviations in the degree of saturation and oxidation allow further differentiation between types of NulOs and the identification of completely new structures. | ||
Nevertheless, identification of the above described conserved ulosonic acid features in mass segments provides very sensitive global maps (channel hits) of the NulO landscape (Fig. 3A and B, ESI-Table, sheet 3‡). The channel hit generation has been automated by a Matlab script, which loops through high-resolution spectra of complete runs within only a few minutes. Nevertheless, molecular studies typically require the exact chemical identity and nature of the NulO. Therefore, we established a chemical filtering and structural evidence scoring pipeline to identify the parent NulO from positively assigned mass segments. In most cases, the small mass segments showed sufficiently low complexity to unambiguously filter for a NulO candidate. To enable higher-throughput studies, the complete process from channel-hit generation to chemical filtering and structural evidence scoring and reporting was automated in a pipeline (ESI‡). Where this provides a very efficient solution to screen classes of NulOs based on the backbone, the closer investigation of fragmentation spectra provides hints on the nature of the modification(s) present.
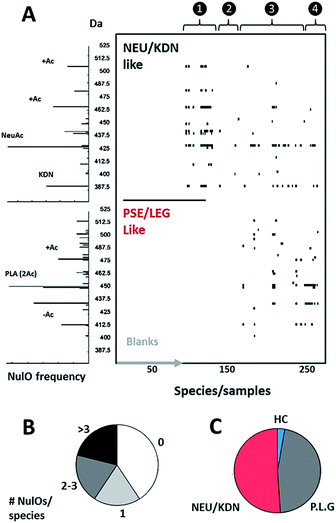 | ||
| Fig. 3 NulO large-scale survey represented as channel hit-map and chemical diversity graphs. (A) shows a hit-map for channels with detected nonulosonic acid (NulO) candidates in samples (duplicates shown separate for samples) individual injections are plotted along the x-axis. The y-axis is binned into 2.5 Da mass channel units. The upper map is filtered for Kdn and Neu-like species and the lower map is filtered for Pse/Leg-type hits. The same data are visualised as binned (0.1 m/z) spectra on the left. The numbers on the top of the hit-map represent: (1) references, spiked E. coli and animal cell samples; (2) plants, fungi and algae; (3) Archaea, alpha-, beta-, gamma-, delta- and epsilon proteobacteria, actinobacteria, firmicutes, cyanobacteria; (4) enrichment/communities. (B) Sialic acids were found at high frequency throughout environmental samples and almost half of all samples showed two or more types per species. (C) Kdn/Neu-type sugars were observed with comparable frequency to bacterial-type NulOs. While the former were found in eukaryotes and prokaryotes, the bacterial types were exclusive to prokaryotes. Potential higher carbon chain ulosonic acids (blue) were only detected in rare cases. (see also ESI-Table, sheets 1–3‡). | ||
A molecular level survey across eukaryotes and prokaryotes
Using our newly established pipeline, we performed a first molecular level survey on prokaryotic NulOs where we included a range of well-characterised commercial standards, animal cells, model plants and algae (Fig. 3A and B, ESI-Table, sheet 3‡). To qualify our approach, we first analysed the well-characterised animal-derived cell materials such as CHO and HeLa cells (cultivated on 2 growth media). Apart from low quantities of Kdn, Neu5Ac was identified in all samples as the predominant species, which appeared further diversified through glycolylation, methylation and higher degrees of acetylation. HeLa cells cultivated on foetal calf serum showed a large increase of the N-glycolylated variant compared to serum-free conditions, which presumably results from uptake and internal release in lysosomes, followed by metabolic incorporation. The well-established model plants Arabidopsis thaliana and Nicotiana benthamiana, as well as baker's yeast, contained lower carbon chain ulosonic acids (not shown), but as expected, did not show any observable nonulosonic acid analogues (9-carbon). Noteworthy, in our experiments, one plant showed misleading artefacts that were difficult to distinguish from Kdn by mass only (‘pseudo-Kdn’, ESI, Section I‡). Such or related other misleading candidates may have fuelled historical debates about the presence of (active) NulO biosynthesis pathways in plants. Furthermore, we included selected algae, which are increasingly considered as production systems in biotechnology. In contrast to a recent report which speculated about the presence of sialylated glycans in Chlamydomonas reinhardtii, nonulosonic acids were not identified in either of the strains analysed. On the other hand, we could confirm the existence of (genuine) Kdn and associated derivatives in some microalgae. In animal tissues, sialic acids provide a barrier which protects from predators. However, some pathogens, including viruses, have adapted to this strategy and developed specific proteins that bind to sialic acids, to use them as an entry point for successful colonisation of the host. Some other pathogens appeared to have adapted to cleave terminal sialic acids and utilise those as a source of energy. Furthermore, many pathogenic or symbiotic bacteria display sialic acids or other NulOs on their own surface, which is regarded as molecular mimicry of the host sialylation. This strategy has been found effective in delaying, or abolishing the host's immune response.24 Hence, we included in our survey a well-studied pathogenic Campylobacter jejuni strain that is able to utilise bacterial-type and animal-type NulOs. As expected, we observed several heavily expressed hits for both animal- and bacterial-type NulOs. The majority of Neu variants are expected to derive from medium uptake (and potential incorporation), but the strain further produced at least 3 distinct bacterial-type NulO variants (ESI-Table, sheet 3‡). Moreover, it is suggested that bacteria apply cell surface diversification strategies (such as through NulOs) to evade recognition by bacteriophages.25 Bacteriophages are viruses that infect and replicate within prokaryotes and attach specifically through bacterial cell surface carbohydrates. For that reason, we selected a range of environmental, non-pathogenic species, spanning most phyla described to date. Surprisingly, the NulOs uncovered from those microbes were rich in diversity and frequency – nonulosonic acid peaks were detected in more than half of the measured species: two-thirds showed more than one hit, and one-half showed three or more NulO hits. The relative sialic acid abundances compared to the mammalian cell lines (normalised to dry weight biomass starting material) was in many cases only slightly lower compared to quantities observed in animal cells. The more rigid bacterial cell walls may, however, make a fair comparison not particularly meaningful (ESI-Table, sheets 1–3‡). The authors also want to emphasize that incubation with acetic acid at higher temperature is required to ensure sufficient release of different types of NulOs from various biological matrices. On the other hand, the release and labelling procedures may introduce additional degradation variants, which impacts on quantitation and comparability. Furthermore, we aimed to ensure excess of labelling reagent over the biological matrix to ensure efficient NulO labelling. An additional reagent mass channel however might be highly useful to monitor the labelling process when dealing with unknown matrices. Nevertheless, as expected, there was a strong trend towards Pse/Leg-type NulOs, but also Kdn and Neu-type acids were found with high frequency. When animal derived media supplements were used (such as casein tryptone) the detected NeuAc may originate from uptake or scavenging rather from neo-biosynthesis. In some cases, additional Neu derivatives were observed indicating that these did result from further cellular processing. Several environmental microbes, isolated from distinctive niches, displayed a remarkable diversity of NulOs, including Streptomyces coelicolor, which is seemingly specialized in abundant Kdn and modified variants thereof, as well as Paracoccus denitrificans harbouring at least 4–5 different bacterial-type sialic acid derivatives (ESI-Table, sheets 1–3‡). The authors also would like to point out, that the investigated microbes were analysed from one particular culturing condition, and the extend of variability between conditions and stimuli remains to be explored.In support of the observed frequency, we analysed the proteomes of selected strains for homologues of known NAB pathways. This largely confirmed observed trends from the molecular identifications, but also underlined the notion that gene and protein sequence databases are still incomplete und selection of comparator sequences is crucial. For example, the homologous genes for Kdn pathways in algae were not identified, although their existence had been confirmed previously in an independent study26 (Fig. 4).
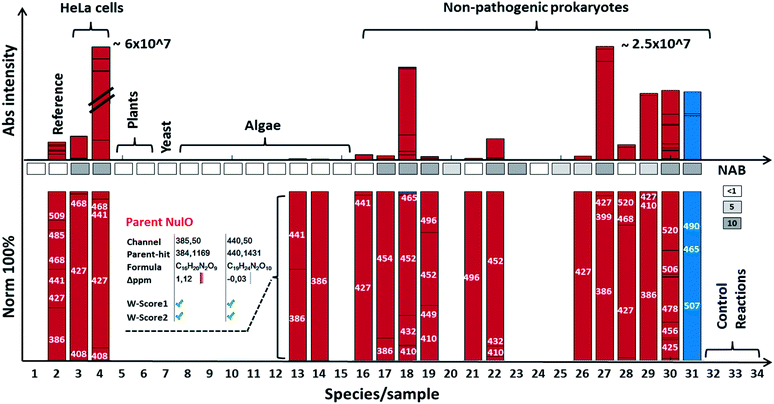 | ||
| Fig. 4 NulO channel-hits and compositions for selected species. The bar graph highlights results for selected species from the NulO survey. The lower bar graphs are normalised to 100%. Individual bars represent a channel hit (annotated by the channel centre mass). The bar graph size correlates to the MS1 intensity of the aligned parent mass peaks. The following samples are shown from left to right: (1) DMB blank incubation; (2) reference sialic acids spiked into E coli, (3) free grown HeLa serum free grown, (4) HeLa grown on serum, (5) A. thaliana, (6) N. benthamiana, (7) Saccharomyces cerevisiae; (8) Chlamydomonas reinhardtii-s1; (9) Chlamydomonas reinhardtii-s2; (10) Cryptomonas; (11) Asterionella formosa, (12) Diatoma, (13) Alexandrium ostenfeldii, (14) Cricophaera carterae, (15) Galdiera sulpharia, (16) Haloferax volcanii, (17) Geitlerinema n., (18) Methanogenic archaeon, (19) Scalindua, (20) Nitrospira moscoviensis; (21) Clostridium S. (22) Thiothrix enr. (23) Pseudomonas putida (24) E. coli BW25113 (25) Myxococcus xanthus, (26) Acidovorax avenae, (27) Streptomyces coelicolor, (28) Mycobacterium smegmatis, (29) Arthrospira platensis, (30) Paracoccus denitrificans, (31) Magnetospirillum gryphiswaldense (MG) acid hydrolysed + labelled, (32) MG non-hydrolysed + labelled, (33) MG acid hydrolysed + unlabelled, and (34) MG non-hydrolysed + unlabelled. The insert for Alexandrium ostenfeldii (13) exemplifies the parent NulO deconvolution from the channel-hit using the chemical and structural evidence scoring and filtering. A comparative gene level NAB pathway search is illustrated in the box between bars (white box = no significant, light grey = <5 and dark grey >5 homologue hits). All survey channel-hits and NulO parent annotations are summarised in the ESI-Table, sheets 1–3,‡ and the NAB pathway search results are summarised in ESI-Table sheet 7.‡ | ||
Carbon chain length diversification: from octulosonic, nonulsonic and higher carbon ulosonic acids
All described nonulosonic acids (9 C) are products of evolutionarily related synthase families. A condensation of a hexose derivative with phosphoenolpyruvate (PEP) takes place by a mechanism comparable to the reaction of the Neu5Ac synthase in humans. Alternative (reversible) routes exist through aldolase activities, which utilize pyruvate instead of PEP. A highly comparable condensation reaction produces other lower carbon ulosonic acids such as heptulosonic (7 C) or octulosonic (8 C) acids. Heptulosonic acids are intermediates of the shikimic-acid pathway (deoxy-D-arabino-heptulosonate phosphate, DHAP), an essential part of the neo-biosynthesis of amino acids. Octulosonic acids (keto-deoxy-octulosonate, KDO) are essential building blocks of cell walls of many prokaryotes and plants. Indeed, the large majority of ulosonic acids (UAs) observed in our survey were of modified 7-, 8-, or 9-carbon nature. However, surprisingly, some rare cases evidenced the existence of higher carbon ulosonic acids (potential 10 C, or other by means of MS indistinguishable variants), such as in Magnetospirillum gryphiswaldense, a Gram-negative magnetotactic alpha-Proteobacterium, capable of orienting and navigating along geomagnetic field lines27 (Fig. 2B). Additional experiments on the unlabelled and fractionated NulO further support the presence of a triply acetylated higher carbon ulosonic acid (ESI, Section H‡). The same species was detected in lower abundance in Methanococcus. In support of this finding, Hsu et al., reported on directed evolution of sialic acid aldolases with low specificity, that accept a variety of (different) monosaccharide substrates for condensation with pyruvate to uncommon NulOs.28 Furthermore, Jacobs et al. demonstrated the efficient incorporation of mannosamine analogues with alternative N-acyl groups.29 The promiscuousness of biosynthetic routes together with substrate availability may be frequent mediators for species/niche diversification processes of sialic acids and other NulOs.Conclusions
In summary, we developed the first universal large-scale survey approach that tackles the enormous chemical diversity of prokaryotic NulOs by using chemical labelling, segmented mass spectrometric scanning and structural evidence filtering and scoring. Comparative large-scale studies will advance the understanding of important molecular level processes relevant to medical applications. Furthermore, we provide a first molecular-level comparative study on selected species representing a large number of the prokaryote phyla described to date, with a focus on the largely unexplored environmental niches. In this study, we observed NulOs at a high frequency and diversity, which challenges the current model of evolution and utilisation of sialic acids and other NulOs being predominantly driven by mechanistic advantages during host–pathogen interactions. This supports a broader utilisation of these compounds, such as for diversification of cell surface attachment points and protection from bacteriophages, which are a major driver of bacterial evolution in the laboratory and in nature.30 Furthermore, our large-scale data evidence potential higher carbon ulosonic acids, which would further expand the boarders on the chemical diversity of natural NulOs. At the same time, we emphasise, although the approach presented here serves as an ideal tool to survey for known and completely new UAs, the latter require orthogonal analysis, such as by NMR, to completely unravel and confirm their chemical structures.Nevertheless, chemical synthesis of sialic acid and other NulO derivatives is highly challenging and often only achieved at low yields.31 The exploration of non-pathogenic microbes for novel biosynthetic routes gives access to new NulO derivatives that have thus far been difficult to produce by chemical synthesis.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful to all colleagues who shared material or provided valuable discussions and advice which are Dennis Claessen, Gijs Kuenen, Sonja Albers, Sebastian Lückner, Jules Beekwilder, Paula van den Brink, Diana Machado de Sousa, Jules Rombouts, Karel Oliveira, Florence Lip, Eleni Vasilaku, Danny de Graeff, Marta Ceruti, Laura Valk, Jure Zoplasa, Michel Mulder, David Calderon, Anna-Maria Zetty, Lina Bird, Denis Grouzdev, Guylaine Nuijten, Michele Laureni, and to the SIAM consortium grant for funding.Notes and references
- A. L. Lewis, N. Desa, E. E. Hansen, Y. A. Knirel, J. I. Gordon, P. Gagneux, V. Nizet and A. Varki, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 13552–13557 CrossRef CAS PubMed
.
- L. Warren, Comp. Biochem. Physiol., 1963, 10, 153–171 CAS
.
- T. Angata and A. Varki, Chem. Rev., 2002, 102, 439–469 CrossRef CAS PubMed
.
-
A. Varki, R. L. Schnaar and R. Schauer, in Essentials of Glycobiology, ed. A. Varki, R. D. Cummings, J. D. Esko, P. Stanley, G. W. Hart, M. Aebi, A. G. Darvill, T. Kinoshita, N. H. Packer, J. H. Prestegard, R. L. Schnaar and P. H. Seeberger, Cold Spring Harbor, NY, 2015, DOI:10.1101/glycobiology.3e.015, pp. 179–195
.
- M. Cohen and A. Varki, OMICS, 2010, 14, 455–464 CrossRef CAS PubMed
.
- D. D. Park, G. Xu, M. Wong, C. Phoomak, M. Liu, N. E. Haigh, S. Wongkham, P. Yang, E. Maverakis and C. B. Lebrilla, Chem. Sci., 2018, 9, 6271–6285 RSC
.
- G. Blix, L. Svennerholm and I. Werner, Acta Chem. Scand., 1952, 6, 358–362 CrossRef CAS
.
- G. Blix, Hoppe-Seyler's Z. Physiol. Chem., 1936, 240, 43–54 CrossRef CAS
.
- R. Schauer and J. P. Kamerling, Adv. Carbohydr. Chem. Biochem., 2018, 75, 1–213 CrossRef PubMed
.
- M. J. Morrison and B. Imperiali, Biochemistry, 2014, 53, 624–638 CrossRef CAS PubMed
.
- Y. A. Knirel, A. S. Shashkov, Y. E. Tsvetkov, P. E. Jansson and U. Zahringer, Adv. Carbohydr. Chem. Biochem., 2003, 58, 371–417 CrossRef CAS PubMed
.
- J. J. Kenyon, A. M. Marzaioli, C. De Castro and R. M. Hall, Glycobiology, 2015, 25, 644–654 CrossRef CAS PubMed
.
- E. Morello, A. Mallet, Y. Konto-Ghiorghi, T. Chaze, M. Y. Mistou, G. Oliva, L. Oliveira, A. M. Di Guilmi, P. Trieu-Cuot and S. Dramsi, PLoS One, 2015, 10, e0138103 CrossRef PubMed
.
- A. I. M. Salah Ud-Din and A. Roujeinikova, Cell. Mol. Life Sci., 2018, 75, 1163–1178 CrossRef CAS
.
- A. F. Carlin, S. Uchiyama, Y. C. Chang, A. L. Lewis, V. Nizet and A. Varki, Blood, 2009, 113, 3333–3336 CrossRef CAS PubMed
.
- T. Angata and A. Varki, Chem. Rev., 2002, 102, 439–470 CrossRef CAS PubMed
.
- A. Klein, S. Diaz, I. Ferreira, G. Lamblin, P. Roussel and A. E. Manzi, Glycobiology, 1997, 7, 421–432 CrossRef CAS PubMed
.
- S. Hara, M. Yamaguchi, Y. Takemori, M. Nakamura and Y. Ohkura, J. Chromatogr., 1986, 377, 111–119 CrossRef CAS PubMed
.
- R. Zeleny, D. Kolarich, R. Strasser and F. Altmann, Planta, 2006, 224, 222–227 CrossRef CAS PubMed
.
- J.-P. Zanetta, A. Pons, M. Iwersen, C. Mariller, Y. Leroy, P. Timmerman and R. Schauer, Glycobiology, 2001, 11, 663–676 CrossRef CAS PubMed
.
- A. C. Schrimpe-Rutledge, S. G. Codreanu, S. D. Sherrod and J. A. McLean, J. Am. Soc. Mass Spectrom., 2016, 27, 1897–1905 CrossRef CAS PubMed
.
- J. J. J. van Der Hooft, J. Wandy, M. P. Barrett, K. E. V. Burgess and S. Rogers, Proc. Natl. Acad. Sci., 2016, 113, 13738–13743 CrossRef PubMed
.
- D. C. P. Lozano, R. Gavard, J. P. Arenas-Diaz, M. J. Thomas, D. D. Stranz, E. Mejía-Ospino, A. Guzman, S. E. Spencer, D. Rossell and M. P. Barrow, Chem. Sci., 2019, 10, 6966–6978 RSC
.
- G. Sekot, G. Posch, P. Messner, M. Matejka, X. Rausch-Fan, O. Andrukhov and C. Schäffer, J. Dent. Res., 2011, 90, 109–114 CrossRef CAS PubMed
.
- R. Louwen, D. Horst-Kreft, A. De Boer, L. Van Der Graaf, G. de Knegt, M. Hamersma, A. Heikema, A. Timms, B. Jacobs and J. Wagenaar, Eur. J. Clin. Microbiol. Infect. Dis., 2013, 32, 207–226 CrossRef CAS PubMed
.
- B. A. Wagstaff, M. Rejzek and R. A. Field, J. Biol. Chem., 2018, 293, 16277–16290 CrossRef CAS PubMed
.
- K. H. Schleifer, D. Schüler, S. Spring, M. Weizenegger, R. Amann, W. Ludwig and M. Köhler, Syst. Appl. Microbiol., 1991, 14, 379–385 CrossRef
.
- C.-C. Hsu, Z. Hong, M. Wada, D. Franke and C.-H. Wong, Proc. Natl. Acad. Sci., 2005, 102, 9122–9126 CrossRef CAS PubMed
.
- C. L. Jacobs, S. Goon, K. J. Yarema, S. Hinderlich, H. C. Hang, D. H. Chai and C. R. Bertozzi, Biochemistry, 2001, 40, 12864–12874 CrossRef CAS PubMed
.
- B. Koskella and M. A. Brockhurst, FEMS Microbiol. Rev., 2014, 38, 916–931 CrossRef CAS PubMed
.
- S. Matthies, P. Stallforth and P. H. Seeberger, J. Am. Chem. Soc., 2015, 137, 2848–2851 CrossRef CAS PubMed
.
Footnotes |
| † The Matlab pipeline is freely available upon request. Mass spectrometry raw data (in generic.mzXML format) are openly accessible through https://researchdata.4tu.nl/en/. DOI: 10.4121/uuid:d2f8970b-f992-43ba-a244-8e916fe3814d. |
| ‡ Electronic supplementary information (ESI) available: Experimental methods, supporting figures, and supplemental tables. See DOI: 10.1039/c9sc06406k |
| § Following Lewis et al., (2009)1 we restrict the terminology ‘sialic acid’ (Sia) to Neu and Kdn derivatives and use ‘nonulosonic acids’ (NulOs) otherwise. |
| This journal is © The Royal Society of Chemistry 2020 |
