Open Access Article
Open Access ArticleDevelopment of a “single-click” analytical platform for the detection of cannabinoids in hemp seed oil†
Roberta Risoluti a,
Giuseppina Gullifaa,
Alfredo Battistinib and
Stefano Materazzi
a,
Giuseppina Gullifaa,
Alfredo Battistinib and
Stefano Materazzi *a
*a
aDepartment of Chemistry, Sapienza University of Rome, p.le A. Moro 5, 00185 Rome, Italy. E-mail: stefano.materazzi@uniroma1.it; Fax: +390649387137; Tel: +390649913616
bConsiglio per la ricerca in agricoltura e l'analisi dell'economia agraria, Centro di Politiche e Bioeconomia, via Pò 14, 00198, Italy
First published on 9th December 2020
Abstract
In this work, an innovative screening platform is developed and validated for the on site detection of cannabinoids in hemp seed oil, for food safety control of commercial products. The novelty of this completely automated tool consists of a miniaturized NIR spectrometer operating in a wireless mode that permits processing samples in a rapid and accurate way and to obtain in a single click the early detection of a residual amount of cannabinoids in oil, including cannabidiol (CBD), the psychoactive Δ9-tetrahydrocannabinol (THC) and the Δ9-tetrahydrocannabinolic acid (THCA). Simulated samples were realized to instruct the platform and prediction models were developed by chemometric analysis of the NIR spectra using partial least square regression algorithms. Once calibrated, the platform was used to predict samples acquired in the market and on websites. Validation of the system was achieved by comparing results with those obtained from GC-MS analyses and a good correlation was observed.
Introduction
Hemp seed oil is the most commercialized hemp based product worldwide, because of its interesting nutritional value:1 in fact, about 100 g of this oil may contain around 63% of the daily recommended amount of protein and unsaturated fatty acids with a very low amount of cholesterol.2 Another important reason that makes the hemp seed oil very attractive is related to the presence of omega-3 and omega-6, flavonoids, mono and sesqui-terpenes, steroids3 and, lastly, of cannabinoids, an important class of substances frequently used for their interesting properties for nutritional and medicinal uses.4 Nevertheless, despite the interesting properties of hemp seed oils, researchers reported on the presence of the illicit Δ9-tetrahydrocannabinol (THC) in urine after hemp seed oil consumption5 and positivity to screening tests.6 As a consequence, a critical issue is actually represented by the dosage and exposure to potentially psychoactive substances when the product is consumed. In fact, despite cannabinoids of the hemp plat are not distributed in the seeds but in the leaves, contamination during harvesting may occur with a consequent possibility to recover phytocannabinoids in the oil. To this aim, the monitoring of cannabinoids content in the hemp seed oil, is strictly required to avoid consumer using or abusing the psychoactive and illicit THC.7The increasing demand of consumers for hemp based products has globally led to an increasing request of procedures and methods to detect the cannabinoids content in food products. Therefore, the ability of a method in assessing the quantitation of illicit or potential illicit substances in food matrices at the first level test represents an important issue when dealing with the monitoring of food quality or human health, especially in forensic field. For this reason, innovative screening systems, able to rapidly identify illegal products without requiring sample pretreatment or clean up in a non destructive way, are more and more recommended.8
Confirmatory analyses of cannabinoids in hemp derivative products usually require chromatographic techniques as High Performance Liquid Chromatography associated to Mass Spectrometry (HPLC-MS)9 and Gas Chromatography coupled to Mass Spectrometry (GC-MS).10 In addition, high resolution Nuclear Magnetic Resonance (NMR) was also proposed for the analysis of cannabinoids.11 Spectroscopic techniques are widely recognized as solvent-less, fast and easy-to-use tools to perform the chemical investigation of different matrices without destroying sample.12 In particular, NIR spectroscopy associated to chemometric analysis proved its high potential in multicomponent analyses, at law costs and without requiring specialized personnel to be used.13
In addition, NIR spectroscopy allows users moving out of the laboratory and performing prediction of analytes in complex matrices directly on site14 by mean of portable instruments that permit to transfer validated methods for the application to real situations.
Based on these considerations, this work proposes a novel method based on a miniaturized spectrometer, the MicroNIR On-Site, for the monitoring of cannabinoids in hemp seed oil. This platform uses chemometric tools to develop models of prediction that once validated, provide the fast and accurate evaluation of the cannabinoids content in the oil.
Results and discussion
Cannabinoids concentration in hemp seed oil strictly depends on the contamination of the seeds during harvesting; in fact, these molecules are mainly recovered in the leaves, in the top 15 cm of the plant. In addition, as a function of the different kind of hemp plant (THC type or CBD type), a higher amount of each cannabinoid may be found. Moreover, hemp plants containing THC and THCA may led to a contaminated oil that can be used as seasoning for food without restrictions. In addition, oils may be used at high temperature during cooking and residual amount of the non psychoactive THCA (most abundant in the hemp plant with respect to the THC) may be recovered as well, as a consequence of the decarboxylation process of THCA. In this view, new screening tools for a rapid and non-destructive analysis of oil specimens would permit the monitoring of psychoactive molecules and in the meanwhile, might address confirmatory analyses on the same specimen if required.The feasibility of innovative techniques to specific issues, strictly relies to the standardization of the method on simulated samples as representative as possible of those to be processed. To this aim, a reference dataset of samples was prepared by fortifying commercially available hemp oils with increasing amount of each cannabinoid, in order to evaluate the effect of the matrix on the detection of the molecules. Spectra in the NIR region were recorded by the MicroNIR On-Site device, as reported in Fig. 1a, and chemometric pre-treatments were investigated in order to separate samples according to the different molecules recovered. The molecular structures of the investigated cannabinoids are reported in Fig. S1.†
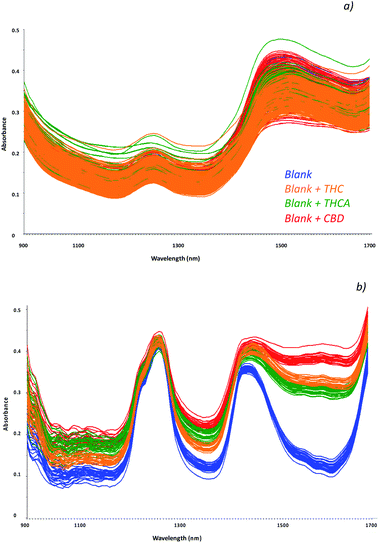 | ||
| Fig. 1 Collected spectra of blank samples (unspiked oils, blue) and fortified samples with THC (orange line), THCA (green lines) and CBD (red lines) as raw data (a) and chemometric treated data (b). | ||
Mathematical transformations usually recommended for spectroscopic data15 were evaluated: in particular, scatter-correction methods were applied such as Standard Normal Variate transform (SNV),16 Multiplicative Scatter Correction (MSC), Mean Centering (MC)17 and normalization.18 In addition, spectral derivation techniques including Savitzky–Golay (SG) polynomial derivative filters19 were considered.
Among the investigated spectra pre-treatments, combination of the baseline correction, first derivative transform and Multiplicative Scatter Correction (MSC) permitted to highlight differences among spectra and thus to separate samples according to the different molecule (Fig. 1b).
This step permits to instruct the method to simultaneously recognize the cannabinoids and not to obtain interferences of each cannabinoid to one another as shown from the spectra of fortified hemp oils with a mixed solution of THC, THCA and CBD (Fig. S2†).
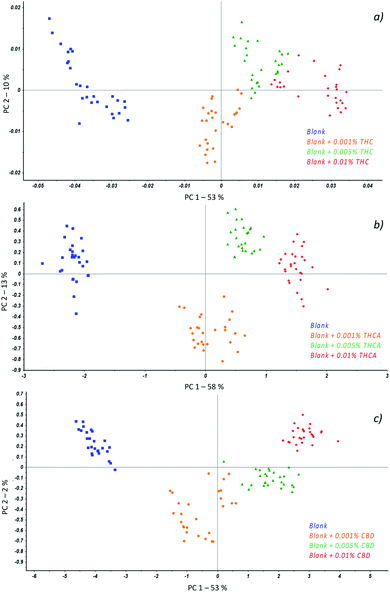
Partial Least Square Discriminant Analysis (PLS-DA) was used as regression algorithm in order to develop a model of prediction of cannabinoids in oil that permits to rapidly evaluate if contamination of seeds was occurred during harvesting of seeds. As reported in Fig. 2, the model allows users to simultaneously differentiate spiked from unspiked samples and to identify the recovered molecule. In fact, all the investigated classes were found to be significantly different to permit an accurate estimation of the positive samples. As a consequence, the model may recognize the cannabinoids in real samples as a function of the belonging cluster of the spectra and preliminarily addresses the subsequent analyses.
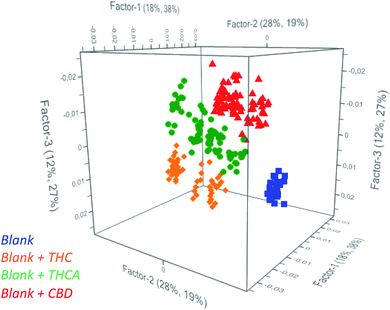 | ||
| Fig. 2 Scores plot from PCA of unspiked oils (blue) and spiked oils with THC (orange), THCA (green) and CBD (red). | ||
The prediction ability of the model was evaluated by estimating the figures of merit in detecting the presence of THC, THCA and CBD in hemp seed oil: in particular, Non Error Rate (NER%), specificity (Sp.%) and the Root Mean Square Error (RMSE) were calculated after dividing the entire dataset into training set (75% of the processed samples) and evaluation set (25% of the samples) and by considering 3 latent variables. Results are summarized in Table 1.
| Calibration | Validation | Prediction | |||||||
|---|---|---|---|---|---|---|---|---|---|
| NER (%) | Sp. (%) | RMSE (%) | NER (%) | Sp. (%) | RMSE (%) | NER (%) | Sp. (%) | RMSE (%) | |
| Blank | 100 | 100 | 0.19 | 100 | 100 | 0.20 | 100 | 100 | 0.18 |
| Blank + THC | 100 | 100 | 0.19 | 100 | 98.9 | 0.19 | 100 | 100 | 0.19 |
| Blank + THCA | 70.1 | 69.3 | 0.29 | 70.1 | 69.3 | 0.30 | 66.7 | 100 | 0.30 |
| Blank + CBD | 97.7 | 75.9 | 0.28 | 96.5 | 75.0 | 0.29 | 100 | 75.0 | 0.24 |
Satisfactory performances of the model may be observed for each molecule, leading to errors in prediction not higher than 0.30% and accuracy values, expressed as non error rate%, not lower than 77% for all the molecules. Specificity of the method was also calculated and suitable outcomes were obtained resulting in Sp. values about 100% for THC and THCA and 75% for CBD. As a consequence of the promising results, specific models of regression were assessed for each cannabinoid in order to provide a tool able to accurate quantify THC, THCA and CBD in hemp seed oils.
For each molecule, spectra of reference blank samples and fortified samples with 0.001, 0.05 and 0.01% w/v of cannabinoids, were collected in the range 900–1700 nm and processed by multivariate statistical analysis.
Principal Component Analysis (PCA) was applied to preliminarily investigate correlations among data and different spectra pre-treatments were evaluated with the aim of improving the signal of cannabinoids with respect to the matrix. In fact, the association of chemometrics to MicroNIR spectra is not only mandatory for interpretation of results but it is of help in the model optimization as it permits to consider the contribution of more than two parameters simultaneously.
In particular, regardless to THC (Fig. 3a), the best separation of samples according to the amount of cannabinoid was achieved when spectra were baseline corrected followed by the first derivative transform while for THCA (Fig. 3b), an additional application of the Standard Normal Variate (SNV) to previous treatments was required. At last, with respect to CBD, the spectra were baseline corrected and SNV was applied resulting in a scores plot reported in Fig. 3c.
The first interesting result consists of the separation of the samples as a function of the class they belong to; in fact, all the measurements resulted well grouped in different colours.
Such a behavior suggested to develop a quantitation model for each molecule by considering Partial Least Square Regression algorithms. As required for the validation of analytical methods, all the collected spectra were divided into training set (about 75% of samples) and evaluation set (about 25% of samples) and a number of parameters were assessed in order to calculate the model's performances.
Among these, the Root Mean Squared Errors (RMSEs) as well as the correlation coefficient (R2) were estimated in calibration and validation by using 7 latent variables, while precision and sensitivity were calculated for each molecule to provide fast and accurate outcomes when dealing with real samples analysis.
Results of the PLS models are reported in Table 2, where satisfactory outcomes may be observed for each molecule: in fact, the correlation values were always not lower than 0.9772 in calibration and higher than 0.9313 in validation. Moreover, the model for THC and THCA prediction provided the best results in terms of RMSE (0.003% w/w), compared to CBD (0.005% w/w).
| Figures of merit | THC | THCA | CBD |
|---|---|---|---|
| a Latent variables.b Minimum detection concentration. | |||
| RMSEC | 0.001 | 0.002 | 0.001 |
| RMSECV | 0.003 | 0.003 | 0.005 |
| LVa | 7 | 7 | 7 |
| R2 calibration | 0.9772 | 0.9814 | 0.9823 |
| R2 validation | 0.9313 | 0.9735 | 0.9710 |
| MDCb (%) | 0.001 | 0.001 | 0.001 |
| Precision (%) | 1.71 | 1.49 | 1.65 |
| Sensitivity (%) | 0.10 | 0.30 | 0.10 |
In addition, the developed models proved to be sensitive (about 0.10% for THC and CBD, 0.30% for THCA) and precise (about 1.71%, 1.49% and 1.65% for THC, THCA and CBD respectively) to ensure a feasible innovative tool to be used as the first level test of cannabinoids in hemp seeds oil.
Feasibility of the platform
Prediction of the real samples was performed by processing a number of 15 hemp seed oil specimens commercially available in the Italian markets, in order to evaluate the platform performances. The resulting chromatogram of the reference standards used to optimize the model is reported in Fig. S3.† Samples were analyzed by the MicroNIR On-Site and the reference method by GC-MS in order to compare results. Among the investigated samples, only one contained a residual amount of about 0.03% of CBD detected by GC-MS, while the remaining 14 samples were found to be negative. Good accordance among measurements was observed from MicroNIR outcomes: in fact, the predicted percentage of CBD recovered in the same sample was about 0.05%. In addition, the remaining 14 samples were correctly predicted by the model as negative (estimated value lower than the detection limit of the platform), confirming the promising application of the platform.Experimental
Analytical workflow
Hemp seed oil specimens (30 samples) were provided by the markets and the producers in Italy, including 4 samples from the websites. For each sample, about 1 ml of the oil was directly analysed by the MicroNIR equipped by a special accessory for liquids and no sample pre-treatment was necessary.In order to calibrate the model, simulated samples containing increasing amounts of cannabinoids in hemp oils from 0.01% to 0.001% w/v were realized by spiking hemp seed oils and processed both by the MicroNIR platform and GC-MS. Reference standards of cannabidiol (CBD), Δ9-tetrahydrocannabinol (THC) and Δ9-tetrahydrocannabinolic acid (THCA) were purchased from Sigma-Aldrich (St. Louis, Missouri United States) as methanolic solution at the concentration of 1 mg ml−1 and were pure at 99%.
Calibration and validation of the platform was obtained by dividing the data set of measurements in training set and evaluation set, while the prediction of real samples was achieved by processing 15 additional samples of hemp seed oil not previously included in the dataset and thus processed as independent batch. This step is strictly required, in order to guarantee the results are not bath-dependent and to ensure reproducibility and effectiveness of the platform.
In addition, the oil samples were analyzed by GC-MS before being fortified, to make a model as representative as possible of real samples and correctly estimate the quantity of cannabinoids before and after the spiking.
MicroNIR On-Site spectrometer
The MicroNIR On-Site is a portable spectrometer device operating in the NIR region of the spectrum (900–1700 nm) and distributed by Viavi Solutions (JDSU Corporation, Milpitas, USA). In particular, it is the latest version of the ultracompact MicroNIR from Viavi and represents the real update in the field of the miniaturized device, moving out of the laboratory. In fact, it is provided by two different software (JDSU Corporation, Milpitas, USA): the first is the MicroNIR Pro software that allows trained users collecting samples and developing a model of prediction; the second, is the MicroNIR On-Site-W system for real time prediction of samples and it is suitable even for not trained users. The technology is based on the Linear Variable Filter (LVF) that is a one dimensional continuously bandpass filter, fabricated on glass using stable inorganic materials for long life and environmental stability. This technology enables the spectrometer being compact, lightweight and robust without moving optic components.Calibration of the instrument was obtained prior to the acquisition of the sample, by the mean of a special accessory that permitted the registration of a dark reference (total absorbance) and a white reference (total reflectance) using Spectralon.
The instrumental settings included: a nominal spectral resolution of the acquisitions at 6.25 nm, an integration time of 10 ms, for a total measurement time of 2.5 s per sample. Chemometric analysis was performed by V-JDSU Unscrambler Lite (Camo software AS, Oslo, Norway).
Ten spectra for each sample were collected in order to ensure heterogeneity of the measurement and the mean was considered for the chemometric analysis. The investigation of samples correlation was first performed by principal component analysis and the models of prediction were developed by the mean of partial least square regression algorithms.
Validation by GC-MS analyses
Gas chromatography coupled to mass spectrometer detector (GC-MS) was used as reference official method to process all the investigated samples and to detect and accurately quantify the cannabinoids. The extraction procedure of 1 ml of oil included three successive cycles of extraction in ethanol 96% (v/v) under magnetic agitation; the extracts were centrifuged, filtered and diluted prior to injection.A PerkinElmer (Waltham, MA) instrument with a mass selective detector (GC-MS) was used for confirmatory analyses. The capillary separation column was a HP-5MS 30 m × 0.25 mm × 0.25 mm; the carrier gas was helium delivered at a constant flow of 1 ml min−1. The oven program was as follows: 120 °C for 1 min, ramping temperature to 240 °C at 30 °C min−1 and then increasing to 290 °C at 10 °C min−1 for 10 min. A post run was made at 300 °C for 5 min. Electron impact (EI) ionization was employed at a voltage of 70 eV. The interface temperature was 250 °C, and the temperature of the ion source and quadrupole were 230 °C and 150 °C, respectively. Collection of mass spectral data was performed in the scan mode from 44 to 450 m/z and the identification of the analyte was achieved in Selected Ion Monitoring (SIM). A split ratio of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was used for injection.20
1 was used for injection.20
The calculated Limit of Detection (LOD) and Limit of Quantification (LOQ) of the method were 0.05 ng ml and 0.1 ng ml, respectively while the correlation coefficient R2 was 0.9989.
Conclusions
In this work, a novel analytical platform based on NIR spectroscopy and chemometrics is proposed for the monitoring of the cannabinoid residues in hemp seed oil, in particular the psychoactive THC and the potential psycotrope THCA. In fact, despite the THCA does not exhibit a binding affinity for CB receptors, it is usually subjected to decarboxylation when exposed to light or temperature (cooking or storage conditions). The novelty of the platform is strictly related to the innovative MicroNIR On-Site device that permits to collect samples and to perform the prediction in few seconds even by not trained personnel by an automated platform. Reliability of this novel test was assessed by processing all the simulated and real samples by the reference official method (GC-MS). In addition, the model was validated by estimating the characteristic figures of merit such as the minimum detection limit, sensitivity, precision and the RMSE, demonstrating to be suitable for application as screening test by law enforcement for forensic controls.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Notes and references
- L. L. Romano and A. Hazekamp, Cannabinoids, 2013, 1(1), 11 Search PubMed; O. Aizpurua-Olaizola, J. Omar, P. Navarro, M. Olivares, N. Etxebarria and A. Usobiaga, Anal. Bioanal. Chem., 2014, 406, 7549–7560 CrossRef CAS.
- United States Department of Agriculture, National Nutrient Database for Standard Reference Full Report (AllNutrients): 12012, Seeds, Hemp Seed, Hulled, 2019 CrossRef CAS; S. B. Karch, Forensic Drug Abuse Advisor Newsletter, 1997, vol. 9, p. 22 CrossRef CAS; L. Bartella, L. Di Donna, A. Napoli, C. Siciliano, G. Sindona and F. Mazzotti, Food Chem., 2019, 278, 261–266 CrossRef CAS.
- O. Werz, J. Seegers, A. M. Schaible, C. Weinigel, D. Barz, A. Koeberle, G. Allegrone, F. Pollastro, L. Zampieri, G. Grassi and G. Appendino, PharmaNutrition, 2014, 2, 53–60 CrossRef CAS; D. Aiello, S. Materazzi, R. Risoluti, H. Thangavel, L. Di Donna and L. Mazzotti, Mol. BioSyst., 2015, 11, 2373–2382 RSC; M. Petrovic, Z. Dbaljak, N. Kezic and P. Dzidara, Food Chem., 2015, 170, 218–225 CrossRef.
- M. M. Radwan, M. A. ElSohly, A. T. El-Alfy, S. A. Ahmed, D. Slade, A. S. Husni, S. P. Manly, L. Wilson, S. Seale, S. J. Cutler and S. A. J. Ross, J. Nat. Prod., 2015, 78, 1271–1276 CrossRef CAS; L. Marchetti, M. G. Sabbieti, M. Menghi, S. Materazzi, M. M. Hurley and G. Menghi, Histol. Histopathol, 2002, 17(4), 1061–1066 Search PubMed; M. A. ElSohly and D. Slade, Life Sci., 2005, 78, 539–548 CrossRef; C. Bretti, F. Crea, C. De Stefano, C. Foti, S. Materazzi and G. Vianelli, J. Chem. Eng. Data, 2013, 58, 2835–2847 CrossRef.
- A. C. Hayley, L. A. Downey, G. Hansen, A. Dowell, D. Savins, R. Buchta, R. Catubig, R. Houlden and C. K. K. Stough, Forensic Sci. Int., 2018, 284, 101–106 CrossRef CAS; F. Crea, G. Falcone, C. Foti, O. Giuffrè and S. Materazzi, New J. Chem., 2014, 38, 3973–3983 RSC.
- S. Baeck, B. Kim, B. Cho and E. Kim, Forensic Sci. Int., 2019, 305, 109997 CrossRef CAS.
- F. Grotenthermen, Curr. Drug Targets: CNS Neurol. Disord., 2005, 4, 507–530 Search PubMed; K. Kurdziel, T. Głowiak, S. Materazzi and J. Jezierska, Polyhedron, 2003, 22, 3123–3128 CrossRef CAS; R. Mechoulam and L. Hanus, Chem. Phys. Lipids, 2000, 108, 1–13 CrossRef.
- A. S. Tsagkaris, J. L. D. Nelis, G. M. S. Ross, S. Jafari, J. Guercetti, K. Kopper, Y. Zhao, K. Rafferty, J. P. Salvador, D. Migliorelli, G. I. J. Salentijn, K. Campbell, M. P. Marco, C. T. Elliot, M. W. F. Nielen, J. Pulkrabova and J. Hajslova, Trends Anal. Chem., 2019, 121, 115688 CrossRef CAS; J. D. Dunn, C. M. Gryniewicz-Ruzicka, J. F. Kauffman, B. J. Westenberger and L. F. Buhse, J. Pharm. Biomed. Anal., 2011, 54, 469–474 CrossRef; R. Risoluti, S. Pichini, R. Pacifici and S. Materazzi, Talanta, 2019, 202, 546–553 CrossRef; R. Risoluti, G. Gullifa, A. Battistini and S. Materazzi, Anal. Chem., 2019, 91(10), 6435–6439 Search PubMed; R. Risoluti, M. A. Fabiano, G. Gullifa, S. Vecchio Ciprioti and S. Materazzi, Appl. Spectrosc. Rev., 2017, 52(1), 39–72 CrossRef; V. D'Elia, G. Montalvo, C. Garc, a Ruiz, V. V. Ermolenkov, Y. Ahmed and I. K. Lednev, Spectrochim. Acta, Part A, 2018, 188, 338–340 CrossRef; S. De Angelis Curtis, K. Kurdziel, S. Materazzi and S. Vecchio, J. Therm. Anal. Calorim., 2008, 92(1), 109–114 CrossRef.
- M. Hadener, S. Konig and W. Weinmann, Forensic Sci. Int., 2019, 299, 142–150 CrossRef CAS; L. A. Ciolino, T. L. Ranieri and A. M. Taylor, Forensic Sci. Int., 2018, 289, 438–447 CrossRef; V. Migliorati, P. Ballirano, L. Gontrani, S. Materazzi, F. Ceccacci and R. Caminiti, J. Phys. Chem. B, 2013, 117, 7806–7818 CrossRef; B. Patel, D. Wene and Z. T. Fan, J. Pharm. Biomed. Anal., 2017, 146, 15–23 CrossRef; A. Barattucci, M. L. Di Gioia, A. Leggio, L. Minuti, T. Papalia, C. Siciliano, A. Temperini and P. Bonaccorsi, Eur. J. Org. Chem., 2014, 10, 2099–2104 CrossRef.
- M. Pellegrini, E. Marchei, R. Pacifici and S. Pichini, J. Pharm. Biomed. Anal., 2005, 36, 939–946 CrossRef CAS; L. Minuti, E. Ballerini, A. Barattucci, P. M. Bonaccorsi, M. L. Di Gioia, A. Leggio, C. Siciliano and A. Temperini, Tetrahedron, 2015, 71(21), 3253–3262 CrossRef.
- W. Peschel and M. Politi, Talanta, 2015, 140, 150–165 CrossRef CAS; O. Jovic, K. Piculjan, T. Hrenar, T. Smolic and I. Primozic, Chemom. Intell. Lab. Syst., 2019, 185, 41–46 CrossRef.
- J. Véstia, J. M. Barros, H. Ferreira, L. Gaspar and A. E. Rato, Food Chem., 2019, 276, 71–76 CrossRef CAS; S. Materazzi, R. Risoluti, S. Pinci and F. S. Romolo, Talanta, 2017, 174, 673–678 CrossRef; P. Oliveri, V. Di Egidio, T. Woodcock and G. Downey, Food Chem., 2011, 125, 1450–1456 CrossRef; S. Materazzi, A. Gregori, L. Ripani, A. Apriceno and R. Risoluti, Talanta, 2017, 166, 328–335 CrossRef.
- C. Meesa, F. Souard, C. Delported, E. Deconinck, P. Stoffelen, C. Stévigny, J. F. Kauffmann and K. De Braekeleer, Talanta, 2018, 177, 4–11 CrossRef CAS; R. Risoluti, D. Piazzese, A. Napoli and S. Materazzi, J. Anal. Appl. Pyrolysis, 2016, 117, 82–87 CrossRef; S. Materazzi and R. Risoluti, Appl. Spectrosc. Rev., 2014, 49(8), 635–665 CrossRef; B. Kordi, M. Kovacevi, T. Slobod, A. Vidovi and B. Jovi, J. Mol. Struct., 2017, 1144, 159–165 CrossRef.
- E. M. Paiva, J. J. R. Rohwedder, C. Pasquini, M. F. Pimentel and C. F. Pereira, Fuel, 2015, 160, 57–63 CrossRef CAS; R. Risoluti, A. Gregori, S. Schiavone and S. Materazzi, Anal. Chem., 2018, 90, 4288–4292 CrossRef; S. Modrono, A. Soldado, A. Martínez-Fernández and B. de la Roza Delgado, Talanta, 2017, 162, 597–603 CrossRef; S. Materazzi, S. Vecchio, L. W. Wo and S. De Angelis Curtis, Thermochim. Acta, 2012, 543, 183–187 CrossRef; R. Risoluti and S. Materazzi, Front. Chem., 2018, 6, 228 CrossRef; K. N. Basri, M. N. Hussain, J. Bakar, M. F. A. Khir and A. S. Zoolfakar, Spectrochim. Acta, Part A, 2017, 173, 335–342 CrossRef; N. C. da Silva, C. J. Cavalcanti, F. A. Honorato, J. M. Amigo and M. F. Pimentel, Anal. Chim. Acta, 2017, 954, 32–42 CrossRef.
- R. J. Barnes, M. S. Dhanoa and S. J. Lister, Appl. Spectrosc., 1989, 43, 772–777 CrossRef CAS; M. A. Navarra, S. Materazzi, S. Panero and B. Scrosati, J. Electrochem. Soc., 2003, 150, A1528–A1532 CrossRef.
- P. Geladi, D. MacDougall and H. Martens, Appl. Spectrosc., 1985, 39, 491–500 CrossRef.
- S. Wold and M. Sjöström, in Chemometrics: Theory and Applications, ed. B. R. Kowalski, American Chemical Society Symposium Series, 1977, vol. 52, pp. 243A–282A Search PubMed.
- M. J. Savitzky and E. Golay, Anal. Chem., 1964, 36, 1627–1639 CrossRef.
- A. Rinnan, F. van den Berg and S. B. Engelsen, Trends Anal. Chem., 2009, 28, 10 CrossRef.
- S. Pichini and R. Pacifici, Linee guida per la determinazione di sostanza d'abuso nella saliva, Istituto Superiore di Sanità, 2013 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra07142k |
| This journal is © The Royal Society of Chemistry 2020 |