Open Access Article
Open Access ArticleThe importance of Asn52 in the structure–function relationship of human cytochrome c†
Dan Lou‡
a,
Xi-Chun Liu‡a,
Xiao-Juan Wanga,
Shu-Qin Gaob,
Ge-Bo Wenb and
Ying-Wu Lin *ab
*ab
aSchool of Chemistry and Chemical Engineering, University of South China, Hengyang 421001, China. E-mail: ywlin@usc.edu.cn
bLaboratory of Protein Structure and Function, University of South China Medical School, Hengyang 421001, China
First published on 18th December 2020
Abstract
The function of the highly conserved residue Asn52 in human cytochrome c (H-Cyt c) is not fully understood. Herein, we show that the naturally occurring variant N52S H-Cyt c has a perturbed secondary structure, with a small fraction of high-spin species. Remarkably, it exhibits an enhanced peroxidase activity by 3–8-fold at neutral pH, as well as self-oxidation in reaction with H2O2. This study suggests that the H-bond network mediated by Asn52 is essential to suppress the apoptotic activity of H-Cyt c under physiological conditions.
Cytochrome c (Cyt c) is an important member of a widely distributed group of redox metalloproteins.1–4 Human mitochondrial cytochrome c (H-Cyt c) is a small heme protein (104 residues) with a covalently attached heme group via two thioester bonds. The central iron is coordinated by His18 on the proximal side and Met80 on the distal side. The main function of H-Cyt c is to transfer electrons between Cyt c and Cyt c oxidase in the respiratory chain.5 In addition to playing an indispensable role in substance metabolism and energy conversion, Cyt c is engaged in an early stage of the intrinsic apoptosis pathway.6,7 Cyt c exhibits peroxidase activity when it interacts with the lipid cardiolipin (CL) in the mitochondrial inner membrane, resulting in the oxygenation of CL and the release of Cyt c into the cytoplasm, which subsequently induces the formation of the apoptosome complex and ultimately leads to cell death.8,9
The obtain peroxidase activity, Cyt c undergoes a conformational change. Typically, the distal ligand Met80 dissociates from the hexa-coordinated heme iron while the proximal ligand His18 is essential to maintain protein structure. A large number of studies have shown that perturbation of the heme pocket can modulate the stability and activity by mutating the key residues in Cyt c.10 The peroxidase activity of Cyt c can be enhanced under alkaline condition by conformational change called “alkaline transition” with a typical pKa between 9 and 9.5.4,11 Protein crystallography studies show a hydroxide or the Nε atom of Lys73/79 residue in the Ω loop 70–85 can replace the distal Met80 ligand of the ferric Cyt c.12,13 In recent years, it has been discovered that alkaline transition can occur at physiological pH with certain post-translation modifications in Cyt c, such as Tyr74 nitration and Tyr48 phosphorylation.14–16 Bowler and co-workers also reported that when Cyt c interacts with detergents, it loses Met80 ligation and forms a well-defined pocket for binding of hydrocarbons, which is a structural mimic for Cyt c mediated cardiolipin oxidation.17 These conformers with enhanced peroxidase activity are useful models for studying the conformational change of Cyt c on the onset of instinct apoptotic process.
Up to date, four pathogenic mutations (G41S,18 Y48H,19 A51V,20 and Lys100del21) have been reported to associate with a low platelet count disease, an autosomal dominant thrombocytopenia 4 (THC4). Three of the four variants are located on the Ω loop 40–57 and show enhanced peroxidase activity.22–27 In 2016, a massive sequencing study identified a N52S mutation in the H-Cyt c.4,28 Although the disease-relevance of the N52S missense mutation is still unclear, the Asn52 residue is one of most conserved residues in eukaryotic Cyt c, as revealed by sequence alignment and structure analysis.10,29 Protein crystallography studies showed that Asn52 forms H-bonds with heme propionate-7 and a structural water molecule, which further interacts with Tyr67 and Thr78 in the ferric state (Fig. 1).30,31 Recent NMR characterization revealed that Asn52 also forms dynamic H-bond with heme propionate-6 in the ferrous state.32 These observations indicate that Asn52 is critical for protein stability, folding and the crosstalk between the Ω loops 40–57 and 70–85. In addition, Asn52 is proposed to interact with CL and involved in the formation of the hydrocarbon binding pocket.33–35 Moreover, the studies of the iso-1-cytochrome c suggest Asn52 mutation affects the electron transfer.36
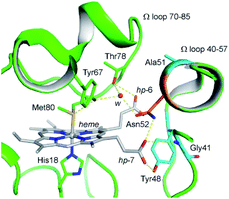 | ||
| Fig. 1 Close up view of the H-bond networks around Asn52 and the heme crevice in the ferric WT H-Cyt c (PDB code 3ZCF31). Asn52 was shown as orange stick. The heme iron and heme cofactor were shown as grey ball and sticks, respectively. H-bonds were shown as yellow dash lines. A structural water molecule (w) was shown as red sphere. The heme propionates 6 and 7 were referred to as hp-6 and hp-7. The residues Gly41, Tyr48, and Ala51 linked to the disease THC4 were shown as cyan sticks. | ||
In the present study, we constructed both N52S and N52A variants to ascertain the importance of highly conserved residue Asn52 in H-Cyt c. The protein was expressed in the E. coli BL21(DE3) stain and the purified protein was certificated by electrospray ionization mass spectrometer (ESI-MS). It showed a molecular weight of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 207.0 Da and 12
207.0 Da and 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 191.0 Da for the ferric N52S and N52A H-Cyt c, respectively, which is in agreement with the calculated mass (N52S, 12
191.0 Da for the ferric N52S and N52A H-Cyt c, respectively, which is in agreement with the calculated mass (N52S, 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 206.9 Da, and N52A, 12
206.9 Da, and N52A, 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 190.9 Da, Fig. S1†). The UV-Vis spectra of both Ser52 and Ala52 variants are highly similar with that of the wild-type (WT) H-Cyt c (Fig. S2†). The absorptions at 415, 521 and 550 nm in ferrous state increased slightly in intensity compared to those of WT H-Cyt c. These spectral features indicate that the first coordination sphere of the heme iron is roughly preserved in the N52S and N52A variants at pH 7.0, where Met80 still persists as a distal ligand.
190.9 Da, Fig. S1†). The UV-Vis spectra of both Ser52 and Ala52 variants are highly similar with that of the wild-type (WT) H-Cyt c (Fig. S2†). The absorptions at 415, 521 and 550 nm in ferrous state increased slightly in intensity compared to those of WT H-Cyt c. These spectral features indicate that the first coordination sphere of the heme iron is roughly preserved in the N52S and N52A variants at pH 7.0, where Met80 still persists as a distal ligand.
In order to investigate whether there were any secondary structure changes induced by the Asn-to-Ser/Ala substitution in position 52, we performed circular dichroism (CD) spectroscopy studies. The far-UV CD spectra of N52S, N52A, and WT H-Cyt c all showed a characteristic α-helical structure with a positive peak at 190 nm and broad negative peaks at 208 nm and 222 nm (Fig. 2A). Meanwhile, the ellipticity (θ) value from 200 to 240 nm increased for the N52S and N52A variants, which suggests that the protein might loss some of the α-helical structure as a result of Asn-to-Ser/Ala substitution, likely due to local conformational changes. Moreover, the CD spectra in the Soret band region changed as well (Fig. 2B). The N52S and N52A variants showed a stronger positive peak at 404 nm and a weaker negative peak at 418 nm compared to those of WT H-Cyt c. These data show that the Ser/Ala substitution in position 52 alters the geometry/strength of the heme coordination state, as well as the local micro-environment, which is presumably due to the perturbation of the H-bond network mediated by Asn52.
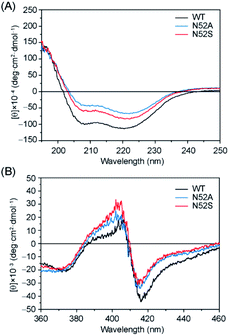 | ||
| Fig. 2 Comparison of the CD spectra in the (A) far-UV region and (B) visible region. Spectra were colored as black, blue, and red for the ferric WT, N52A, and N52S H-Cyt c, respectively. | ||
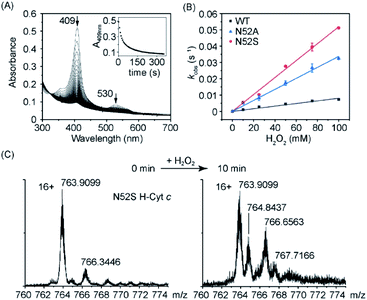
To further reveal the heme micro-environment as a result of conformational changes in the N52S and N52A variants, we performed electron paramagnetic resonance (EPR) studies. The spin states of the ferric heme iron were monitored at 20 K (Fig. 3). Control experiment showed that WT H-Cyt c was totally in a low-spin state (gz = 3.09, gy = 2.17) at neutral pH due to the coordination of Met80/His18. The N52A variant was also primarily in a low-spin state with typical EPR signals (gz = 3.06 and gy = 2.20). In contrast to WT and N52A H-Cyt c, a small fraction of high-spin species (g⊥ = 5.95) was observed in EPR spectra for N52S H-Cyt c, which is similar to those observed for other heme proteins, such as myoglobin and neuroglobin variants.37–39 Note that this characteristic could not be detected using UV-Vis spectroscopy. Instead, the EPR technology revealed a small fraction of N52S H-Cyt c in high-spin state, which is likely due to the dissociation of Met80 from the heme iron as a result of structural perturbation by the N52S mutation.
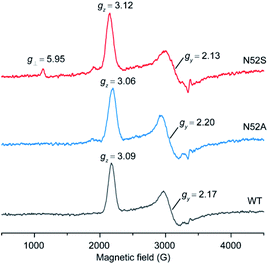 | ||
| Fig. 3 EPR spectra of ferric WT (black), N52A (blue), and N52S (red) H-Cyt c (0.25 mM), collected at 20 K, 2 mW power and 9.4 GHz. | ||
As aforementioned, high-spin species of Cyt c typically exhibits peroxidase activity. According to EPR observation, the N52S H-Cyt c might exhibit an enhanced peroxidase activity compared to the WT and N52A H-Cyt c. To confirm this speculation, we performed a series of kinetic studies to measure the peroxidase activity. First, the reaction with hydrogen peroxide was carried out, in the absence of substrates H-Cyt c catalyzes the oxidation of itself, resulting in heme degradation, as indicated by the decrease of the Soret band over time (Fig. 4A and S3†). The heme degradation rates (kobs) were obtained at various the concentrations of H2O2. The rate constants were calculated to be 0.52 ± 0.01 M−1 s−1 for N52S variant and 0.34 ± 0.01 M−1 s−1 for N52A variant, respectively, by linear fitting of the data (Fig. 4B), which is ∼6 times and ∼4 times faster than that of WT H-Cyt c (0.08 ± 0.01 M−1 s−1). Moreover, the self-oxidation of N52S H-Cyt c was monitored by using mass spectrometry (Fig. 4C).40 The mass spectrum of N52S variant showed that a series of oxidized products (+16 and +32 Da) were formed with incubation of H2O2 (0.1 mM) for 10 minutes, which was more obvious than those for N52A and WT H-Cyt c (Fig. S4†). These observations indicate that the N52S variant is more sensitive to H2O2, even with a low concentration.
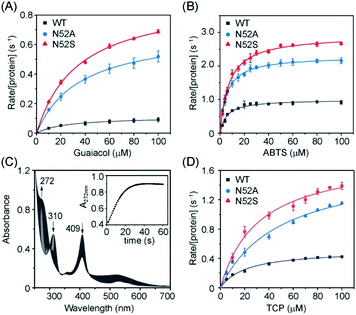
In addition to the heme degradation assay, we measured the peroxidase activities of H-Cyt c using guaiacol and 2,2′-azinobis-3-ethylbenzthiazoline-6-sulphonate (ABTS) as typical substrates and H2O2 as an oxidant (Fig. 5A and B). The optimal concentration of H2O2 was found to be 200 mM and 100 mM for guaiacol and ABTS, respectively, based on the catalysis reaction at varies concentrations of H2O2 (Fig. S5†). With the optimal concentration of H2O2, the catalytic reactions at different substrate concentrations were performed (Fig. S6 and S7†), and the kinetic parameters were obtained by fitting data to the Mickaelis–Menten equation. As shown in Table 1, the Km values were not largely affected by the N52A and N52S mutations. Meanwhile, these mutations lead to a significant increase in catalytic rate. The kcat values of N52S and N52A variants for guaiacol oxidation are ∼8-fold and ∼6-fold higher than that of WT H-Cyt c, respectively. For ABTS oxidation, the kcat values of N52S and N52A variants are ∼3-fold and ∼2-fold higher than that of WT H-Cyt c, respectively.
| Protein | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) |
|---|---|---|---|
| Guaiacol | |||
| WT Cyt c | 0.11 ± 0.1 | 24.94 ± 2.13 | 4400 |
| N52A Cyt c | 0.7 ± 0.1 | 35.87 ± 1.64 | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 500 |
| N52S Cyt c | 0.92 ± 0.2 | 34.22 ± 1.74 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| ABTS | |||
| WT Cyt c | 1.00 ± 0.02 | 5.38 ± 0.53 | 185![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900 |
| N52A Cyt c | 2.29 ± 0.02 | 5.30 ± 0.21 | 432![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
| N52S Cyt c | 2.92 ± 0.03 | 7.34 ± 0.32 | 397![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| 2,4,6-TCP | |||
| WT Cyt c | 0.52 ± 0.02 | 21.52 ± 2.21 | 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 200 |
| N52A Cyt c | 1.70 ± 0.12 | 49.88 ± 7.97 | 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
| N52S Cyt c | 1.8 ± 0.08 | 31.58 ± 4.05 | 57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Moreover, we determined the dehaloperoxidase activity using 2,4,6-trichlorophenol (TCP) as a typical substrate. As shown in Fig. 5C, N52S H-Cyt c was able to catalyze the dehalogenation of TCP. Upon addition of H2O2, the absorbance of TCP at 310 nm decreased rapidly and a characteristic absorption peak (272 nm) of 2,6-dichloroquinone (DCQ) was produced. Steady-state kinetic studies further showed that in dehalogenation of TCP, the kcat values of N52S and N52A H-Cyt c were similar, with both ∼3-fold higher than that of WT H-Cyt c (Table 1). These results thus confirm that the N52S and N52A variants exhibit enhanced peroxidase activities.
It should be noted that there did not seem to be strong correlation between the catalytic properties and the high-spin population of Cyt c, since although only N52S Cyt c showed a small population of high-spin species in EPR spectroscopy (Fig. 3), rate-enhancements were also observed for N52A Cyt c (Table 1). To further provide evidence for the lability of Met80, we performed kinetic studies of azide binding.24 As shown in Fig. S8,† the observed rate constants were calculated to 8.4 ± 1.5 × 10−3 s−1 and 4.8 ± 0.6 × 10−3 s−1 for N52S and N52A variants, respectively, which is 2–3-fold higher than that of WT H-Cyt c (2.6 ± 0.9 × 10−3 s−1), indicating the weakness of Met80 binding in both variants, especially for N52S Cyt c.
In conclusion, our study on the naturally occurring N52S variant of H-Cyt c provided valuable insight into the structure–function of the highly conserved residue of Asn52. The mutation of Asn to Ser in position 52 enhanced the peroxidase activity of H-Cyt c by 3–8-fold, depending on the substrates, which was comparable to those disease-relevant mutations (G41S, Y48H, and A51V) located on the Ω loop 40–57.27 In particular, a high-spin species was detected for N52S H-Cyt c variant at neutral pH by EPR studies. Structural perturbation of the Ω loops 40–57 and 70–85 might weak the coordination bond of Fe(III)-Met80 and subsequently promote the formation of the high-spin species. Therefore, this study suggests that a conserved proper residue of Asn in position 52 is essential to suppress the peroxidase activity of H-Cyt c.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21977042). EPR spectra were collected on the steady high magnetic field facilities, high magnetic field laboratory (Hefei, China).References
- I. Bertini, G. Cavallaro and A. Rosato, Chem. Rev., 2006, 106, 90–115 Search PubMed.
- L. J. Smith, A. Kahraman and J. M. Thornton, Proteins, 2010, 78, 2349–2368 Search PubMed.
- J. Liu, S. Chakraborty, P. Hosseinzadeh, Y. Yu, S. L. Tian, I. Petrik, A. Bhagi and Y. Lu, Chem. Rev., 2014, 114, 4366–4469 Search PubMed.
- D. Alvarez-Paggi, L. Hannibal, M. A. Castro, S. Oviedo-Rouco, V. Demicheli, V. Tortora, F. Tomasina, R. Radi and D. H. Murgida, Chem. Rev., 2017, 117, 13382–13460 Search PubMed.
- S. Yoshikawa and A. Shimada, Chem. Rev., 2015, 115, 1936–1989 Search PubMed.
- Y. L. P. Ow, D. R. Green, Z. Hao and T. W. Mak, Nat. Rev. Mol. Cell Biol., 2008, 9, 532–542 Search PubMed.
- A. V. Kulikov, E. S. Shilov, I. A. Mufazalov, V. Gogvadze, S. A. Nedospasov and B. Zhivotovsky, Cell. Mol. Life Sci., 2012, 69, 1787–1797 Search PubMed.
- V. E. Kagan, V. A. Tyurin, J. Jiang, Y. Y. Tyurina, V. B. Ritov, A. A. Amoscato, A. N. Osipov, N. A. Belikova, A. A. Kapralov, V. Kini, I. I. Vlasova, Q. Zhao, M. Zou, P. Di, D. A. Svistunenko, I. V. Kurnikov and G. G. Borisenko, Nat. Chem. Biol., 2005, 1, 223–232 Search PubMed.
- P. P. Parui, Y. Sarakar, R. Majumder, S. Das, H. Yang, K. Yasuhara and S. Hirota, Chem. Sci., 2019, 10, 9140–9151 Search PubMed.
- S. Zaidi, M. I. Hassan, A. Islam and F. Ahmad, Cell. Mol. Life Sci., 2014, 71, 229–255 Search PubMed.
- H. Theorell and Å. Åkesson, J. Am. Chem. Soc., 1941, 63, 1818–1820 Search PubMed.
- L. J. McClelland, T. C. Mou, M. E. Jeakins-Cooley, S. R. Sprang and B. E. Bowler, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 6648–6653 Search PubMed.
- J. F. Amacher, F. F. Zhong, G. P. Lisi, M. Q. Zhu, S. L. Alden, K. R. Hoke, D. R. Madden and E. V. Pletneva, J. Am. Chem. Soc., 2015, 137, 8435–8449 Search PubMed.
- J. M. Garcia-Heredia, I. Diaz-Moreno, P. M. Nieto, M. Orzaez, S. Kocanis, M. Teixeira, E. Perez-Paya, A. Diaz-Quintana and M. A. De la Rosa, Biochim. Biophys. Acta, Bioenerg., 2010, 1797, 981–993 Search PubMed.
- I. Diaz-Moreno, J. M. Garcia-Heredia, A. Diaz-Quintana, M. Teixeira and M. A. De la Rosa, Biochim. Biophys. Acta, Bioenerg., 2011, 1807, 1616–1623 Search PubMed.
- J. M. Garcia-Heredia, I. Diaz-Moreno, A. Diaz-Quintana, M. Orzaez, J. A. Navarro, M. Hervas and M. A. De la Rosa, FEBS Lett., 2012, 586, 154–158 Search PubMed.
- L. J. McClelland, H. B. Steele, F. G. Whitby, T. C. Mou, D. Holley, J. B. Ross, S. R. Sprang and B. E. Bowler, J. Am. Chem. Soc., 2016, 138, 16770–16778 Search PubMed.
- I. M. Morison, E. M. Cramer Borde, E. J. Cheesman, P. L. Cheong, A. J. Holyoake, S. Fichelson, R. J. Weeks, A. Lo, S. M. K. Davies, S. M. Wilbanks, R. D. Fagerlund, M. W. Ludgate, F. Tatley, M. S. A. Coker, N. A. Bockett, G. Hughes, D. A. Pippig, M. P. Smith, C. Capron and E. C. Ledgerwood, Nat. Genet., 2008, 40, 387–389 Search PubMed.
- D. De Rocco, C. Cerqua, P. Goffrini, G. Russo, A. Pastore, F. Meloni, E. Nicchia, C. T. Moraes, A. Pecci, L. Salviati and A. Savoia, Biochim. Biophys. Acta, Mol. Basis Dis., 2014, 1842, 269–274 Search PubMed.
- L. Ong, I. M. Morison and E. C. Ledgerwood, Br. J. Haematol., 2017, 176, 268–279 Search PubMed.
- Y. Uchiyama, K. Yanagisawa, S. Kunishima, M. Shiina, Y. Ogawa, M. Nakashima, J. Hirato, E. Imagawa, A. Fujita, K. Hamanaka, S. Miyatake, S. Mitsuhashi, A. Takata, N. Miyake, K. Ogata, H. Handa, N. Matsumoto and T. Mizuguchi, Clin. Genet., 2018, 94, 548–553 Search PubMed.
- T. M. Josephs, M. D. Liptak, G. Hughes, A. Lo, R. M. Smith, S. M. Wilbanks, K. L. Bren and E. C. Ledgerwood, J. Biol. Inorg Chem., 2013, 18, 289–297 Search PubMed.
- A. I. Karsisiotis, O. M. Deacon, M. T. Wilson, C. Macdonald, T. M. A. Blumenschein, G. R. Moore and J. A. R. Worrall, Sci. Rep., 2016, 6, 30447 Search PubMed.
- O. M. Deacon, A. I. Karsisiotis, T. Moreno-Chicano, M. A. Hough, C. Macdonald, T. M. A. Blumenschein, M. T. Wilson, G. R. Moore and J. A. R. Worrall, Biochemistry, 2017, 56, 6111–6124 Search PubMed.
- M. D. Liptak, R. D. Fagerlund, E. C. Ledgerwood, S. M. Wilbanks and K. L. Bren, J. Am. Chem. Soc., 2011, 133, 1153–1155 Search PubMed.
- H. Lei and B. E. Bowler, J. Phys. Chem. B, 2019, 123, 8939–8953 Search PubMed.
- O. M. Deacon, R. W. White, G. R. Moore, M. T. Wilson and J. A. R. Worrall, J. Inorg. Biochem., 2020, 203, 110924 Search PubMed.
- M. Lek, K. J. Karczewski, E. V. Minikel, K. E. Samocha, E. Banks, T. Fennell, A. H. O'Donnell-Luria, J. S. Ware, A. J. Hill, B. B. Cummings, T. Tukiainen, D. P. Birnbaum, J. A. Kosmicki, L. E. Duncan, K. Estrada, F. M. Zhao, J. Zou, E. Pierce-Hollman, J. Berghout, D. N. Cooper, N. Deflaux, M. DePristo, R. Do, J. Flannick, M. Fromer, L. Gauthier, J. Goldstein, N. Gupta, D. Howrigan, A. Kiezun, M. I. Kurki, A. L. Moonshine, P. Natarajan, L. Orozeo, G. M. Peloso, R. Poplin, M. A. Rivas, V. Ruano-Rubio, S. A. Rose, D. M. Ruderfer, K. Shakir, P. D. Stenson, C. Stevens, B. P. Thomas, G. Tiao, M. T. Tusie-Luna, B. Weisburd, H. H. Won, D. M. Yu, D. M. Altshuler, D. Ardissino, M. Boehnke, J. Danesh, S. Donnelly, R. Elosua, J. C. Florez, S. B. Gabriel, G. Getz, S. J. Glatt, C. M. Hultman, S. Kathiresan, M. Laakso, S. NcCarroll, M. I. McCarthy, D. McGovern, R. McPherson, B. M. Neale, A. Palotie, S. M. Purcell, D. Saleheen, J. M. Scharf, P. Sklar, P. F. Sullivan, J. Tuomilehto, M. T. Tsuang, H. C. Watkins, J. G. Wilson, M. J. Daly, D. G. MacArthur and C. Exome Aggregation, Nature, 2016, 536, 285–291 Search PubMed.
- L. Banci, I. Bertini, A. Rosato and G. Varani, J. Biol. Inorg Chem., 1999, 4, 824–837 Search PubMed.
- C. M. Lett, A. M. Berghuis, H. E. Frey, J. R. Lepock and J. G. Guillemette, J. Biol. Chem., 1996, 271, 29088–29093 Search PubMed.
- B. S. Rajagopal, A. N. Edzuma, M. A. Hough, K. L. I. M. Blundell, V. E. Kagan, A. A. Kapralov, L. A. Fraser, J. N. Butt, G. G. Silkstone, M. T. Wilson, D. A. Svistunenko and J. A. R. Worrall, Biochem. J., 2013, 456, 441–452 Search PubMed.
- M. Imai, T. Saio, H. Kumeta, T. Uchida, F. Inagaki and K. Ishimori, Biochem. Biophys. Res. Commun., 2016, 469, 978–984 Search PubMed.
- M. Rytomaa and P. K. Kinnunen, J. Biol. Chem., 1994, 269, 1770–1774 Search PubMed.
- Y. L. Deng, F. F. Zhong, S. L. Alden, K. R. Hoke and E. V. Pletneva, Biochemistry, 2018, 57, 5827–5840 Search PubMed.
- Y. L. Deng, M. L. Weaver, K. R. Hoke and E. V. Pletneva, Inorg. Chem., 2019, 58, 14085–14106 Search PubMed.
- T. B. Karpishin, M. W. Grinstaff, S. Komar-Panicucci, G. McLendon and H. B. Gray, Structure, 1994, 2, 415–422 Search PubMed.
- P. K. Witting, A. G. Mauk and P. A. Lay, Biochemistry, 2002, 41, 11495–11503 Search PubMed.
- H.-X. Liu, L.-Z. Li, B. He, S.-Q. Gao, G.-B. Wen and Y.-W. Lin, Dalton Trans., 2018, 47, 10847–10852 Search PubMed.
- P. Zhang, H. Yuan, J.-K. Xu, X.-J. Wang, S.-Q. Gao, X.-S. Tan and Y.-W. Lin, ACS Catal., 2020, 10, 891–896 Search PubMed.
- V. Yin, G. S. Shaw and L. Konermann, J. Am. Chem. Soc., 2017, 139, 15701–15709 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details, UV-Vis, mass spectra and kinetic studies. See DOI: 10.1039/d0ra09961a |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2020 |