 Open Access Article
Open Access ArticleEnantioselective sulfoxidation using Streptomyces glaucescens GLA.0†
Sara Salama a,
Tarek Dishisha
a,
Tarek Dishisha b,
Mohamed H. Habib
b,
Mohamed H. Habib c,
Ahmed Z. Abdelazem
c,
Ahmed Z. Abdelazem a,
Walid Bakeer
a,
Walid Bakeer b,
Mahmoud Abdel-Latif
b,
Mahmoud Abdel-Latif d and
Yasser Gaber
d and
Yasser Gaber *be
*be
aBiotechnology and Life Sciences Department, Faculty of Postgraduate Studies for Advanced Sciences, Beni-Suef University, Beni-Suef, 62517, Egypt. E-mail: sarasalama@psas.bsu.edu.eg; aznagi@psas.bsu.edu.eg
bDepartment of Microbiology and Immunology, Faculty of Pharmacy, Beni-Suef University, Beni-Suef, 62511, Egypt. E-mail: tarek.dishisha@pharm.bsu.edu.eg; walid.bakeer@yahoo.com; yasser.gaber@pharm.bsu.edu.eg
cDepartment of Microbiology and Immunology, Faculty of Pharmacy, Cairo University, Cairo, 11562, Egypt. E-mail: mohamed.habib@pharma.cu.edu.eg
dImmunity Division, Zoology Department, Faculty of Science, Beni-Suef University, Beni-Suef, 62511, Egypt. E-mail: mahmoudsayed@science.bsu.edu.eg
eDepartment of Pharmaceutics and Pharmaceutical Technology, College of Pharmacy, Mutah University, Al-Karak, 61710, Jordan
First published on 1st September 2020
Abstract
Asymmetric oxidation of prochiral sulfides is a direct means for production of enantiopure sulfoxides which are important in organic synthesis and the pharmaceutical industry. In the present study, Streptomyces glaucescens GLA.0 was employed for stereoselective oxidation of prochiral sulfides. Growing cells selectively catalyzed the oxidation of phenyl methyl sulfide to the corresponding sulfoxide. Only very little overoxidation was observed, resulting in minor amounts of the unwanted sulfone. Addition of isopropyl alcohol as a co-solvent, time of substrate addition and composition of the reaction media resulted in enhanced phenyl methyl sulfide biotransformation. The concentration of the undesired by-product (sulfone) was as low as 4% through the reaction course under optimal reaction conditions. The results show that S. glaucescens GLA.0 is a promising whole-cell biocatalyst for preparing highly enantiopure (R)-phenyl methyl sulfoxide in high yield (90%) with an enantiomeric excess (ee) exceeding 99%.
1. Introduction
Chiral sulfoxides possess several applications in chemical and pharmaceutical industries. They are useful synthons, valuable reagents and precursors for bioactive compounds. However, it is necessary to obtain them in an enantiomeric pure form, since usually only one enantiomer exerts the desired biological activity.1,2 Esomeprazole is one of the most important examples of the biocatalyzed sulfoxidation using whole cells in the pharmaceutical industry. In this case, only the S-enantiomer acts as a proton pump inhibitor and is used to treat gastric ulcers and gastroesophageal disorders.3 Therefore, the stereoselective oxidation of organic sulfides, the precursors for chiral sulfoxides, is receiving a growing interest nowadays.4,5Optically pure sulfoxides can be prepared via chemical or biological catalysis. Chemical catalysis uses transition metals or organic catalysts and is limited by high cost of isolation and poor enantioselectivity. Likewise, biological catalysis using purified enzymes is costly (enzyme purification and co-factor dependency) and often suffers from poor protein stability. On the other hand, the use of whole cells, lysed cells, or cellular extracts as catalysts for asymmetric sulfoxidation reactions is a more attractive and a cheaper alternative since it avoids the need for adding expensive cofactors.3 Fungi are widely used for catalyzing oxidation reactions compared to bacteria.6–8 However, it was indicated that bacteria belonging to the genus Gordonia, Rhodococcus and Pseudomonas possess fairly high activity and enantioselectivity for a large number of sulfides.3,9–11 Actinobacteria have been exploited successfully in the search for novel biocatalysts.12,13 However, as reported earlier, the ability of enantioselective oxidation of organic sulfides was only studied in a limited number of Actinobacteria.9,14–17
Enantioenriched phenyl methyl sulfoxide (PMS) could be produced by various biotechnological approaches. These approaches include the use of isolated enzymes or biotransformation in microbial cultures using whole cells either of native hosts or recombinant systems. These methods depend on either asymmetric oxidation of PMS or kinetic resolution of the racemic sulfoxide.18 The formation of R-phenyl methyl sulfoxide was favored through biotransformation of the respective substrate (PMS) by the growing cells of Aspergillus japonicus ICFC 744/11.6 Two examples of microbial transformations of PMS were recorded using resting cells of Pseudomonas monteilii CCTCC M2013683 and using immobilized cells of Gordonia terrae IEGM 136. The first could catalyze the formation of R-PMSO in a yield of 99.0% with an ee >99.0%. The latter, however, produced a yield of 100% with an ee >95.0%.11,19 S-PMSO formation was effectively catalyzed by using whole-cells of Rhodococcus sp. CCZU10-1 in biphasic systems of n-octane/water using fed-batch fermentation. This conversion produced high yield of the S-sulfoxide (91%) with an ee >99.0%.20 The isooctane-aqueous system was applied using the resting cells of Rhodococcus sp. ECU0066 to successfully catalyze S-PMSO production in high yield (82%) and ee >99.0%.21 In addition, resting cells of Rhodococcus sp. ECU0066 catalyzed the bio-resolution of the racemic PMSO to produce S-PMSO in a concentration of 37.8 mM and ee of 93.7% in fed-batch reactions.22 Concerning recombinant whole-cell catalysis, recently, AbIMO, a flavoprotein monooxygenase from Acinetobacter baylyi ADP1 was heterologously expressed and purified in E. coli and then examined for its sulfoxidation activity.23 The system produced S-PMSO at 73% yield and high ee (>99.0%) via continuous substrate feeding strategy.
Regarding biotransformation using isolated enzymes, various types of oxidoreductase enzymes such as monooxygenases, dioxygenases and Baeyer–Villiger monooxygenases (BVMOs) have been exploited in the synthesis of enantioenriched PMSO.24 The R isomer was prepared in high yield (90.0%) and high ee (>98.0%) by toluene dioxygenase (TDO EC 1.4.12.11) purified from Pseudomonas putida F1.18 It was also prepared in high yield (97.0%) and high ee (>91.0%) from polycyclic ketone monooxygenase (PockeMO) isolated from Thermothelomyces thermophila.25 One of the BVMOs expressed from Rhodococcus jostii RHA1 converted PMS to R-PMSO in ee of 82.0% while four other BVMOs catalyzed the formation of the S-isomer in ee of 90.0%.26 The BVMO 4-hydroxyacetophenone monooxygenase purified from Pseudomonas fluorescens ACG is one of the enzymes that catalyzed the formation of S-isomer in ee exceeding 99.0%.27
In this work, S. glaucescens GLA.0, a soil actinobacterium that is not fully characterized in terms of its biocatalytic properties, was shown to be a valuable whole-cell biocatalyst with the ability to enantioselectively oxidize aromatic prochiral sulfides. Phenyl methyl sulfide was used during the experiments as a model substrate. Moreover, we studied some of the influencing factors on the reaction yield and selectivity. This included the use of isopropyl alcohol as a co-solvent, the time of substrate addition to the biotransformation media, and the composition of the biotransformation media.
2. Materials and methods
2.1 Chemicals
Phenyl methyl sulfide (99%) was purchased from Alfa Aesar (Kandel, Germany), cyclohexanol and malt extract were purchased from Loba-Chemie (Mumbai, India), while yeast extract (YE) was from Difco (Detroit, MI, USA). Ammonium sulfate, magnesium sulfate heptahydrate, ferrous sulfate heptahydrate, disodium hydrogen phosphate dihydrate, dipotassium hydrogen phosphate, potassium dihydrogen phosphate and calcium chloride dihydrate were obtained from El Nasr Pharmaceutical Chemicals Company (Cairo, Egypt). Isopropyl alcohol (IPA), ethyl alcohol, ethyl acetate, glycerol and glucose were procured from Sigma-Aldrich (St. Louis, MO, USA). The enzymes phosphite dehydrogenase, cyclohexanone monooxygenase and phenylacetone monooxygenase were obtained from GECCO-biotech (Groningen, The Netherlands).Phenyl methyl sulfoxide (PMS) and phenyl methyl sulfone (PMSO) were synthesized by chemical oxidation of phenyl methyl sulfide as described elsewhere28 and purified by silica column chromatography. A mixture of n-hexane and ethyl acetate at a ratio (1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and (2
1) and (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) were used for the elution of PMS and PMSO, respectively. Both purified compounds were used as reference standards to identify the products by retention times and characteristic fragmentation patterns. Also, FTIR analysis (VERTEX 70v, Bruker, Ettlingen, Germany) was performed on the synthesized standards to confirm their identity.
3) were used for the elution of PMS and PMSO, respectively. Both purified compounds were used as reference standards to identify the products by retention times and characteristic fragmentation patterns. Also, FTIR analysis (VERTEX 70v, Bruker, Ettlingen, Germany) was performed on the synthesized standards to confirm their identity.
2.2 Culture media
ISP2 medium was prepared as follows (per liter): 4 g YE, 4 g glucose, 10 g malt extract, 2 g CaCO3 and 20 g agar, pH 7.2. The medium was used for initial culturing and maintenance. In case of the preparation of liquid medium, agar and CaCO3 were excluded. Medium 2 was prepared as follows (per liter): 2 g (NH4)2SO4, 2 g K2HPO4, 4 g YE, 50 mL of 10% glycerol, 0.01 g CaCl2·2H2O, 0.01 g FeSO4·7H2O and 0.1 g MgSO4·7H2O, pH 7.5. Medium 3 contained per liter: 3 g (NH4)2SO4, 5 g Na2HPO4·2H2O, 2 g KH2PO4, 0.2 g YE, 50 mL of 10% glycerol, 0.1 g CaCl2·2H2O, 5 g MgSO4·7H2O and 1 g FeSO4·7H2O, pH 7.1. These media were used for biotransformation experiments. Salts were sterilized separately through 0.22 μm syringe filters and added later to the autoclaved culture media.2.3 Microorganism, storage and culture conditions
Streptomyces glaucescens GLA.0 (DSM 14766) was purchased, in a lyophilized form, from DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen), Germany. The lyophilized powder was resuspended in 1 mL of sterile distilled water and then 200 μL of the bacterial suspension was spread over ISP2 agar plate. The cultures were incubated at 30 °C for 4 days. Subsequently, two colonies on the agar plates were aseptically transferred to 10 mL of ISP2 medium in 50 mL Falcon tubes and then incubated at 30 °C for 4 days. The resulting culture was used to prepare glycerol stocks (50% v/v) that were stored at −20 °C.For refreshment of the microorganism prior to biotransformation process, 50 μL of the glycerol stock was aseptically transferred to 5 mL of ISP2 liquid medium and incubated at 30 °C and 150 rpm on a shaker incubator (SCILOGEX, SK-O330-PRO, USA) for 48 h. 50 μL of the resulting culture were spread on ISP2 agar plates, supplemented with 2.5 mL L−1 of Nystatin (2.5 × 105 IU L−1) as a selection agent, and incubated at 30 °C for 3 days. Subsequently, 2 colonies were transferred to 5 mL of sterile ISP2 liquid medium supplemented with Nystatin and incubated at 30 °C for 24 hours. 50 μL were again transferred to ISP2 agar plate supplemented with Nystatin and incubated at 30 °C for 3 days. The fresh ISP2 agar plates were used directly for subsequent biotransformation processes.
2.4 Biotransformation of phenyl methyl sulfide
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10). The medium was inoculated with 2 colonies from a freshly prepared ISP2 agar plate and incubated at 30 °C and 150 rpm in a shaker incubator (Lab TECH LSI-30 16R, Korea) for 48 h. The OD600 nm after 48 h was 0.6. Cells were harvested by centrifugation at 10
10). The medium was inoculated with 2 colonies from a freshly prepared ISP2 agar plate and incubated at 30 °C and 150 rpm in a shaker incubator (Lab TECH LSI-30 16R, Korea) for 48 h. The OD600 nm after 48 h was 0.6. Cells were harvested by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min at room temperature and washed twice with 50 mM sodium phosphate buffer, pH 7.4. Cells were then resuspended in the same buffer to a final optical density of 3.8 at 600 nm (equivalent to 0.14 gww mL−1).
000g for 10 min at room temperature and washed twice with 50 mM sodium phosphate buffer, pH 7.4. Cells were then resuspended in the same buffer to a final optical density of 3.8 at 600 nm (equivalent to 0.14 gww mL−1).| Reaction | Substrate conc. (mM) | IPA conc. (% v/v) | Reaction medium | RPM | Time of sub. additiona (day) | Sampling time (h) | PMSOb (%) | PMSO/sulfone | ee (%), configurationc |
|---|---|---|---|---|---|---|---|---|---|
| a Addition of the substrate at zero time of inoculation or on the 3rd day of inoculation.b Calculated as the relative peak area of PMSO to the sum of the peak areas of PMS, PMSO and sulfone as calculated by GC-MS.c Configurations and (ee%) enantiomeric excess values were analyzed by chiral GC-FID; ND – not determined. | |||||||||
| A | 1 | 0.2 | Medium 3 | 150 | 3rd | 168 | 78 | 4 | ND |
| B | 1 | 0 | Medium 3 | 150 | 3rd | 168 | 75 | 3 | ND |
| C | 1 | 0 | Medium 3 | 180 | 3rd | 120 | 83 | 5 | >99 (R) |
| D | 1 | 0 | Medium 3 | 180 | Zero | 120 | 83 | 6 | >99 (R) |
| E | 1 | 0.2 | Medium 2 | 150 | Zero | 72 | 79 | 7 | ND |
| F | 1 | 0.2 | Medium 3 | 150 | Zero | 144 | 71 | 3 | ND |
| G | 5 | 1 | Medium 3 | 150 | Zero | 72 | 76 | 5 | ND |
| H | 5 | 1 | Medium 2 | 150 | Zero | 120 | 83 | 7 | ND |
| I | 1 | 0.2 | Medium 3 | 180 | Zero | 120 | 81 | 4 | >99 (R) |
| J | 1 | 0.2 | Medium 2 | 180 | Zero | 96 | 90 | 23 | >99 (R) |
| K | 5 | 1 | Medium 2 | 180 | Zero | 120 | 89 | 9 | >99 (R) |
The different optimization experiments were performed under the following conditions: (1) bacterial cells at a relatively low starting inoculum concentration (2.65 ± 0.2 gww L−1), (2) PMS pre-dissolved in IPA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 v/v) and (3) PMS added at inoculation time. These conditions were selected based on the results of the preliminary experiments. The preculture was used to inoculate the biotransformation medium, as described above, and PMS pre-dissolved in IPA (1
10 v/v) and (3) PMS added at inoculation time. These conditions were selected based on the results of the preliminary experiments. The preculture was used to inoculate the biotransformation medium, as described above, and PMS pre-dissolved in IPA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) was subsequently added. The concentration of the co-solvent differed (0.2–1%) according to the initial concentrations of PMS (1–5 mM), respectively. For reactions E–H, the flasks were incubated at 30 °C and 150 rpm in a shaker incubator for 168 h. However, reactions I–K were incubated at 180 rpm for 120 h. Samples were collected at different intervals and analyzed for product yield and percentage conversion by GC-MS.
10) was subsequently added. The concentration of the co-solvent differed (0.2–1%) according to the initial concentrations of PMS (1–5 mM), respectively. For reactions E–H, the flasks were incubated at 30 °C and 150 rpm in a shaker incubator for 168 h. However, reactions I–K were incubated at 180 rpm for 120 h. Samples were collected at different intervals and analyzed for product yield and percentage conversion by GC-MS.
In all cases, the product yield and substrate conversion were calculated using the corrected peak areas obtained from the GC/MS Selective Ion Monitoring (SIM) mode analysis using the following equations:
 | (1) |
 | (2) |
Enantioselectivity (configuration and enantiomeric excess) was analyzed by using a chiral GC column. The enantiomeric excess (ee) was calculated using the peak areas according to the following equation:
 | (3) |
2.5 GC-MS analyses
The products resulted from the biotransformation reaction were extracted twice with ethyl acetate (2 × 0.5 mL), dried over anhydrous Na2SO4, filtered and used directly for GC analysis. A gas chromatography-mass spectrometry analysis of the products was performed using an Agilent 7890A GC equipped with a 5975 Inert MS with Triple Axis Detector using a (0.25 mm × 30 m) HP-5MS capillary column with 0.25 μm film thickness (Agilent, Germany). The MS was fixed at 70 eV ionization energy with mass electron (m/z) range of 50–550. The split-less mode was used in the injection of 1 μL sample solution and helium was used as a carrier gas with a flow rate of 1 mL min−1. The GC oven temperature was programmed to hold at 70 °C for 2 min, then heated to 140 °C with a rate of 25 °C min−1. Finally, the column was heated to 180 °C with a rate of 5 °C min−1. Retention times of phenyl methyl sulfide, phenyl methyl sulfoxide and phenyl methyl sulfone were 5.1 min, 7.5 min and 8.3 min, respectively. For confirmation of GC-MS analysis, a copy of samples from experiments performed once (reactions C, D, I, J and K) were analyzed with HP-1 column (Agilent, 30 m × 0.25 mm × 0.25 μm). The GC oven was programmed to hold at 40 °C for 2 min with an injection temperature of 300 °C. It was then heated to 100 °C with a rate of 5 °C min−1. Finally, the temperature was ramped at 10 °C min−1 until it reached 250 °C and held for 10 min. The split ratio was 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Retention times of phenyl methyl sulfide, phenyl methyl sulfoxide and phenyl methyl sulfone were 13.1 min, 19 min and 20.2 min, respectively.
1. Retention times of phenyl methyl sulfide, phenyl methyl sulfoxide and phenyl methyl sulfone were 13.1 min, 19 min and 20.2 min, respectively.
2.6 Chiral GC analyses
Chiral GC analyses were performed using a Chiraldex G-TA (Grace Alltech number 4139, 30 m × 0.25 mm × 0.125 μm) column equipped with FID-Detector (Max temp: 180 °C). The initial temperature was adjusted at 50 °C. The column was then heated to 170 °C with a rate of 10 °C min−1 and the temperature was held for 17 min. The temperature was finally ramped at 10 °C min−1 until it reached 50 °C at a final working time of 40 min. The split ratio was 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with a flow rate of 3 mL min−1. The retention time of R-enantiomer, S-enantiomer and phenyl methyl sulfone were 15.7 min, 18.5 min and 18.9 min, respectively. To confirm the identity of the enantiomers obtained in the whole-cell biotransformation reactions with PMS as a substrate, the latter was oxidized to the corresponding sulfoxide by enzymatic reaction using phenylacetone monooxygenase (PAMO) and cyclohexanone monooxygenase (CHMO). The peaks corresponding to the products formed were used as positive controls to identify different enantiomers produced by this reaction.29,30
1 with a flow rate of 3 mL min−1. The retention time of R-enantiomer, S-enantiomer and phenyl methyl sulfone were 15.7 min, 18.5 min and 18.9 min, respectively. To confirm the identity of the enantiomers obtained in the whole-cell biotransformation reactions with PMS as a substrate, the latter was oxidized to the corresponding sulfoxide by enzymatic reaction using phenylacetone monooxygenase (PAMO) and cyclohexanone monooxygenase (CHMO). The peaks corresponding to the products formed were used as positive controls to identify different enantiomers produced by this reaction.29,30
2.7 Standard enzymatic synthesis of R-phenyl methyl sulfoxide
Synthesis of standard PMSO was done according to Dudek et al.30 Briefly, phenyl acetone monooxygenase (PAMO) or cyclohexanone monooxygenase (CHMO) (2 μM), phenyl methyl sulfide (3.4 mM), NADPH (150 μM), phosphite (20 mM) and phosphite dehydrogenase (PTDH) (2 μM) in 50 mM Tris–HCl at pH 7.0 were added to a 20 mL glass vial. The vial was placed in a 24 °C/135 rpm shaker for 18 h. The reaction volume was extracted twice using 500 μL ethyl acetate and the organic layer separated. The ethyl acetate fraction was collected and dried over anhydrous MgSO4. The samples were then injected into a GC using a chiral column for separation as detailed above.2.8 Statistical analysis
All the experiments were performed as analytical replicates. The presented data are the mean of two measurements. Control reactions and analyses were performed (Fig. S4–S6†).3. Results and discussion
3.1 Parameters affecting the biotransformation reaction
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 v/v).31 The cells were then transferred to biotransformation medium (Medium 3) where the main product was detected after 15 h (Fig. S9†) and 6 h (Fig. S10†) in experiments using 3 mM and 5 mM of PMS, respectively. Subsequently, a set of experiments (Table 1: reactions A, B, C and D) were run and followed for up to 120 h.
10 v/v).31 The cells were then transferred to biotransformation medium (Medium 3) where the main product was detected after 15 h (Fig. S9†) and 6 h (Fig. S10†) in experiments using 3 mM and 5 mM of PMS, respectively. Subsequently, a set of experiments (Table 1: reactions A, B, C and D) were run and followed for up to 120 h. | ||
| Fig. 1 Time courses of phenyl methyl sulfide biotransformation with the growing cells of Streptomyces glaucescens GLA.0 under different conditions; reactions D, E, F, G, H, I, J and K were performed as described in materials and methods and differences in conditions among reactions are stated in Table 1. The ee values are shown only if it is in favor of R-isomer formation. Values >99% are shown as 99% in the plot. The detailed reaction conversion values are provided in the ESI.† | ||
 | ||
| Fig. 2 Chiral GC chromatograms showing, (A) standard phenyl methyl sulfide (Rt = 9.4 min), (B) standard R-phenyl methyl sulfoxide (Rt = 15.69), (C) R-phenyl methyl sulfoxide (Rt = 15.7 min) produced after 96 hours of the substrate addition to the reaction media. PMS (1 mM) was added at zero time of inoculation to Medium 2 and mixing speed was kept at 180 rpm throughout the whole process (reaction J, Table 1). The analysis details are found in the material and methods under Section (2.6). | ||
The relation of bacterial cell density to PMSO/sulfone ratio was analyzed based on the data recorded for some reactions (Fig. 3). The wet weights of bacterial biomass were determined for each sampling time for the different reactions. Several factors are expected to contribute to the PMSO/sulfone ratio including the initial concentration of substrate used, the composition of medium used and hence the bacterial cell density obtained. Reactions G and H differed in the biotransformation medium used. Reaction H was run using yeast-containing medium (Medium 2) and as a result, high bacterial cell concentration and better PMSO/sulfone ratio was obtained compared to reaction G (Fig. 3). The yield of sulfoxide produced in reaction H was 83%. When Medium 3 was used in reaction G, the cell concentration dropped significantly. This led to a lower PMSO yield (54% at 168 h). This drop in sulfoxide level coincides with the reduction of cell concentration over time. Due to the possible toxic effect of using a high initial substrate concentration, reactions starting with 5 mM substrate resulted in cell concentrations not exceeding 0.059 g mL−1. The highest cell density recorded was for reactions that started with 1 mM substrate concentration; reactions B, E and F (Fig. 3). Cell density around 0.07 g mL−1 seems to be the optimum value for getting high PMSO/sulfone ratio as was noticed in reactions B and F.
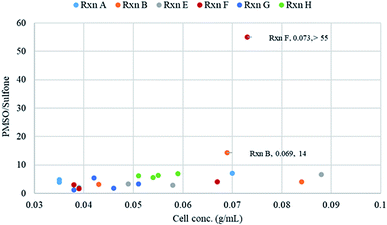 | ||
| Fig. 3 Effect of final concentrations of S. glaucescens GLA.0 growing cells on PMSO/sulfone ratio. Reactions A, B, E, F, G and H were performed in duplicates as stated in materials and methods and differences in conditions among reactions are stated in Table 1. Wet weights of cells were determined in 1 mL samples taken at each interval. Relative amount of phenyl methyl sulfoxide and phenyl methyl sulfone in reaction samples was calculated using GC-MS. | ||
In literature, the use of high cell concentration of Rhodococcus sp. ECU0066 favored the overoxidation of sulfoxide to sulfone.21 No sulfone reduction activity was recorded in control reactions.22 It was reported that PMSO shows lower biotoxicity compared to PMS which may be due to the hydrophobic nature of the latter as stated before.22,37,38 Also, phenyl methyl sulfone was reported to be one of the pesticide metabolites that contribute to potential toxicity to soil bacteria.39 As a result, it is thought to induce the reductase system and undergo deoxygenation once configured. Notably, such a bio-reduction reaction has been reported in literature for some sulfone compounds.40 S. glaucescens GLA.0 bacterial cells maintained their viability and growth by triggering high level of PMSO production at the expense of both the substrate and the by-product. This can be explained by the fact that PMSO is not as toxic compared to the substrate (PMS) and the by-product (phenyl methyl sulfone). In addition, (S)-PMSO could preferentially be oxidized to sulfone and that enabled the R enantiomer to be accumulated predominantly, hence retained in high ee (Scheme 1).
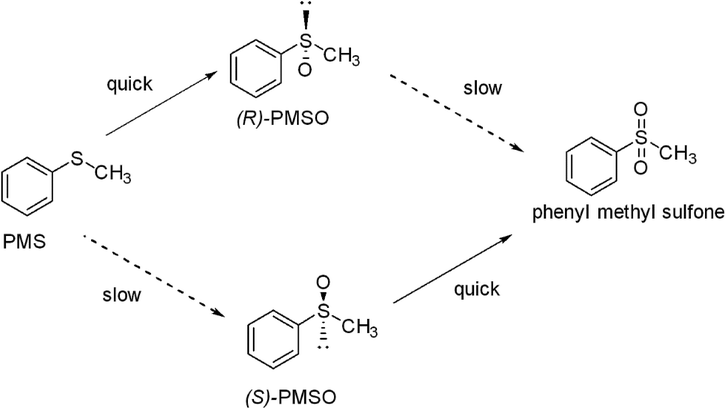 | ||
| Scheme 1 The putative pathway for the enantioselective bio-oxidation of phenyl methyl sulfide (PMS) to R-phenyl methyl sulfoxide and S-phenyl methyl sulfoxide with formation of phenyl methyl sulfone as a by-product as described in Zambianchi et al. 2007 (ref. 50) with modification.19 PMS was predominantly oxidized to R-phenyl methyl sulfoxide utilizing growing cells of Streptomyces glaucescens GLA.0 under standard conditions. Dashed arrows indicate slow reaction rate. | ||
3.2 In silico analysis of putative BVMOs in S. glaucescens GLA.0
To identify the putative BVMO in the genome of S. glaucescens GLA.0, we used the sequence of Streptomyces coelicolor BVMO Q9RKB5 to do a BLAST search. The BLAST search retrieved one putative BVMO in the genome of S. glaucescens.41–43 The putative sequence Uniprot Accession number is A0A089YZ45 (Fig. S1†). The sequence is composed of 514 amino acids, which lies in the typical known BVMOs length range (500–550 amino acids).44 The alignment revealed the presence of Rossmann fold domains (GXGXXG, GG, ATG) required for binding of FAD and NADPH.41,45 The two BVMO-specific fingerprints could be identified: FXGXXXHXXXWD and GGXWXXXXYPGXXXD. The conserved regions are flanked by the two Rossmann fold sequence motifs (GXGXX[G/A]).37 This strongly suggests that this protein is a typical Type I BVMO. The alignment showed that it possesses 86% sequence identity with a BVMO from Streptomyces coelicolor A3(2) (UniProt Accession No. Q9RKB5). From BVMOs with known structures, it displays the highest sequence identity with steroid monooxygenase from Rhodococcus rhodochrous, another actinobacterium. Notably, BVMOs are well recognized as one of the enzymes catalyzing asymmetric sulfoxidation.183.3 Overview on S. glaucescens GLA.0
4. Conclusion
The current study provides a green method for the enantioselective sulfide oxidation using Streptomyces glaucescens GLA.0 as a whole-cell biocatalyst. The R enantiomeric sulfoxide was obtained almost as a single enantiomer using Streptomyces glaucescens GLA.0. The developed process involved the use of whole cells which eliminated the need for cofactor recycling. The process provided almost pure R-enantiomer of the corresponding sulfoxide (ee > 99%) with a very good yield (up to 90%). Moreover, the by-product (sulfone) was produced in small amounts. The optimal conditions involved the use of a tuned medium for cell growth and conversion, the addition of isopropyl alcohol as a co-solvent and a tuned timing of substrate addition. However, further optimization processes are required for biotransformation of high concentrations of PMS and for detection of substrate specificity. This also includes organic sulfides which possess importance as substrates in industrial and pharmaceutical applications. Other ideas such as bacterial cell immobilization, employment of resting cultures grown on carbon sources such as glycerol or hexadecane, application of batch fermentation and/or using biphasic systems are recommended for further research. The performance of Streptomyces glaucescens GLA.0 towards stereoselective sulfoxidation can rank this species among potential biocatalysts for the synthesis of optically pure sulfoxides on large scale application. This is due to S. glaucescens being one of the characteristic Actinobacteria allowing for targeted enantiomerically enriched (R)-phenyl methyl sulfoxide formation which will direct the focus of research to exploit bio-oxidative characteristics of Streptomyces strains that have not been fully investigated yet. This will result in a potentially wide spectra of biocatalysts conducting such bio-oxidative transformations under mild conditions.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to acknowledge Dr Milos Trajkovic (University of Groningen, The Netherlands) for support and help in chiral GC analysis.References
- I. Fernández and N. Khiar, Chem. Rev., 2003, 103, 3651–3706 CrossRef PubMed.
- P. Babiak, E. Kyslíková, V. Stěpánek, R. Valešová, A. Palyzová, H. Marešová, J. Hájíček and P. Kyslík, Bioresour. Technol., 2011, 102, 7621–7626 CrossRef CAS PubMed.
- F. Garzón-Posse, L. Becerra-Figueroa, J. Hernández-Arias and D. Gamba-Sánchez, Molecules, 2018, 23, 1265 CrossRef PubMed.
- E. Wojaczyńska and J. Wojaczyński, Chem. Rev., 2010, 110, 4303–4356 CrossRef.
- J. Barluenga and C. Valdés, Chem. Commun., 2005, 4891–4901 RSC.
- M. L. Mascotti, A. A. Orden, F. R. Bisogno, G. de Gonzalo and M. Kurina-Sanz, J. Mol. Catal. B: Enzym., 2012, 82, 32–36 CrossRef CAS.
- L. C. Ricci, J. V. Comasseto, L. H. Andrade, M. Capelari, Q. B. Cass and A. L. M. Porto, Enzyme Microb. Technol., 2005, 36, 937–946 CrossRef CAS.
- B. J. Auret, D. R. Boyd, H. B. Henbest, C. G. Watson, K. Balenović, V. Polak, V. Johanides and S. Divjak, Phytochemistry, 1974, 13, 65–68 CrossRef CAS.
- A. A. Elkin, T. I. Kylosova, V. V. Grishko and I. B. Ivshina, J. Mol. Catal. B: Enzym., 2013, 89, 82–85 CrossRef CAS.
- A.-T. Li, J.-D. Zhang, J.-H. Xu, W.-Y. Lu and G.-Q. Lin, Appl. Environ. Microbiol., 2009, 75, 551–556 CrossRef CAS PubMed.
- Y. Chen, J. Zhuo, D. Zheng, S. Tian and Z. Li, J. Mol. Catal. B: Enzym., 2014, 106, 100–104 CrossRef CAS.
- A. Gran-Scheuch, M. Trajkovic, L. Parra and M. W. Fraaije, Front. Microbiol., 2018, 9, 1609 CrossRef PubMed.
- D. Tischler, W. J. H. van Berkel and M. W. Fraaije, Front. Microbiol., 2019, 10, 800 CrossRef PubMed.
- H. Ohta, Y. Okamoto and G. Tsuchihashi, Chem. Lett., 1984, 13, 205–208 CrossRef.
- H. Ohta, Y. Okamoto and G. Tsuchihashi, Agric. Biol. Chem., 1985, 49, 671–676 CAS.
- H. L. Holland, F. M. Brown, A. Kerridge, P. Pienkos and J. Arensdor, J. Mol. Catal. B: Enzym., 2003, 22, 219–223 CrossRef CAS.
- M. L. Mascotti, M. A. Palazzolo, E. Lewkowicz and M. Kurina-Sanz, Biocatal. Agric. Biotechnol., 2013, 2, 399–402 CrossRef.
- W. Mączka, K. Wińska and M. Grabarczyk, Catalysts, 2018, 8, 624 CrossRef.
- T. I. Kylosova, A. A. Elkin, V. V. Grishko and I. B. Ivshina, J. Mol. Catal. B: Enzym., 2016, 123, 8–13 CrossRef CAS.
- Y.-C. He, C.-L. Ma, Z.-X. Yang, M. Zhou, Z. Xing, J.-T. Ma and H.-L. Yu, Appl. Microbiol. Biotechnol., 2013, 97, 10329–10337 CrossRef CAS PubMed.
- A.-T. Li, J.-D. Zhang, H.-L. Yu, J. Pan and J.-H. Xu, Process Biochem., 2011, 46, 689–694 CrossRef CAS.
- A.-T. Li, H.-L. Yu, J. Pan, J.-D. Zhang, J.-H. Xu and G.-Q. Lin, Bioresour. Technol., 2011, 102, 1537–1542 CrossRef CAS PubMed.
- C. Willrodt, J. A. D. Gröning, P. Nerke, R. Koch, A. Scholtissek, T. Heine, A. Schmid, B. Bühler and D. Tischler, ChemCatChem, 2020, 12, 1–9 CrossRef.
- G. de Gonzalo and A. Franconetti, Enzyme Microb. Technol., 2018, 113, 24–28 CrossRef CAS PubMed.
- G. de Gonzalo, M. J. L. J. Fürst and M. W. Fraaije, Catalysts, 2017, 7, 288 CrossRef.
- A. Riebel, H. M. Dudek, G. de Gonzalo, P. Stepniak, L. Rychlewski and M. W. Fraaije, Appl. Microbiol. Biotechnol., 2012, 95, 1479–1489 CrossRef CAS PubMed.
- G. de Gonzalo, D. E. T. Pazmino, G. Ottolina, M. W. Fraaije and G. Carrea, Tetrahedron: Asymmetry, 2006, 17, 130–135 CrossRef CAS.
- H. Firouzabadi, N. Iranpoor, A. A. Jafari and E. Riazymontazer, Adv. Synth. Catal., 2006, 348, 434–438 CrossRef CAS.
- E. Romero, J. R. G. Castellanos, A. Mattevi and M. W. Fraaije, Angew. Chem., Int. Ed., 2016, 55, 15852–15855 CrossRef CAS PubMed.
- H. M. Dudek, G. de Gonzalo, D. E. T. Pazmiño, P. Stępniak, L. S. Wyrwicz, L. Rychlewski and M. W. Fraaije, Appl. Environ. Microbiol., 2011, 77, 5730–5738 CrossRef CAS PubMed.
- A. A. El'kin, V. V. Grishko and I. B. Ivshina, Appl. Biochem. Microbiol., 2010, 46, 586–591 CrossRef.
- G. de Gonzalo, G. Ottolina, F. Zambianchi, M. W. Fraaije and G. Carrea, J. Mol. Catal. B: Enzym., 2006, 39, 91–97 CrossRef CAS.
- M. Sayed, T. Dishisha, W. F. Sayed, W. M. Salem, H. M. Temerk and S.-H. Pyo, Process Biochem., 2017, 63, 1–7 CrossRef CAS.
- M. Sayed, T. Dishisha, W. F. Sayed, W. M. Salem, H. A. Temerk and S.-H. Pyo, J. Biotechnol., 2016, 221, 62–69 CrossRef CAS PubMed.
- C. V. F. Baldwin and J. M. Woodley, Biotechnol. Bioeng., 2006, 95, 362–369 CrossRef CAS PubMed.
- C. Zhang, D. Wu and H. Ren, Sci. Rep., 2020, 10, 5959 CrossRef CAS PubMed.
- M. J. L. J. Fürst, A. Gran-Scheuch, F. S. Aalbers and M. W. Fraaije, ACS Catal., 2019, 9, 11207–11241 CrossRef.
- A. Amanullah, C. J. Hewitt, A. W. Nienow, C. Lee, M. Chartrain, B. C. Buckland, S. W. Drew and J. M. Woodley, Biotechnol. Bioeng., 2002, 80, 239–249 CrossRef CAS PubMed.
- L. Somasundaram, J. R. Coats, K. D. Racke and H. M. Stahr, Bull. Environ. Contam. Toxicol., 1990, 44, 254–259 CrossRef CAS PubMed.
- J. Kazumi and D. G. Capone, Appl. Environ. Microbiol., 1995, 61, 2820–2829 CrossRef CAS PubMed.
- M. W. Fraaije, J. Wu, D. P. H. M. Heuts, E. W. van Hellemond, J. H. L. Spelberg and D. B. Janssen, Appl. Microbiol. Biotechnol., 2005, 66, 393–400 CrossRef CAS PubMed.
- J. Park, D. Kim, S. Kim, J. Kim, K. Bae and C. Lee, J. Microbiol. Biotechnol., 2007, 17, 1083–1089 CAS.
- M. W. Fraaije, N. M. Kamerbeek, W. J. H. van Berkel and D. B. Janssen, FEBS Lett., 2002, 518, 43–47 CrossRef CAS.
- C. Szolkowy, L. D. Eltis, N. C. Bruce and G. Grogan, ChemBioChem, 2009, 10, 1208–1217 CrossRef CAS PubMed.
- O. Vallon, Proteins: Struct., Funct., Bioinf., 2000, 38, 95–114 CrossRef CAS.
- T. Gul, M. Krzek, H. P. Permentier, M. W. Fraaije and R. Bischoff, Drug Metab. Dispos., 2016, 44, 1270–1276 CrossRef CAS.
- D. C. Lamb, M. R. Waterman and B. Zhao, Expert Opin. Drug Metab. Toxicol., 2013, 9, 1279–1294 CrossRef CAS PubMed.
- T. Matsui, Y. Dekishima and M. Ueda, Appl. Microbiol. Biotechnol., 2014, 98, 7699–7706 CrossRef CAS.
- B. Shen and C. R. Hutchinson, Biochemistry, 1993, 32, 6656–6663 CrossRef CAS.
- F. Zambianchi, M. W. Fraaije, G. Carrea, G. de Gonzalo, C. Rodríguez, V. Gotor and G. Ottolina, Adv. Synth. Catal., 2007, 349, 1327–1331 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra05838f |
| This journal is © The Royal Society of Chemistry 2020 |
