Open Access Article
Open Access ArticleLinear consecutive hexaoxazoles as G4 ligands inducing chair-type anti-parallel topology of a telomeric G-quadruplex†
Shogo Sasakia,
Yue Mab,
Takumi Ishizuka c,
Hong-Liang Bao
c,
Hong-Liang Bao c,
Takatsugu Hirokawadef,
Yan Xu
c,
Takatsugu Hirokawadef,
Yan Xu c,
Masayuki Teraa and
Kazuo Nagasawa
c,
Masayuki Teraa and
Kazuo Nagasawa *a
*a
aDepartment of Biotechnology and Life Science, Tokyo University of Agriculture and Technology, 2-24-16 Naka-cho, Koganei, Tokyo 184-8588, Japan. E-mail: knaga@cc.tuat.ac.jp
bInstitute of Global Innovation Research, Tokyo University of Agriculture and Technology, 2-24-16 Naka-cho, Koganei, Tokyo 184-8588, Japan
cDivision of Chemistry, Department of Medical Sciences, Faculty of Medicine, University of Miyazak, 5200 Kihara, Kiyotake, Miyazaki 889-1692, Japan
dTransborder Medical Research Center, University of Tsukuba, 1-1-1 Tennodai, Tsukuba, 305-8575, Japan
eDivision of Biomedical Science, University of Tsukuba, 1-1-1 Tennodai, Tsukuba, 305-8575, Japan
fCellular and Molecular Biotechnology Research Institute, National Institute of Advanced Industrial Science and Technology, 2-4-7 Aomi, Koto-ward, Tokyo 135-0064, Japan
First published on 7th December 2020
Abstract
G-quadruplex structures (G4s) in guanine-rich regions of DNA play critical roles in various biological phenomena, including replication, translation, and gene expression. There are three types of G4 topology, i.e., parallel, anti-parallel, and hybrid, and ligands that selectively interact with or stabilize a specific topology have been extensively explored to enable studies of topology-related functions. Here, we describe the synthesis of a new series of G4 ligands based on 6LCOs (6-linear consecutive oxazoles), i.e., L2H2-2M2EA-6LCO (2), L2A2-2M2EAc-6LCO (3), and L2G2-2M2EG-6LCO (4), which bear four aminoalkyl, acetamidealkyl, and guanidinylalkyl side chains, respectively. Among them, ligand 2 stabilized telomeric G4 and induced anti-parallel topology independently of the presence of cations. The anti-parallel topology induced by 2 was identified as chair-type by means of 19F NMR spectroscopy and fluorescence experiments with 2-aminopurine-labeled DNA.
Guanine-rich sequences in single-stranded DNA or RNA form a characteristic secondary structure known as the G-quadruplex (G4).1 G4 structures were originally found in telomeres2 and some gene promoter regions,3 and next-generation sequencer analysis suggested the existence of over 700
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 kinds of G4-forming sequences in the human genome.4 G4s have been reported to regulate critical biological phenomena, including transcription of oncogenes5 and inhibition of ribosome biogenesis in cancer cells,6 by targeting specific proteins.7 The G4 structure can adopt a variety of topologies depending upon the sequence and the conditions (especially the presence of cations), and these can be classified into three groups, i.e., parallel, hybrid, and anti-parallel topologies.8 In particular, telomere sequences with TTAGGG repeats are structurally diverse and can form all three topologies.9 Control of the topology in G4 is of great interest as a means of potentially regulating the activities of G4s by modulating their specific interactions with their target proteins. For example, HP1 (heterochromatin protein 1), which is involved in the maintenance of heterochromatin, binds only to the parallel form of telomeric G4.10 Further, Rif1, a protein controlling the replication timing in yeast, specifically interacts with its target G4, which adopts parallel topology.11 Thus, ligands that selectively interact with or control each topology in G4 are expected to be useful tools for elucidating the biological functions of G4 structures.
000 kinds of G4-forming sequences in the human genome.4 G4s have been reported to regulate critical biological phenomena, including transcription of oncogenes5 and inhibition of ribosome biogenesis in cancer cells,6 by targeting specific proteins.7 The G4 structure can adopt a variety of topologies depending upon the sequence and the conditions (especially the presence of cations), and these can be classified into three groups, i.e., parallel, hybrid, and anti-parallel topologies.8 In particular, telomere sequences with TTAGGG repeats are structurally diverse and can form all three topologies.9 Control of the topology in G4 is of great interest as a means of potentially regulating the activities of G4s by modulating their specific interactions with their target proteins. For example, HP1 (heterochromatin protein 1), which is involved in the maintenance of heterochromatin, binds only to the parallel form of telomeric G4.10 Further, Rif1, a protein controlling the replication timing in yeast, specifically interacts with its target G4, which adopts parallel topology.11 Thus, ligands that selectively interact with or control each topology in G4 are expected to be useful tools for elucidating the biological functions of G4 structures.
G4 contains three characteristic motifs, i.e., the G-quartet plane, the loops and the grooves. The G-quartet is a common structure in each type of G4, while the loops and grooves are specific to each topology. Thus, most of the G4 ligands developed to date have targeted not only the G-quartet plane through π–π interaction, but also the loops or the grooves. We have developed a series of macrocyclic hexaoxazole compounds (6OTDs) as G4 ligands.12 The 6OTDs are similar in size to the planar structure of the G-quartet, and interact efficiently with the G-quartet through π–π interaction. However, they lack topological selectivity. Recent efforts to develop topology-selective G4 ligands have targeted the loops or grooves in G4.13,14 In 2011, Teulade-Fichou and co-workers reported a groove binder, TOxaPy, which selectively interacts with anti-parallel-type telomeric G4.15 The TOxaPy-related linear pyridine-oxazole ligands developed by several groups can induce anti-parallel topology of telomeric G4 in the presence of K+.16 In addition, Chang and co-workers developed a carbazole-type ligand, BMCV-8C3O, which induces parallel-type topology of telomeric G4 from the hybrid form in the presence of K+.17 Although, some topology-selective interacting ligands have been reported so far, details of the relevant topologies, such as hybrid-1 or hybrid-2, or anti-parallel with chair-type or basket-type, remain unclear.
In this study, we designed linear-type hexaoxazole compounds of 6LCO (6-linear consecutive oxazole) as possible G4 ligands based on the hypothesis that one set of trioxazoles would interact with the G-quartet and the other set would recognize the grooves in a specific topology. Specifically, we synthesized L2H2-2M2EA-6LCO (2), L2A2-2M2EAc-6LCO (3), and L2G2-2M2EG-6LCO (4), which have four aminoalkyl, acetamidealkyl, and guanidinylalkyl side chains, respectively (Fig. 1).18 We evaluated the ability of these ligands to interact with G4 structure as well as their selectivity for the topologies in G4. For this purpose, we used the telomere sequence, which is known to form all three typical topologies of G4 under various conditions.
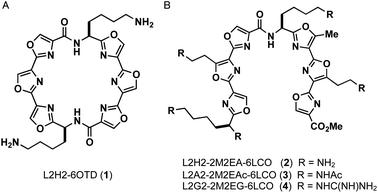 | ||
| Fig. 1 Structures of ligands L2H2-6OTD (1), L2H2-2M2EA-6LCO (2), L2A2-2M2EAc-6LCO (3), L2G2-2M2EG-6LCO (4). | ||
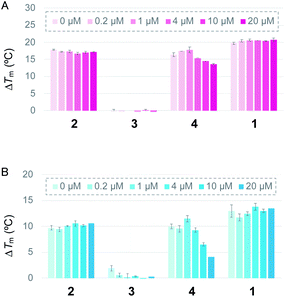
First, the ability of the linear ligands 6LCOs 2–4 and L2H2-6OTD (1)12a to interact with and stabilize telomeric G4 (Flu-telo21; Table S1†) was examined by means of FRET (Förster resonance energy transfer) melting experiments19 in the presence of K+ cation (Fig. 2A). In the case of ligands 2 and 4, which have cationic functional groups (amine or guanidine) in their side chain, the telomeric G4 was efficiently stabilized with ΔTm values of 18 °C and 15 °C, respectively, which are the same level with that of the compound 1. Notably, ligand 2 was selective for telomeric G4 over duplex DNA even in the presence of 100 equivalents of double-stranded DNA (Fig. 2A). Titration experiments showed that ligand 4 interacts with both telomeric G4 and dsDNA. Ligand 3 with the acetamidealkyl, group in its side chain did not show any interaction with or stabilization of telomeric G4. Similar tendencies for stabilization of telomeric G4 by ligands 2–4 were observed in the presence of Na+ (Fig. 2B).
Next, the topologies of G4 formed in the presence of ligands 2–4 were evaluated by means of CD titration analysis, using telomeric sequences of 24TTA in the presence of K+ and Na+ (Fig. S1†). We found that ligand 2 changed the hybrid topology of telomeric G4 into an anti-parallel topology in the presence of K+. Ligand 4 did not change the topology of telomeric G4, but reduced the intensity of the CD spectra upon titration. Since ligand 4 interacts with telomeric G4 as well as dsDNA, as determined by FRET melting analysis (Fig. 2), the ligand seems to interact non-specifically with DNAs, presumably via its guanidine functional group. Ligand 3 did not change the topology of telomeric G4 induced by K+, as determined by CD titration experiments. Based on these observations, the characteristic anti-parallel structure-inducing ability of 2 can be attributed to both the linear-type structure and the amine functional groups in the ligand.
Next, the characteristic topology-inducing ability of the ligand 2 was further investigated by using telomeric DNAs with various sequences and lengths: 22AG,8a,20 23TAG,21 24TTA,9d and 25TAG.22 22AG and 24TTA form both hybrid-1 and hybrid-2 structures, while 23TAG and 25TAG preferentially form hybrid-1 and hybrid-2, respectively, in the presence of K+ cation (Table S1†). Upon titration of 2 into these sequences (one to five equivalents), we found that the CD spectra changed, and anti-parallel topologies were efficiently induced in all cases (Fig. 3). We further examined the CD titration of 2 with 22AG in the presence of Na+. Under these conditions, the anti-parallel topology of 22AG was maintained (Fig. S2†). These results indicate that L2H2-2M2EA-6LCO (2) interact with telomeric G4s and tend to switch their topology to anti-parallel form regardless of the presence or absence of K+ and Na+.
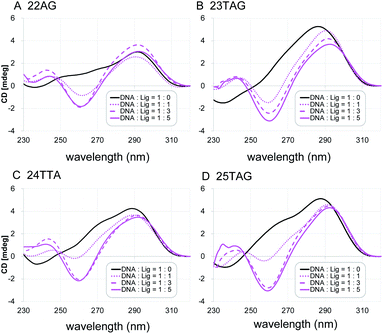 | ||
| Fig. 3 The CD spectra of telomeric DNA (2 μM; (A) 22AG, (B) 23TAG, (C) 24TTA, (D) 25TAG in 50 mM Tris–HCl with 100 mM KCl in the presence of 0–5 equivalents of L2H2-2M2EA-6LCO (2). | ||
Since it is difficult to determine whether a complex has a single topology or not by means of CD spectral analysis, the structure of 22AG induced by ligand 2 was further examined by NMR analysis of the 19F-labeled telomere sequence (19F–22AG; Table S1†).23 The 19F NMR spectrum showed two characteristic singlet peaks at −63.05 and −62.95 ppm (green circle), which correspond to the topologies of hybrid-1 and hybrid-2, in the presence of K+. When ligand 2 was added (0.5, 1.0, and 1.5 equivalents), the two singlet peaks assigned to hybrid-1 and hybrid-2 disappeared, and a broad singlet appeared at −63.17 ppm (cyan circle), as shown in Fig. 4. Since the CD experiments demonstrated that the sequence of 22AG was changed from hybrid to anti-parallel topology upon titration with 2 (Fig. 3), the singlet peak at −63.17 ppm observed in the 19F NMR spectrum suggests the presence of the anti-parallel form as a single topology under these conditions. So far, two types of anti-parallel structures, i.e., chair and basket types, have been characterized in telomere sequences.24 The telomeric DNA of 19F–22AG is reported to form anti-parallel topology of basket type in the presence of Na+, featuring a signal at −62.96 ppm (magenta circle) in 19F NMR.25 The chemical shift of the peak of 19F–22AG in the presence of the ligand 2 was observed at −63.17 ppm, which suggested the topology of 19F–22AG would be an anti-parallel form with a chair type.
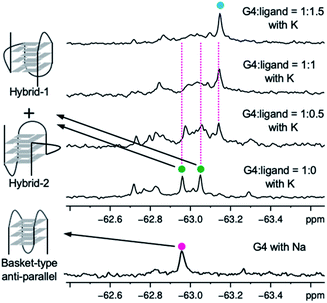 | ||
| Fig. 4 19F NMR spectra of 19F–22AG (200 μM) with K+ during titration with ligand 2 (0–1.5 equiv.), and spectra for 19F–22AG in the presence of Na+. | ||
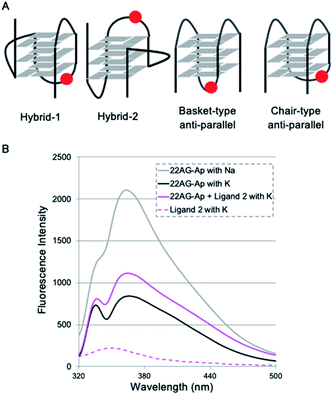
To identify the specific type of anti-parallel topology in 22AG induced by ligand 2, we next performed fluorescence experiments using a 2-aminopurine (Ap)-containing telomere sequence, whose adenine residue at the 13th position was replaced with 2-Ap (22AG–Ap; Fig. 5A and Table S1†). In the case of the hybrid or chair type of the anti-parallel form, 2-Ap in the 22AG will be located at the lateral loop, while in the basket type it will be in the diagonal loop. Since 2-Ap in the diagonal loop is reported to show stronger fluorescence, we can distinguish the topological change from the hybrid to the chair and basket forms of anti-parallel topology by following the fluorescence intensity of the 2-aminopurine.26
We confirmed that the modification with Ap did not influence the formation of topologies in the presence of K+ or Na+ cations as detected by CD analysis (Fig. S4†). Indeed, topology change to the anti-parallel form was observed in 22AG–Ap upon addition of ligand 2 in a similar manner to that of unmodified 22AG (Fig. 3 and Fig S2†). The ligand alone showed no significant fluorescence in the presence of K+ (Fig. 5B, dotted line). Then, the fluorescence intensity of 22AG–Ap was investigated in the presence of K+ or Na+ (Fig. 5B). In the presence of K+, weak fluorescence was observed, which indicates that 22AG–Ap forms a hybrid-type topology with Ap located in the lateral loop (black line). The fluorescence increased in the presence of Na+, showing that 22AG–Ap forms a basket-type anti-parallel topology with the aminopurine located in the diagonal loop (gray line; black to gray: ca. 2-fold). When we added ligand 2 in the presence of K+, the intensity was much weaker than that with Na+ alone, but stronger than that with K+ alone, as shown with a magenta line in Fig. 5B (black to magenta: 1.3-fold).
The 2-Ap quenching analysis addresses two questions, i.e., (1) whether 2-Ap is flipped out from the G-quartet plane, and (2) whether 2-Ap is located in the lateral or diagonal loop. Regarding the first point, there is generally a fluorescence enhancement of over ca. 3-fold when 2-AP is flipped out of the G4 plane.27 Indeed, we observed 3.1-fold fluorescent enhancement upon addition of L2H2-6OTD (1) to 22AG in the presence of K+ (black to cyan: 3.1-fold in Fig. S5†), because L2H2-6OTD (1) is known to stack with the G-quartet plane and to displace the neighboring 2-Ap.12c Regarding the second point, there is generally a ca. 2-fold difference in fluorescence intensity between the cases where 2-Ap is located at the lateral and diagonal loops. We observed a ca. 2-fold fluorescence difference for 22AG–Ap in the presence of K+ and in the presence of Na+, where the topology takes hybrid (i.e., Ap is located laterally), and basket (i.e., Ap is located diagonally) forms, respectively. Since the ligand 2 induced little fluorescent change of 22AG–Ap (black to magenta: 1.3-fold in Fig. 5), it seems likely that 2-Ap in the 22AG is still located at the lateral loop in the presence of ligand 2 and K+. Thus, the results of the CD experiments and 2-Ap quenching analysis indicate that 2 induces chair-type anti-parallel topology in the presence of K+.
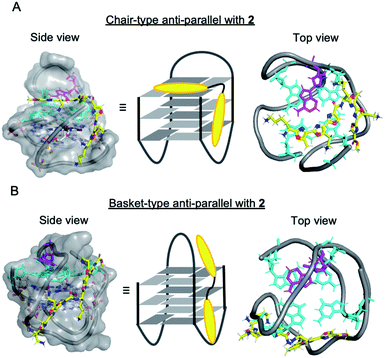
It is not yet clear why the linear polyoxazole induces chair-type anti-parallel topology of telomeric G4, regardless of the presence of cations. Preliminary docking simulation studies of L2H2-2M2EA-6LCO (2) with chair-type (Fig. 6A) and basket-type structures (Fig. 6B) of telomeric G4 gave values of the ligand binding free energy for chair- and basket-types of −78.98 kcal mol−1 and −61.23 kcal mol−1, respectively (Fig S6†). Interaction of the trioxazole moiety with the groove may favor the chair-type structure (Fig. 6A). According to the docking models, ligand 2 does not influence the location of the 13th adenine (colored magenta, Fig. 6A) in the chair-form G4, which is consistent with the observation that ligand 2 did not enhance the fluorescence in a manner that would indicate flipping out of 2-Ap (Fig. 5B). In terms of the recently proposed folding pathways of G4,28 the topological change from the hybrid to the chair-type anti-parallel structure takes fewer steps than the change to basket-type structure, and this could be a factor.
Conclusions
We have developed a new series of G4 ligands based on linear consecutive polyoxazole structure. The ligands with cationic functional groups preferentially interact with telomeric G4. Among them, L2H2-2M2EA-6LCO (2), with aminoalkyl side chains, interacts with telomeric G4s and induces anti-parallel topology regardless of the presence of cations (K+ and Na+). Furthermore, ligand 2 is suggested to induce “chair-type” anti-parallel topology, based on the results of 19F NMR and 2-Ap quenching studies.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by Grants-in-Aid for Scientific Research, Japan Society for the Promotion of Science (JSPS) (Scientific Research (B) 20H02876 to K. N.; Scientific Research (C) 19K05743 to M. T.; Early-Career Scientists 20K15411 to Y. M.), Grants-in-Aid for Scientific Research on Innovative Areas “Middle Molecular Strategy” (18H04387 to K. N.), Japan Science and Technology Agency (JST) ACT-X JPMJAX191E to Y. M., Inamori Grants from the Inamori Foundation to M. T. and grants from the Nippon Foundation and The Translational Research programand, and a Grant-in-Aid for a JSPS Research Fellow (JP 20J13814 to S. S.), as well as funds from the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from the Japan Agency for Medical Research and Development (AMED, JP18am0101114 to TH).Notes and references
- J. T. Davis, Angew. Chem., Int. Ed., 2004, 43, 668–698 CrossRef CAS.
- (a) T. M. Bryan, Molecules, 2020, 25, 3686 CrossRef CAS; (b) H. Seimiya, Cancer Sci., 2020, 111, 3089–3099 CrossRef CAS.
- (a) J. L. Huppert and S. Balasubramanian, Nucleic Acids Res., 2007, 35, 406–413 CrossRef CAS; (b) J. L. Huppert, A. Bugaut, S. Kumari and S. Balasubramanian, Nucleic Acids Res., 2008, 36, 6260–6268 CrossRef CAS; (c) A. Verma, V. K. Yadav, R. Basundra, A. Kumar and S. Chowdhury, Nucleic Acids Res., 2009, 37, 4194–4204 CrossRef CAS.
- V. S. Chambers, G. Marsico, J. M. Boutell, M. D. Antonio, G. P. Smith and S. Balasubramanian, Nat. Biotechnol., 2015, 33, 877–881 CrossRef.
- (a) G. W. Collie and G. N. Parkinson, Chem. Soc. Rev., 2011, 40, 5867–5892 RSC; (b) S. J. Adam, C. L. Grand, D. J. Bearss and L. H. Hurley, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 11593–11598 CrossRef; (c) S. Kumari, A. Bugaut, J. L. Huppert and S. Balasubramanian, Nat. Chem. Biol., 2007, 3, 218–221 CrossRef CAS; (d) L. A. Cahoon and H. S. Seifert, Science, 2009, 325, 764–767 CrossRef CAS; (e) A. Bugaut and S. Balasubramanian, Nucleic Acids Res., 2012, 40, 4727–4741 CrossRef CAS.
- (a) D. Sun, B. Thompson, B. E. Cathers, M. Salazar, S. M. Kerwin, J. O. Trent, T. C. Jenkins, S. Neidle and L. H. Hurley, J. Med. Chem., 1997, 40, 2113–2116 CrossRef CAS; (b) T. Tauchi, K. Shin-ya, G. Sashida, M. Sumi, A. Nakajima, T. Shimamoto, J. H Ohyashiki and K. Ohyashiki, Oncogene, 2003, 22, 5338–5347 CrossRef CAS; (c) C. M. Incles, C. M. Schultes, H. Kempski, H. Koehler, L. R. Kelland and S. Neidle, Mol. Cancer Ther., 2004, 3, 1201–1206 CAS; (d) H. Tahara, K. Shin-ya, H. Seimiya, H. Yamada, T. Tsuruo and T. Ide, Oncogene, 2006, 25, 1955–1966 CrossRef CAS; (e) M. Gunaratnam, O. Greciano, C. Martins, A. P. Reszk, C. M. Schultes, H. Morjani, J.-F. Riou and S. Neidle, Biochem. Pharmacol., 2007, 74, 679 CrossRef CAS.
- (a) V. Brázda, L. Hároníková, J. C. C. Liao and M. Fojta, Int. J. Mol. Sci., 2014, 15, 17493–17517 CrossRef; (b) M. M. Fay, S. M. Lyons and P. Ivanov, J. Mol. Biol., 2017, 429, 2127–2147 CrossRef CAS.
- (a) S. Burge, G. N. Parkinson, P. Hazel, A. K. Todd and S. Neidle, Nucleic Acids Res., 2006, 34, 5402–5415 CrossRef CAS; (b) T.-M. Ou, Y.-J. Lu, J.-H. Tan, Z.-S. Huang, K.-Y. Wong and L.-Q. Gu, ChemMedChem, 2008, 3, 690–713 CrossRef CAS; (c) R. Del Villar-Guerra, J. O. Trent and J. B. Chaires, Angew. Chem., Int. Ed., 2018, 57, 7171–7175 CrossRef CAS; (d) S. Amrane, R. W. Lin Ang, Z. M. Tan, C. Li, J. K. Lim, J. M. Lim, K. W. Lim and A. T. Phan, Nucleic Acids Res., 2009, 37, 931–938 CrossRef CAS.
- (a) Y. Wang and D. J. Petal, Structure, 1993, 1, 263–282 CrossRef CAS; (b) G. N. Parkinson, M. P. H. Lee and S. Neidle, Nature, 2002, 417, 876–880 CrossRef CAS; (c) J. Y. Lee, B. Okumus, D. S. Kim and T. Ha, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 18938–18943 CrossRef CAS; (d) A. Ambrus, D. Chen, J. Dai, T. Bialis, R. A. Jones and D. Yang, Nucleic Acids Res., 2006, 34, 2723–2735 CrossRef CAS; (e) A. T. Phan, FEBS J., 2010, 277, 1107–1117 CrossRef CAS; (f) B. Heddi and A. T. Phan, J. Am. Chem. Soc., 2011, 133, 9824–9833 CrossRef CAS.
- R. J. Roach, M. Garavís, C. González, G. B. Jameson, V. V. Filichev and T. K. Hale, Nucleic Acids Res., 2020, 48, 682–693 CrossRef CAS.
- H. Masai, R. Fukatsu, N. Kakusho, Y. Kanoh, K. Moriyama, Y. Ma, K. Iida and K. Nagasawa, Sci. Rep., 2019, 9, 8618 CrossRef.
- (a) M. Tera, H. Ishizuka, M. Takagi, M. Suganuma, K. Shin-ya and K. Nagasawa, Angew. Chem., Int. Ed., 2008, 47, 5557–5560 CrossRef CAS; (b) K. Iida and K. Nagasawa, Chem. Rec., 2013, 13, 539–548 CrossRef CAS; (c) W. J. Chung, B. Heddi, M. Tera, K. Iida, K. Nagasawa and A. T. Phan, J. Am. Chem. Soc., 2013, 135, 13495–13501 CrossRef CAS.
- (a) Y. Ma, K. Iida, S. Sasaki, T. Hirokawa, B. Heddi, A. T. Phan and K. Nagasawa, Molecules, 2019, 24, 263 CrossRef; (b) Y. Ma, Y. Tsushima, M. Sakuma, S. Sasaki, K. Iida, S. Okabe, H. Seimiya, T. Hirokawa and K. Nagasawa, Org. Biomol. Chem., 2018, 16, 7375–7382 RSC.
- (a) Y. Ma, K. Iida and K. Nagasawa, Biochem. Biophys. Res. Commun., 2020, 531, 3–17 CrossRef CAS; (b) S. Asamitsu, T. Bando and H. Sugiyama, Chem.–Eur. J., 2019, 25, 417–430 CrossRef CAS; (c) M. P. O'Hagan, J. C. Morales and M. C. Galan, Eur. J. Org. Chem., 2019, 31, 4995–5017 CrossRef.
- F. Harmon, E. Largy, A. G. B.-Beaurepaire, M. R.-Dagois, A. Sidibe, D. Monchou, J.-L. Mergny, J.-F. Riou, C.-H. Nguyen and M.-P. Teulade-Fichou, Angew. Chem., Int. Ed., 2011, 50, 8745–8749 CrossRef.
- (a) F. Doria, V. Pirota, M. Petenzi, M.-P. Teulade-Fichou, D. Verga and M. Freccero, Molecules, 2018, 23, 2162 CrossRef; (b) N, Rizeq and S. N. Georgiades, Molecules, 2017, 22, 2160 CrossRef; (c) M. Petenzi, D. Verga, E. Largy, F. Hamon, F. Doria, M.-P. Teulade-Fichou, A. Guédin, J.-L. Mergny, M. Mella and M. Freccero, Chem.–Eur. J., 2012, 18, 14487–14496 CrossRef CAS.
- Z.-F. Wang, M.-H. Li, W.-W. Chen, S.-T. Danny Hsu and T.-C. Chang, Nucleic Acids Res., 2016, 44, 3958–3968 CrossRef CAS.
- The synthesis of the ligands 2–4 was shown in ESI.†.
- (a) A. De Rache and J.-L. Mergny, Biochimie, 2015, 115, 194–202 CrossRef; (b) A. De Cian, L. Guittat, M. Kaiser, B. Saccá, S. Amrane, A. Bourdoncle, P. Alberti, M. P. Teulade-Fichou, L. Lacroix and J. L. Mergny, Methods, 2007, 42, 183–195 CrossRef CAS.
- (a) R. Buscaglia, R. D. Gray and J. B. Chaires, Biopolymer, 2013, 99, 1006–1018 CAS; (b) M. C. Miller, R. Buscaglia, J. B. Chaires, A. N. Lane and J. O. Trent, J. Am. Chem. Soc., 2010, 132, 17105–17107 CrossRef CAS; (c) Y. Xu, Y. Noguchi and H. Sugiyama, Bioorg. Med. Chem., 2006, 14, 5584–5591 CrossRef CAS.
- A. T. Phan, V. Kuryavyi, K. N. Luu and D. J. Patel, Nucleic Acids Res., 2007, 35, 6517–6525 CrossRef CAS.
- K. W. Lim, S. Amrane, S. Bouaziz, W. Xu, Y. Mu, D. J. Patel, K. N. Luu and A. T. Phan, J. Am. Chem. Soc., 2009, 131, 4301–4309 CrossRef CAS.
- (a) H.-L. Bao, H.-shan Liu and Y. Xu, Nucleic Acids Res., 2019, 47, 4940–4947 CrossRef CAS; (b) H. Chen, S. Viel, F. Ziarellic and L. Peng, Chem. Soc. Rev., 2013, 42, 7971–7982 RSC.
- (a) C. Liu, B. Zhou, Y. Geng, D. Y. Tam, R. Feng, H. Miao, N. Xu, X. Shi, Y. You, Y. Hong, B. Z. Tang, P. K. Lo, V. Kuryavyi and G. Zhu, Chem. Sci., 2019, 10, 216 Search PubMed; (b) K. W. Lim, P. Alberti, A. Guedin, L. Lacroix, J.-F. Riou, N. J. Royle, J.-L. Mergny and A. T. Phan, Nucleic Acids Res., 2009, 37, 6239–6248 CrossRef CAS; (c) A. Matsugami, H. Tsuchibayashi, Y. Xu, Y. Noguchi, H. Sugiyama and M. Katahira, Nucleic Acids Symp. Ser., 2006, 50, 45–46 CrossRef.
- H.-L. Bao, T. Ishizuka, A. Iwanami, T. Oyoshi and Y. Xu, ChemistrySelect, 2017, 2, 4170 CrossRef CAS.
- (a) R. D. Gray, L. Petraccone, R. Buscaglia and J. B. Chaires, Methods Mol. Biol., 2010, 608, 121–136 CrossRef CAS; (b) R. Buscaglia, D. M. Jameson and J. B. Chaires, Nucleic Acids Res., 2012, 40, 4203–4215 CrossRef CAS.
- (a) T. Kimura, K. Kawai, M. Fujitsuka and T. Majima, Tetrahedron, 2007, 63, 3585–3590 CrossRef CAS; (b) T. Kimura, K. Kawai, M. Fujitsuka and T. Majima, Chem. Commun., 2006, 401–402 RSC.
- (a) T. Frelih, B. Wang, J. Plavec and P. . Sket, Nucleic Acids Res., 2020, 48, 2189–2197 CrossRef CAS; (b) P. Stadlbauer, P. Kührova, L. Vicherek, P. Banàš, M. Otyepka, L. Trantìrek and J. Sponer, Nucleic Acids Res., 2019, 47, 7276 CrossRef CAS; (c) X.-M. Hou, Y.-B. Fu, W.-Q. Wu, L. Wang, F.-Y. Teng, P. Xie, P.-Y. Wang and X.-G. Xi, Nucleic Acids Res., 2017, 45, 11401–11412 CrossRef CAS; (d) A. Marchand and V. Gabelica, Nucleic Acids Res., 2016, 44, 10999–11012 CrossRef CAS; (e) D. Koirala, C. Ghimire, C. Bohrer, Y. Sannohe, H. Sugiyama and H. Mao, J. Am. Chem. Soc., 2013, 135, 2235–2241 CrossRef CAS; (f) M. Boncǐna, J. Lah, I. Prislan and G. Vesnaver, J. Am. Chem. Soc., 2012, 134, 9657–9663 CrossRef; (g) T. Mashimo, H. Yagi, Y. Sannohe, A. Rajendran and H. Sugiyama, J. Am. Chem. Soc., 2010, 132, 14910–14918 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra09413g |
| This journal is © The Royal Society of Chemistry 2020 |