Open Access Article
Open Access ArticleTemplates direct the sequence-specific anchoring of the C-terminus of peptido RNAs†
Biswarup
Jash
 and
Clemens
Richert
and
Clemens
Richert
 *
*
Institute of Organic Chemistry, University of Stuttgart, 70569 Stuttgart, Germany. E-mail: lehrstuhl-2@oc.uni-stuttgart.de; Fax: +49 711 608 64321; Tel: +49 711 608 64311
First published on 2nd March 2020
Abstract
When amino acids and ribonucleotides react in aqueous condensation buffer, they form peptido RNA with a phosphoramidate bond between the N-terminus of the peptide and the 5′-terminal phosphate of a ribonucleotide. If peptido RNA was the product of spontaneous reactions of amino acids and nucleotides, there must have been a transition to peptidyl tRNAs, where the C-terminus of the peptide is ester-linked to the 2′,3′-terminus of an oligonucleotide. Here we report how short peptido RNAs react with the 3′-terminus of oligodeoxynucleotides, templated by RNA strands. In our model system, the rate and yield of the anchoring of the C-terminus of the dipeptido dinucleotides to an amino group was found to depend on the sequence of the peptide, the 5′-terminal nucleotide of the dinucleotide and the RNA template. In all cases tested, highest yields were found for dinucleotides hybridizing next to the primer terminus. For the most reactive species, GlyPro-AA, anchoring yields ranged from 8–99%, depending on the template. When LeuLeu-AA, PhePhe-AA and GlyGly-AA were allowed to compete for anchoring on 3′-UUC-5′ as templating sequence, they gave a product ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6, and this selectivity was almost independent of the terminal base of the primer. Our results show the control that a simple duplex context has over the covalent anchoring of peptido RNAs at a position known from peptidyl tRNAs. Processes of this type may have bridged the gap between untemplated condensation reactions and the highly specific processes of ribosomal protein synthesis.
6, and this selectivity was almost independent of the terminal base of the primer. Our results show the control that a simple duplex context has over the covalent anchoring of peptido RNAs at a position known from peptidyl tRNAs. Processes of this type may have bridged the gap between untemplated condensation reactions and the highly specific processes of ribosomal protein synthesis.
Introduction
The synthesis of polypeptides in the cell is an RNA-directed process.1 The sequence information encoded in a gene is transcribed to a messenger RNA, which is then translated by the ribosomal machinery. It is not clear how this system arose from simple precursors that were formed spontaneously during an earlier phase of evolution. There have been detailed proposals for how the first nucleic acids2 and the first oligopeptides may have been formed,3,4 but how this system then evolved into the RNA-controlled processes found in extant biology is far from certain.5There is reason to believe that amino acids were formed early.6 If so, ribonucleotides or RNA strands must have found a way to capture them, and to induce controlled and eventually highly sequence specific peptide syntheses.7 One of the most likely avenues to RNA-controlled peptide synthesis is the formation of covalently linked molecular species that contain an RNA portion and a peptide portion.8 With a covalent bond, it is easier to steer a molecule toward a desired reaction. Covalent linking is not only found in ribosomal polypeptide synthesis, but also in non-ribosomal peptide synthesis.9,10 Either form of chain assembly uses an ester or thioester link to the carboxy group to hold on to the chain.
While the individual stages of the emergence of the translational apparatus remain challenging to elucidate,5 there has been progress in studies focused on the formation of covalently linked peptide-RNA species. Classical work on the products of the mixed condensation of amino acids and ribonucleotides dates back to the 1950s,11 and aspects of prebiotic chemistry of such condensation reactions have been explored in the past.12,13 Pascal, Sutherland and colleagues have focused on cyclic intermediates in pathways to ribonucleotide-driven peptide syntheses,14–16 Yarus and coworkers showed that short phenylalanine oligomers can form via ester chemistry, when the amino acid is preactivated as a mixed anhydride with adenosine monophosphate (AMP), in reactions of a tetranucleotide substrate catalyzed by a pentanucleotide ribozyme.17,18
We have recently reported the spontaneous formation of ‘peptido RNA’ from a mixture of ribonucleotides and amino acids in aqueous condensation buffer.19 Peptido RNAs are covalently linked molecules, where the N-terminus of the peptide chain is connected to the 5′-phosphate. Growth of the peptide chain is much faster than the background oligomerization when the first amino acid has been captured by the 5′-phosphate,20 and the aqueous condensation buffer chemistry is compatible with RNA chain growth and genetic copying.21 Further, the condensation processes are compatible with all of the twenty proteinogenic amino acids and all four canonical ribonucleotides.22 But, the processes leading to the formation of peptido RNA are not template-directed, and it was not clear whether a template would be able to direct them.
There is some literature precedence for template-directed reactions involving amino acids. Transamidation involving 2′/3′-linked amino acids and 5′-phosphoimidazolides templated by a homopolymer has been studied in the 1970s,23 and doubly linked amino acids were recently shown to be more stable hydrolytically than their aminoacyl counterparts.24 More distantly related coupling reactions have been performed with the goal of reading out sequence information of a template,25 but how RNA templates may have directed the anchoring of the carboxy-terminus of N-terminally linked peptides to the 3′-terminus of oligonucleotides sequence-specifically was unclear.
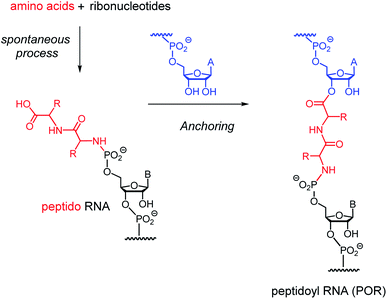
Because the carboxy terminus of the peptide chain is left free in peptido RNA, it may get covalently anchored at the 2′,3′-terminus of another RNA strand, as shown for a dipeptido RNA in Fig. 1. We propose the term ‘peptidoyl RNA’ (POR) for the doubly linked species produced in this reaction. The ending “oyl” is used for acyl groups, as in “benzoyl” or “pivaloyl”, but the name also contains the “peptido” term used to identify peptide chains N-linked as phosphoramidates. The anchoring step may be controlled by a template, as either of the strands involved can engage in base pairing with a complementary sequence. If so, this would be a step toward RNA-controlled polypeptide biosynthesis. Further, if the reaction was strongly dependent on the sequence of the peptide, it could help to select specific peptides from statistical oligomers, again a favorable trait for a biosyntheses. Such control would be most attractive, it was to set in early, before the peptide chain had grown ‘out of control’ and would be a plausible step toward the peptidyl tRNAs found in present-day translation.
Here we report anchoring of dipeptido RNAs at the 3′-terminus of a 3′-aminoterminal oligodeoxynucleotide primer, directed by RNA templates. We established reaction conditions for the anchoring reaction, and we screened different peptide lengths and sequences for their ability to undergo rapid and high-yielding anchoring reactions. The results show that anchoring is readily templated by oligoribonucleotides, with significant sequence selectivity, indicating that the very first dipeptides produced in spontaneous, ribonucleotide-induced processes could have been selected by reactions controlled by RNA.
Results
Experimental system and initial screen
Fig. 2A and Scheme 1 show the reactions studied in this work. They are directed by an RNA template (1cuuu–1cuuc), to which a primer (2a–g) is hybridized. In order to obtain stable amide bonds, rather than labile 2′,3′-esters, 3′-aminoterminal oligodeoxynucleotides were used as primers. To the primer-template duplexes, individual peptido dinucleotides (3gly–9) were added as reaction partners in a condensation buffer21 containing EDC as water-soluble carbodiimide. A dinucleotide was chosen because this is a long enough sequence to hybridize to specific positions of a template. Dissociation constants of dinucleotide-primer: template complexes are in the single-digit millimolar range.26 With low millimolar affinity, only low concentrations of peptido RNAs are needed to achieve sufficient occupancy at the coupling site. Further, since there are 16 different dinucleotides in a four-letter genetic system, this length is sufficient to encode 16 amino acids or dipeptides, which is not far from the number of proteinogenic amino acids. Dipeptido dinucleotides are species that are formed in significant quantities in spontaneous condensation mixtures,19 so we consider them plausible species in a prebiotic setting involving the condensation of amino acids and (oligo)ribonucleotides.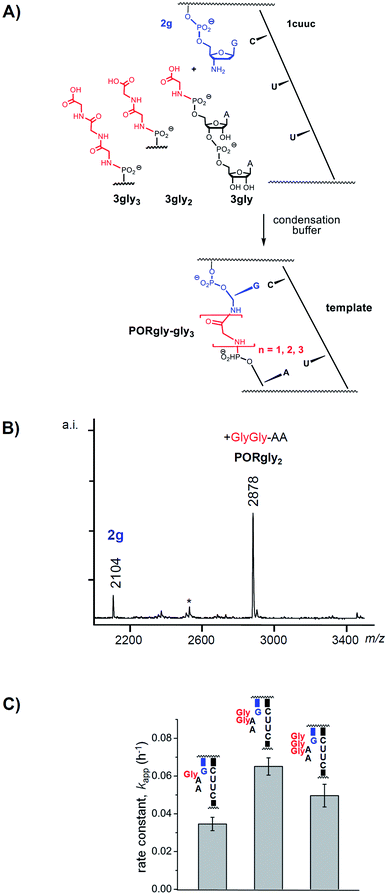 | ||
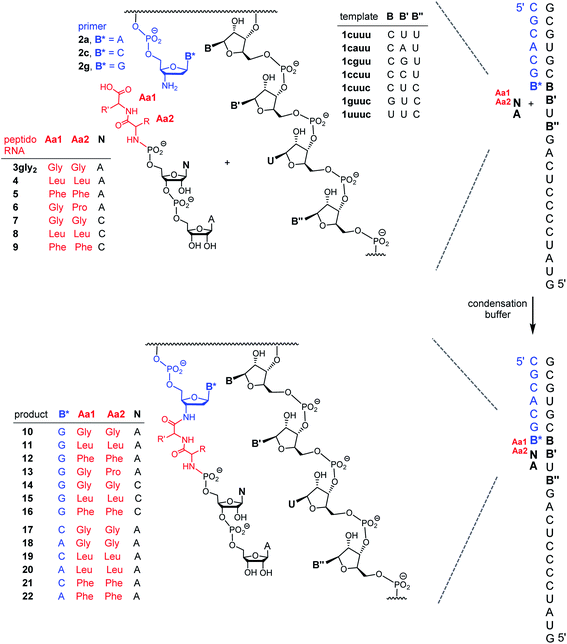
| Fig. 2 Effect of the length of the peptide chain on the rate of formation of PORs. (A) Reaction scheme, (B) representative MALDI-TOF mass spectrum, and (C) apparent rate constants of the anchoring reactions with the peptido dinucleotides with one, two or three glycyl residues. Error bars are standard errors, as calculated by OriginPro, version 8 for monoexponential fits to the experimental data. Replicates of the assay with 3gly2 gave kapp = 0.065 ± 0.002 h−1 (mean ± one SD), with an even smaller error than the one shown here (0.065 ± 0.005), which was derived from a single assay. The full sequences of primer and template are given in Scheme 1. See Fig. S15–S24 and Table S1 of the ESI for additional details.† | ||
 | ||
| Scheme 1 Anchoring reactions: sequences and reactions employed in this study. Conditions: 20 μM primer and RNA template, 200 μM peptido dimer, 50 mM MOPS, 0.2 M EDC, 0.2 M NaCl, 0 °C, pH 7.4. | ||
First, we screened for peptide chain lengths that give rapid coupling to the primer terminus. Fig. 2A shows the three dinucleotide species employed in this phase of the study. They feature one (3gly), two (3gly2) or three glycine residues (3gly3) at the 5′-phosphate terminus. The assay conditions were chosen to allow for rapid anchoring, with a low level of side reactions or decomposition. An aqueous medium containing 50 mM MOPS buffer, pH 7.4, with 200 mM EDC and 200 mM NaCl fulfilled those criteria. Template and primer were 20 μM, and the dinucleotide species was 200 μM in the mixture. Assays were run at 0 °C, as in our earlier study on template-directed reactions in condensation buffer.21 Product formation was monitored by MALDI-TOF mass spectrometry under conditions that allow for quantitative detection.27
All three dinucleotides (3gly–3gly3) showed coupling in the anchoring assays, as evidenced by MALDI-TOF spectra of the reaction mixtures (compare Fig. 2B). Monoexponential functions fit the kinetic data for the different couplings well, with R2 values of 0.989 or better (Table S1 and Fig. S15–S24, ESI†). Such fits provided the apparent rate constants (kapp) shown in Fig. 2C. The highest rate constant was measured for the dipeptido dinucleotide 3gly2, with significantly lower rates for the species with one or three amino acid residues. This made us choose dipeptido dinucleotides for the subsequent study on specificity of the anchoring reaction. The choice of dipeptido chains also appeared reasonable from a prebiotic standpoint because this is the first product of the peptido RNA pathway28 that contains a peptidic amide bond. If the anchoring of such dipeptido species occured with specificity, longer RNA-bound peptides may have been synthesized in RNA-directed processes, even if the formation of the initial dipeptide was driven by intrinsic reactivity of amino acids.22
Effect of template and peptide sequence
To study specificity, the dipeptides to be tested were selected to cover a range of shapes and polarities. The sequences employed are shown in Scheme 1. They include GlyGly as α-unsubstituted species with a flexible chain, LeuLeu as a conformationally more restricted chain with aliphatic side chains, PhePhe as a dipeptide with aromatic side chains that can engage in π-stacking, and GlyPro as a species with a conformationally restricted N-terminal amino acid residue. Synthetic details for 4–9, as well as analytical data for the peptido dinucleotides are presented in the ESI.† A representative calibration plot for the mass spectrometric detection of 13 is provided in Fig. S14 of the ESI.†The right-hand side of Scheme 1 lists the different products of the anchoring reactions with dipeptido dinucleotides (10–22). The results of reactions directed by different templates shed light on the effect of positioning of the dipeptido dinucleotides relative to the primer terminus. Upon hybridization, the AA or CA dinucleotide pairs adjacent to the 3′-terminus of the primer or farther away from it. This results in different spatial proximity and structural arrangements for the anchoring reaction.
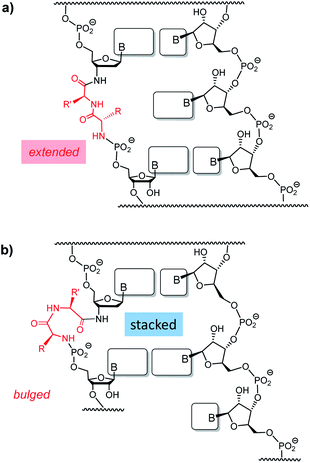
Initially, we assumed that the desired anchoring reaction would be favored if there was an unpaired nucleotide in the RNA template opposite the dipeptide chain of the peptido RNA, leaving nucleotide B′ without base pairing partner. Molecular modeling of the unrestrained conformation of the dipeptide chain in a peptidoyl dinucleotide gave a distance of 6.3 Å between the N-terminus of the dipeptide and the ester group linking it to the 3′-position of the 5′-terminal guanosine (Fig. 4). This is close to the length taken up by a nucleotide residue in the backbone of RNA in A-type duplexes (5.9 Å),29 corroborating this view.
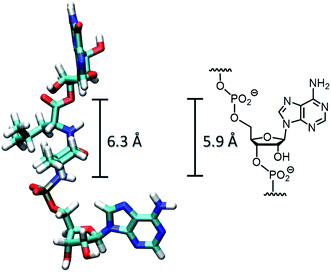 | ||
| Fig. 4 Comparison of the distance between the termini of a dipeptide chain in the peptidoyl species 5′-G-LeuLeu-A-3′, with coordinates generated in Chem3D Pro, 14.0 and visualized in VMD,30 and the phosphate-to-phosphate distance between nucleotide residues known for an A-type duplex.29 | ||
The experiment revealed something different. The yield of the anchoring was low when a “bridging” nucleotide in the template was facing the dipeptide (templates 1cauu, 1cguu, 1ccuu with AA dinucleotides in 3–6). However, when we used RNA templates that lack a non-base pairing, bridging nucleotide opposite the dipeptido chains, so that two base-paired duplex regions are adjacent to each other, yields were much higher for the anchoring reactions (Table 1, and spectra underlying this data shown in Chapter 5 of the ESI†).
| Template | Dipeptido dinucleotide | |||
|---|---|---|---|---|
| GlyGly-AA (3gly2) | LeuLeu-AA (4) | PhePhe-AA (5) | GlyPro-AA (6) | |
| a As detected by MALDI-TOF MS; a correction factor of 1.1 was applied to compensate for differences in desorption/ionization; see the ESI for details. b Conditions: 20 μM DNA 2g, 20 μM template, 200 μM dipeptido dinucleotide, 50 mM MOPS buffer, pH 7.4, 0.2 M EDC, 0.2 M NaCl, 0 °C. c Mean ± one standard deviation from four independent assays. d Quantitative conversion. | ||||
| — | 4 | 4 | 5 | 23 |
| 1cauu | 7 | 3 | 9 | 21 |
| 1cguu | 5 | <1 | 8 | 8 |
| 1ccuu | <1 | <1 | 7 | 10 |
| 1cuuu | 8 | 20 | 16 | 66 |
| 1cuuc | 28 | 82 ± 2c | 56 ± 3c | Quantd |
Overall, the results in Table 1 show several effects that are of interest. The first is that a template can both increase and decrease the yield of the anchoring reaction over that of the background reaction. If the dinucleotide portion is well positioned, the yield increases, if it is positioned unfavorably, it decreases. This demonstrates the control the template exerts over the reaction. Secondly, even if the templating sequence offers a pairing site next to the terminus of the primer:template duplex region, the nucleobase downstream of the templating dinucleotide significantly affects yields. With 1cuuu, which allows “sliding” of the AA dimer along the template, the yields are significantly lower for all peptido RNAs tested than with 1cuuc, which has a mismatched C adjacent to the UU template dimer. Thirdly, there is a significant effect of peptide sequence. For example, GlyPro-AA (6) shows higher reactivity than any of the remaining peptido RNAs tested. Whereas the latter give similar yields in the untemplated reaction (4–5% after 6 h), there is more conversion with 6 in all reaction systems tested. This indicates that the structure of its dipeptido chain improves reactivity, probably due to a preorganizing effect of the prolyl residue. Similarly, anchoring assays with 4 showed the strongest directing effect of the template, i.e. the largest difference between template-mediated (with 1cuuc) and control assays in the formation of PORs, probably because the sterically demanding side chains limit the conformations available to the backbone to reach the transition states that the template allows.
Often, selectivity in biochemical reactions depends on kinetics. To study selectivity in more detail, we next performed a series of kinetic studies with the different dipeptido dinucleotides and their cognate templates. For peptido RNAs 3gly2–6 this was 1cuuc, and for peptido RNAs 7–9, this was 1cguu. In either case, sliding of the dinucleotide along the template would result in a mismatch (A:C or C:U), making it unlikely that alternative hybridization arrangements contribute significantly to the template effect. Again, monoexponential fits gave good agreement with the experimental data, allowing us to extract apparent rate constants and the maximum yield at infinite reaction time (Table 2, Fig. 2C, and S17–S36 in the ESI† for spectra and plots). The data shows how strongly the structure of the dipeptide chain and the 5′-terminal nucleotide of the peptido RNA affect rates. The difference in the half-life times between the fastest and the slowest reactions was 28-fold. Interestingly, the diglycyl RNAs 3gly2 and 7 gave the slowest rates (0.065 h−1 and 0.031 h−1), even though their peptide chains are the most flexible ones. Among the remaining dipeptides, LeuLeu and PhePhe gave comparable results, but GlyPro again stuck out as the most reactive species. Even more unexpected was the effect of the nucleotide with the phosphoramidate bond to the N-terminus of the peptide. Anchoring was several-fold faster and yields were higher when this 5′-terminal nucleotide was A (3gly2–6) than when it was C (7–9). This again shows how delicately the local structure affects the yield of the anchoring reaction, a trait that is favorable for specificity in RNA-directed syntheses of polypeptides.
| Template | Peptido dinucleotide | k (h−1) | t 1/2 (h) | y max (%) |
|---|---|---|---|---|
| a Fits to: y = ymax (1 − e−kt), where y is the yield, k is the rate constant, and t1/2 is the half-life time for formation of the POR. Error values are those obtained by fitting with OriginPro, version 8. See Fig. S17–S36 of the ESI for spectra and kinetic plots. b Conditions: 20 μM DNA primer, 20 μM template, 200 μM dipeptido dinucleotide, 50 mM MOPS buffer, pH 7.4, 0.2 M EDC, 0.2 M NaCl at 0 °C. | ||||
| 1cuuc | 4 | 0.39 ± 0.03 | 1.8 | 93 ± 3 |
| 1cuuc | 5 | 0.24 ± 0.02 | 2.9 | 80 ± 1 |
| 1cuuc | 6 | 0.91 ± 0.06 | 0.76 | 86 ± 2 |
| 1cguu | 7 | 0.031 ± 0.005 | 22 | 30 ± 2 |
| 1cguu | 8 | 0.087 ± 0.007 | 8.0 | 48 ± 1 |
| 1cguu | 9 | 0.087 ± 0.003 | 8.0 | 50 ± 0.4 |
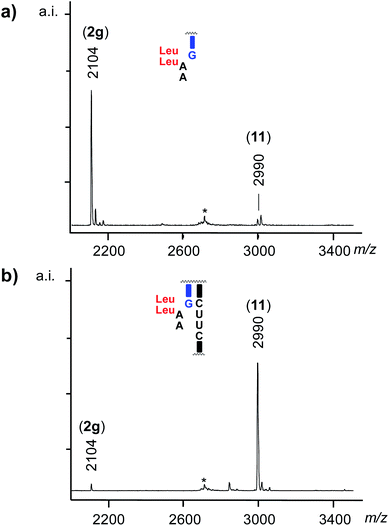
In the final stage of our study, we measured the selectivity of anchoring in assays in which three different dipeptido dinucleotides compete directly with each other, in one aqueous solution. The total concentration of dipeptido dinucleotides was kept the same as in the single-compound assays, but each of the competing species was now present at the same, lower concentration. Results from such “molecular competitions” in the presence of different templates are shown in Fig. 5. Additional data can be found in Fig. S43–S46 and Tables S8–S10 of the ESI.†Fig. 5a shows a MALDI-TOF mass spectrum of the assay with template 1cuuc and 3gly2/4/5, where POR 11 can be identified as the clear winner of the competition. Fig. 5b shows the product distribution obtained in the presence of different templates (or lack thereof) for the same mix of peptido RNAs. Again, the templates with the UU binding site for AA separated from the primer terminus by a bridging nucleotide gave yields lower than the control reaction. In the assays with 1cuuu and 1cuuc, LeuLeu-AA (4) was the clear winner. The highest selectivity was observed in the presence of template 1cuuc, with a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 between the different PORs. This demonstrates that the selectivity observed in individual assays is also realized in the direct competition that mimics the situation in a primordial cell beginning to evolve specific peptide synthesis.
6 between the different PORs. This demonstrates that the selectivity observed in individual assays is also realized in the direct competition that mimics the situation in a primordial cell beginning to evolve specific peptide synthesis.
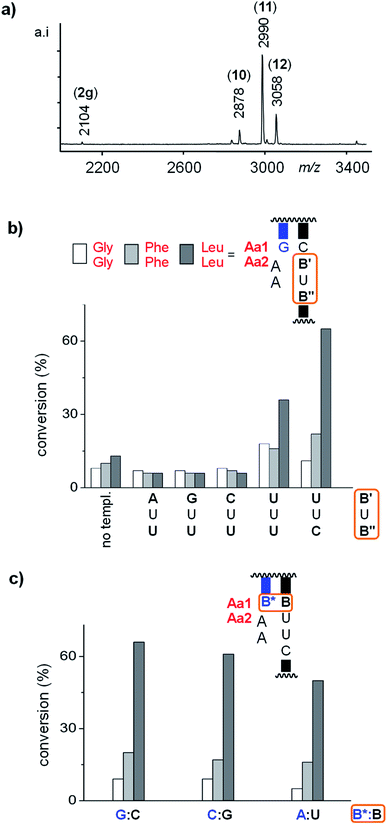 | ||
| Fig. 5 Results from competition assays with dipeptido dinucleotides 3gly2/4/5 (67 μM each) reacting with primer 2g (20 μM), as directed by different templates (20 μM). (a) MALDI-TOF mass spectrum from assay with 1cuuc after 2 d. (b) Extent of anchoring after 2 d for the templating sequences listed below each cluster of bar graphs. The left-most cluster is for an assay without template; (c) effect of the base pair at the terminus of the primer:template duplex, as determined by primer conversion after 1 d. Conditions: 50 mM MOPS buffer, pH 7.4, 0.2 M EDC, 0.2 M NaCl, 0 °C. See footnote (c) to Table 1 for the reproducibility of yields. The results from a read-out at an earlier time point (18 h) are show in Fig. S46 of the ESI.† | ||
We then asked whether the control the templating sequence has over anchoring is unaffected by the duplex upstream of the peptido RNA binding site. For this, we varied the terminal base pair of the duplex, using two other primer-template combinations that possess a C:G or A:U base pair at what is the 3′-terminus of the primer. Fig. 5c shows that there is a small effect on the yield of the anchoring reaction and that the selectivity is essentially unchanged. This suggests that the control that the templating sequence exerts over which dipeptide gets covalently linked to the 3′-position of the primer is almost unaffected by the sequence context. Again, this is a favorable trait for a molecular system that is evolving encoded synthesis, as selectivity should be controlled by the “codon” alone, not any neighboring bases, in order to allow for the encoded synthesis of any given polypeptide sequence.
Discussion
These results shed a light on the structural requirements for coupling the carboxy terminus of a N-terminally bound peptide to the 3′-terminus of a neighboring strand held on an RNA template. One aspect that is worth noting is the positioning of the peptide between the two termini, that is between the 5′-terminus of the peptido RNA and the 3′-nucleophile of the strand to be acylated. An amino acid residue has three atoms in the backbone of a peptide, whereas a nucleotide residue has six. So, one may assume that a bridging ribonucleotide residue in the template would be the best arrangement for placing carboxy group and nucleophile into close proximity (Fig. 3 and 6a). This arrangement would be produced when the dipeptido dinucleotide was in an extended conformation with the dinucleotide hybridized to templates, leaving a non-pairing base between the primer terminus and the binding site for the dinucleotide, such as in 4 hybridizing to 2g:1cauu.The alternative structural arrangement has continuously stacked base pairs, with the dinucleotide:template duplex next to the primer:template miniduplex and the dipeptide in a loop that bulges out of the backbone (Fig. 6b). This is the ‘no gap’ template/dinucleotide assembly of 1cuuu or 1cuuc and 3gly2–6. While this arrangement is less appealing in two-dimensional drawings, the experimental results with templates that either possess or lack an unpaired template base at the coupling site provide a clear picture: the continuously base stacked arrangement is more favorable for anchoring the C-terminus. In other words, base stacking “wins” over nucleotide-amino acid interactions. This is significant for understanding molecular recognition of peptido RNAs, RNA-templated peptide synthesis, and perhaps the origin of the genetic code.31,32 Amino acid-nucleic acid interactions have been the subject of intense research33,34 and the preorienting effect that RNA duplex regions can (or cannot) provide may prove a critical aspect.
A second aspect has to do with which parts of the primer-template-dinucleotide assembly significantly affects the rate of the anchoring reaction and its yield. The evidence from the screen performed by us suggests that while the primer terminus has little effect, the neighboring base pair between 5′-terminal base of the dinucleotide and the first base of the template downstream from the primer binding region has a strong effect (AA 3–6versus CA dinucleotides of 7–9). Further, the nucleobase immediately downstream of the templating region of the template does have such an effect, as evidenced by the differences in rate observed for reactions on 1cuuu and 1cuuc (Table 2), again confirming just how significant the role of the template is.
The third aspect is the dependence of the anchoring on the structure of the dipeptide chain. The three different dipeptides of the peptido dinucleotides (3gly2, 4, 5) differ only in their side chains, and little change in chemical reactivity of the terminal carboxy group should result from these differences. Yet, the rate of their anchoring is quite different (Table 1 and Fig. 5). If the bulged-out arrangement of Fig. 6b is the relevant structure for anchoring, it is not surprising that the effect of side chains on the ability to adopt a loop-like conformation that is productive for coupling can be quite significant.
The enhanced reactivity of 6 is thus most probably the result of the structural effect the of proline residue. Incidentally, it has been reported that proline residues adapt a strikingly different conformation in the ribosomal A site, compared to other amino acids.35 We suspect that proline produces a unique orientation in the dipeptide moiety that facilitates the anchoring of the C-terminus of the glycine in GlyPro-AA (6). Likewise, the low reactivity and reduced template dependence in anchoring 3gly2 can be attributed to the flexibility of its peptide backbone, allowing the carboxy-terminus to adopt many unproductive conformations.
Conclusions
In conclusion, we report that the anchoring of the C-terminus of dipeptido dinucleotides at the 3′-terminus of a primer is a reaction that can readily be controlled by an RNA template. The RNA sequence can modulate the rate of the reaction almost 30-fold for the cases studied, and can make it faster or slower than the background reaction. Base stacking between the terminus of the primer:template duplex and the miniduplex between peptido dinucleotide and templating dinucleotide appear to dominate over peptide-RNA interactions. In turn, the conformation forced on the dipeptide by the side chains of the amino acid residues appears to dominate over other effects, such as molecular interactions via π-stacking, when the selection of dipeptides occurs in the molecular competition between different peptido RNAs on a given template. Neighboring nucleotides have a significant effect in the single-stranded, templating sequence, but not in the duplex at the terminus of primer and template.Overall, the data provide a fascinating glimpse at RNA-controlled molecular recognition beyond that of free amino acids and nucleotides, but not yet in the confinement of a ribosomal reaction center with the proofreading capabilities of a fully developed translational machinery. Once anchored at the 3′-terminus of an RNA, peptidyl transfer through transacylation may occur, as found in translation. While these insights were gained for a model system, we note that this is a close model system, with the RNA template expected to force an A-type structure on the template:primer duplex and the amino group at the primer terminus being isoelectronic and largely isosteric to the hydroxy group that covalently anchors the peptide chain in peptidyl tRNAs in the transacylation-based chemistry of translation.
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors thank Dr Henri-Philippe Mattelaer for sharing results from an exploratory study, Dr Eric Kervio for discussions and help with data analysis, Oliver Bernhard for expert technical assistance, Sebastian Motsch for sharing synthetic protocols, and Daniel Pfeffer for help with kinetics. This work was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – grant RI 1063/16-1 and project-ID 364653263 – TRR 235.References
- M. M. Yusupov, G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. Cate and H. F. Noller, Science, 2001, 292, 883–896 CrossRef CAS PubMed.
- J. P. Ferris, A. R. Hill, R. H. Liu and L. E. Orgel, Nature, 1996, 381, 59–61 CrossRef CAS PubMed.
- G. Danger, R. Plasson and R. Pascal, Chem. Soc. Rev., 2012, 41, 5416–5429 RSC.
- E. T. Parker, M. Zhou, A. S. Burton, D. P. Glavin, J. P. Dworkin, R. Krishnamurthy, F. M. Fernandez and J. L. Bada, Angew. Chem., Int. Ed., 2014, 53, 8132–8136 CrossRef CAS PubMed.
- D. W. Morgens, J. Mol. Evol., 2013, 77, 185–196 CrossRef CAS PubMed.
- K. R-Mirazo, C. Briones and A. de la Escosura, Chem. Rev., 2014, 114, 285–366 CrossRef PubMed.
- R. Pascal, L. Boiteau and A. Commeyras, Top. Curr. Chem., 2005, 259, 69–122 CrossRef CAS.
- J. D. Sutherland and J. M. Blackburn, Chem. Biol., 1997, 4, 481–488 CrossRef CAS PubMed.
- S. A. Sieber and M. A. Marahiel, J. Bacteriol., 2003, 185, 7036–7043 CrossRef CAS PubMed.
- S. A. Sieber and M. A. Marahiel, Chem. Rev., 2005, 105, 715–738 CrossRef CAS PubMed.
- P. Berg, J. Biol. Chem., 1958, 233, 608–611 CAS.
- M. Paecht-Horowitz, J. Berger and A. Katchalsky, Nature, 1970, 228, 636–639 CrossRef CAS PubMed.
- J. R. Hawker and J. Oro, J. Mol. Evol., 1981, 17, 285–294 CrossRef CAS PubMed.
- J.-P. Biron, A. L. Parkes, R. Pascal and J. D. Sutherland, Angew. Chem., Int. Ed., 2005, 44, 6731–6734 CrossRef CAS PubMed.
- D. Beaufils, G. Danger, L. Boiteau, J.-C. Rossi and R. Pascal, Chem. Commun., 2014, 50, 3100–3102 RSC.
- Z. Liu, L. Rigger, J.-C. Rossi, J. D. Sutherland and R. Pascal, Chem.–Eur. J., 2016, 22, 14940–14949 CrossRef CAS PubMed.
- M. T. Rebecca, V. C. Nataliya and M. Yarus, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 4585–4589 CrossRef PubMed.
- R. M. Turk, M. Illangasekare and M. Yarus, J. Am. Chem. Soc., 2011, 133, 6044–6050 CrossRef CAS PubMed.
- M. Jauker, H. Griesser and C. Richert, Angew. Chem., Int. Ed., 2015, 54, 14564–14569 CrossRef CAS PubMed.
- H. Griesser, P. Tremmel, E. Kervio, C. Pfeffer, U. E. Steiner and C. Richert, Angew. Chem., Int. Ed., 2017, 56, 1219–1223 CrossRef CAS PubMed.
- M. Jauker, H. Griesser and C. Richert, Angew. Chem., Int. Ed., 2015, 54, 14559–14563 CrossRef CAS PubMed.
- H. Griesser, M. Bechthold, P. Tremmel, E. Kervio and C. Richert, Angew. Chem., Int. Ed., 2017, 56, 1224–1228 CrossRef CAS PubMed.
- J. L. Shim, R. Lohrmann and L. E. Orgel, J. Am. Chem. Soc., 1974, 96, 5283–5284 CrossRef CAS PubMed.
- Z. Liu, G. Ajram, J.-C. Rossi and R. Pascal, J. Mol. Evol., 2019, 87, 83–92 CrossRef CAS PubMed.
- J. Michaelis, A. Maruyama and O. Seitz, Chem. Commun., 2013, 49, 618–620 RSC.
- M. Sosson, D. Pfeffer and C. Richert, Nucleic Acids Res., 2019, 47, 3836–3845 CrossRef CAS PubMed.
- D. Sarracino and C. Richert, Bioorg. Med. Chem. Lett., 1996, 6, 2543–2548 CrossRef CAS.
- P. Tremmel, H. Griesser, U. E. Steiner and C. Richert, Angew. Chem., Int. Ed., 2019, 58, 13087–13092 CrossRef CAS PubMed.
- W. Saenger, Principles of Nucleic Acid Structure, Springer Publishers, New York, 1984 Search PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- S. Zhang and M. Egli, Origins Life, 1994, 24, 495–505 CrossRef CAS PubMed.
- S. Zhang and M. Egli, Complexity, 1995, 1, 49–56 CrossRef.
- J. C. Lacey and D. W. Mullins, Origins Life, 1983, 13, 3–42 CrossRef CAS PubMed.
- H. Mizutani and C. Ponnamperuma, Origins Life Evol. Biospheres, 1977, 8, 183–219 CrossRef CAS PubMed.
- S. Melnikov, J. Mailliot, L. Rigger, S. Neuner, B.-S. Shin, G. Yusupova, T. E. Dever, R. Micura and M. Yusupov, EMBO Rep., 2016, 12, 1776–1784 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and methods, supplementary text and data. See DOI: 10.1039/c9sc05958j |
| This journal is © The Royal Society of Chemistry 2020 |