Open Access Article
Open Access ArticleCurcumin loaded zinc oxide nanoparticles for activity-enhanced antibacterial and anticancer applications†
W. P. T. D. Perera ab,
Ranga K. Dissanayake
ab,
Ranga K. Dissanayake *bc,
U. I. Ranatungad,
N. M. Hettiarachchiab,
K. D. C. Perera
*bc,
U. I. Ranatungad,
N. M. Hettiarachchiab,
K. D. C. Perera ab,
Janitha M. Unagollae,
R. T. De Silvab and
L. R. Pahalagedarab
ab,
Janitha M. Unagollae,
R. T. De Silvab and
L. R. Pahalagedarab
aAcademy of the Sri Lanka Institute of Nanotechnology, Nanotechnology and Science Park, Mahenwatte, Pitipana, Homagama 10206, Sri Lanka
bSri Lanka Institute of Nanotechnology, Nanotechnology and Science Park, Mahenwatte, Pitipana, Homagama 10206, Sri Lanka
cDepartment of Pharmacy and Pharmaceutical Sciences, Faculty of Allied Health Sciences, University of Sri Jayewardenepura, Gangodawila, Nugegoda 10250, Sri Lanka. E-mail: rangad@sjp.ac.lk
dDepartment of Biochemistry and Molecular Biology, Faculty of Medicine, University of Colombo, 25 Kynsey Road, Colombo 00800, Sri Lanka
eBiomedical Engineering Program, Department of Bioengineering, College of Engineering, University of Toledo, Toledo, OH 43607, USA
First published on 19th August 2020
Abstract
Zinc oxide nanoparticles and curcumin have been shown to be excellent antimicrobial agents and promising anticancer agents, both on their own as well as in combination. Together, they have potential as alternatives/supplements to antibiotics and traditional anticancer drugs. In this study, different morphologies of zinc oxide-grafted curcumin nanocomposites (ZNP–Cs) were synthesized and characterized using SEM, TGA, FTIR, XRD and UV-vis spectrophotometry. Antimicrobial assays were conducted against both Gram negative and Gram-positive bacterial stains. Spherical ZnO–curcumin nanoparticles (SZNP–Cs) and rod-shaped ZnO–curcumin nanoparticles showed the most promising activity against tested bacterial strains. The inhibition zones for these curcumin-loaded ZnO nanocomposites were consistently larger than their bare counterparts or pure curcumin, revealing an additve effect between the ZnO and curcumin components. The potential anticancer activity of the synthesized nanocomposites was studied on the rhabdomyosarcoma RD cell line via MTT assay, while their cytotoxic effects were tested against human embryonic kidney cells using the resazurin assay. SZNP–Cs exhibited the best balance between the two, showing the lowest toxicity against healthy cells and good anticancer activity. The results of this investigation demonstrate that the nanomatrix synthesized can act as an effective, additively-enhanced combination delivery/therapeutic agent, holding promise for anticancer therapy and other biomedical applications.
Introduction
Medicinal plants serve as nature's gift to humanity, aiding in the pursuit for better health; plants and their bioactive metabolites have been used in medicinal practices around the world since antiquity.1 Phytochemicals have seen a steady rise in popularity in the recent past with the introduction of a range of nutraceuticals and herbal medications into the market. There are several phytochemicals that have scientifically proven bioactive potential, which can serve as an alternative strategy for controlling the initiation and progression of common diseases. Among them are compounds such as curcumin, resveratrol, linamarin, cyanidin, apigenin, flavopiridol, epigallocatechin gallate (EGCG), and indole-3-carbinol.2Of the potent bioactive metabolites that have been identified from plant sources, curcumin is one of the most-investigated.2 Named [1,7-bis(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione] under the IUPAC system, it is a polyphenolic, hydrophobic yellow pigment derived from the rhizome of the well-known spice plant turmeric (Curcuma longa L.).3,4 It is a mixture of three curcuminoids: diferuloylmethane (curcumin proper), demethoxycurcumin and bisdemethoxycurcumin. Curcumin exhibits keto–enol tautomerism, having a predominant keto form in acidic/neutral solutions and a stable enol form in alkaline media.5,6 Research over the last two decades has shown it to be a potent antioxidant, anti-inflammatory, antiproliferative, antimetastatic, antiangiogenic, antidiabetic, hepatoprotective, antiatherosclerotic, antithrombotic, and antiarthritic agent in cell- and animal studies.4
Although curcumin has exhibited potent biological activities in vitro, it has managed to show only low activity in various clinical studies. This is mainly due to the poor bioavailability of curcumin, contributed to by its insolubility, instability, poor absorption, and rapid biotransformation.3,7 Curcumin is therefore designated as a typical class IV Biopharmaceutical Classification System (BCS) molecule, which exhibit many characteristics that are problematic for effective oral delivery with poor pharmacokinetics.8,9 Various drug delivery systems such as nanoparticles,10–12 liposomes,12–15 microparticles,13 microemulsion16 and implants17,18 have been shown to significantly enhance preventive/therapeutic efficacy by increasing the drugs' bioavailability and targetability.9,19 The work outlined herein has chosen zinc oxide (ZnO) nanoparticles to work with, predominantly due to their well-understood properties (both physical and biological), and an existing basis in literature for combination with curcumin.
ZnO is biocompatible—its common spherical nanoformulation is considered to be non-toxic and non-immunogenic as well and is a cost-effective starting material.9 It has been found to induce necrotic- and apoptotic pathways in cancer cells via the disruption of the cell membrane and reduction of the mitochondrial membrane potential.20 The latter increases levels of intracellular reactive oxygen species (ROS) and triggers the mitochondria subsidiary apoptotic pathway. ZnO nanoparticles' antibacterial effects are produced in a similar way, enhanced by an abrasive surface created by surface defects.21 The ability of ZnO nanoparticles (ZNPs) to produce ROS and attack pathogenic microorganisms in particular has been well-documented in literature through the years.22–24 Incorporation of curcumin into this system will likely improve these properties via a possible additive effect; extant literature has already shown that ZnO-coupled curcumin has a significantly higher bioavailability than free curcumin. Another point of interest is that ZNPs have been shown to be pH-sensitive, dissociating into Zn2+ at low pH values.21 The acidic tumor microenvironment is expected to produce similar results, aiding in the release of surface-bound curcumin within- or around the tumor.4
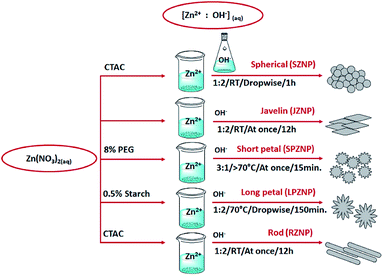
In the present study, the physical-, chemical- and morphological properties, and antibacterial-, anticancer- and cytotoxic effects of a range of ZnO nanoparticle morphologies and their curcumin-grafted counterparts were examined. These were developed to improve the bioavailability, stability, solubility, and dosage of the individual components to enhance their bioactivities. The nanocomposites include spherical- (SZNP–C), rod- (RZNP–C), javelin- (JZNP–C), short petal- (SPZNP–C), and long petal- (LPZNP–C) morphologies. While several works have investigated the delivery properties of multiple morphologies of zinc oxide nanoparticles and nanocomposites for a variety of drugs and nutraceuticals, this is the first time a range of such curcumin-grafted morphologies are being subjected to a standardized method.
Experimental procedure
Materials
Zinc nitrate, sodium hydroxide, cetyltrimethylammonium chloride (CTAC; molecular weight of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), polyethylene glycol (PEG; molecular weight of 4000), starch and curcumin were purchased from Sigma-Aldrich (USA). The antimicrobial compound cloxacillin sodium was gifted by the State Pharmaceutical Manufacturing Corporation of Sri Lanka. Mueller-Hinton agar (Hardy, USA) and phosphate buffered saline (Sigma, USA) were used for microbiology studies. Dulbecco's Modified Eagle Medium (with L-glutamine and nonessential amino acids), Hanks Buffered Salt Solution (HBSS), fetal bovine serum (FBS) and trypsin were purchased from Sigma-Aldrich (USA) for anticancer studies, along with cycloheximide (Himedia, India).
000), polyethylene glycol (PEG; molecular weight of 4000), starch and curcumin were purchased from Sigma-Aldrich (USA). The antimicrobial compound cloxacillin sodium was gifted by the State Pharmaceutical Manufacturing Corporation of Sri Lanka. Mueller-Hinton agar (Hardy, USA) and phosphate buffered saline (Sigma, USA) were used for microbiology studies. Dulbecco's Modified Eagle Medium (with L-glutamine and nonessential amino acids), Hanks Buffered Salt Solution (HBSS), fetal bovine serum (FBS) and trypsin were purchased from Sigma-Aldrich (USA) for anticancer studies, along with cycloheximide (Himedia, India).
Synthesis of ZnO nanoparticles
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 in distilled water (Fig. 1). The zinc nitrate solution was mixed with a CTAC solution and stirred until homogenous. The NaOH was then added dropwise into the zinc nitrate/CTAC solution, and the particles collected by centrifugation (10
2 in distilled water (Fig. 1). The zinc nitrate solution was mixed with a CTAC solution and stirred until homogenous. The NaOH was then added dropwise into the zinc nitrate/CTAC solution, and the particles collected by centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm). The pellet was oven-dried overnight at 60 °C.25
000 rpm). The pellet was oven-dried overnight at 60 °C.25
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar ratio (Fig. 1). The mixture was stirred rapidly at room temperature over 12 hours. The sample was then centrifuged (10
2 molar ratio (Fig. 1). The mixture was stirred rapidly at room temperature over 12 hours. The sample was then centrifuged (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm) and washed thrice with water and once with ethanol. The final product was collected by oven-drying the centrifuged pellet overnight at 60 °C.26
000 rpm) and washed thrice with water and once with ethanol. The final product was collected by oven-drying the centrifuged pellet overnight at 60 °C.26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OH− was 3
OH− was 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 1). This mixture was heated to 70 °C under rapid stirring for 15 minutes. Once cooled to room temperature, the pH of the solution was adjusted to pH 11 using a 1 M NaOH solution. The resultant solution was centrifuged (10
1 (Fig. 1). This mixture was heated to 70 °C under rapid stirring for 15 minutes. Once cooled to room temperature, the pH of the solution was adjusted to pH 11 using a 1 M NaOH solution. The resultant solution was centrifuged (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm) and washed thrice with distilled water and once with ethanol. The final product was collected by oven-drying the centrifuged pellet overnight at 60 °C.27
000 rpm) and washed thrice with distilled water and once with ethanol. The final product was collected by oven-drying the centrifuged pellet overnight at 60 °C.27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar ratio. Starch was added to the Zn(NO3)2 solution to a final concentration of 0.5%, while the NaOH solution was heated to 70 °C (Fig. 1). The zinc nitrate/starch solution was then added dropwise into the base over 30 minutes, followed by rapid stirring for 2 h. The sample was then centrifuged and washed thrice with water and once with ethanol. The final product was collected by oven-drying the centrifuged pellet overnight at 60 °C.28
2 molar ratio. Starch was added to the Zn(NO3)2 solution to a final concentration of 0.5%, while the NaOH solution was heated to 70 °C (Fig. 1). The zinc nitrate/starch solution was then added dropwise into the base over 30 minutes, followed by rapid stirring for 2 h. The sample was then centrifuged and washed thrice with water and once with ethanol. The final product was collected by oven-drying the centrifuged pellet overnight at 60 °C.28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar ratio. The Zn2+ solution was mixed with a CTAC solution, and the OH− solution was added into it at once (Fig. 1). The mixture was stirred rapidly at room temperature overnight, following which the sample was centrifuged and washed thrice with water and once with ethanol. The final product was collected by oven-drying the centrifuged pellet overnight at 60 °C.29
2 molar ratio. The Zn2+ solution was mixed with a CTAC solution, and the OH− solution was added into it at once (Fig. 1). The mixture was stirred rapidly at room temperature overnight, following which the sample was centrifuged and washed thrice with water and once with ethanol. The final product was collected by oven-drying the centrifuged pellet overnight at 60 °C.29Characterization of curcumin-loaded nanoparticles
The thermal stability of both ZNPs and ZNP–Cs was determined by thermogravimetric analysis (TGA; STD Q600 setup) over a temperature range of 25 to 1000 °C at a ramp of 10 °C min−1 in a nitrogen medium.
Assessment of curcumin loading
A standard series of curcumin in acetone (1–8 μg mL−1/1–8 ppm) was made and intensity values at a λmax of 419 nm collected. This was used for the construction of a calibration plot for the purpose of curcumin quantification.10 mg of each ZNP–C sample was first resuspended in acidified acetone (1 M HCl), then sonicated for 20 minutes at room temperature in order to encourage complete dissociation of curcumin from ZnO. Aliquots of the resultant suspension were drawn in triplicate, diluted as appropriate in acetone and subjected to UV-vis spectrophotometry; absorbance data was collected at a λmax of 419 nm. Concentrations of previously ZnO-bound curcumin were calculated by use of the calibration plot.30
Antibacterial study
The antibacterial activities of ZNPs and ZNP–Cs were tested against three Gram-positive bacteria (Staphylococcus aureus (ATCC 25923), Staphylococcus epidermidis (clinical isolate), Bacillus cereus (ATCC 9027)) and one Gram-negative bacterium (Escherichia coli (ATCC 35218)) using well- and drop diffusion methods. The well diffusion method was performed as described by Ratnasooriya et al. with slight modifications.31–33 Briefly, the organisms were subcultured on Mueller-Hinton agar, and a lawn culture prepared by spreading a fresh 100 μL bacterial solution (having 105 CFU mL−1 (complying with McFarland 0.5)) of each test organism on Mueller-Hinton agar plates. A 50 μL aliquot of briefly-sonicated nanoparticle suspension (5 mg mL−1) was poured into the prepared wells (6 mm diameter). For drop diffusion, the same solutions were added as 10 μL drops onto fresh agar surfaces in the same positions as in the well diffusion assay, in place of cutting wells.In both cases, overnight incubation at 37 °C followed, after which the different zones of inhibition were measured. Distilled water was used as the negative control, while gentamicin was used as the positive control throughout.34
Assessment of anticancer activity and cytotoxicity
The anticancer effect of ZNPs, ZNP–Cs and pure curcumin was assayed using the standard MTT (3-(4,5-dimethyl thiazol-2yl)-2,5-diphenyl tetrazolium bromide) assay. Briefly, the wells were carefully emptied and filled with 100 μL MTT (5 mg mL−1 in PBS). The cells were then incubated at 37 °C for 4 hours and the excess MTT reagent was carefully removed without disturbing formazan crystals. The formazan crystals were then dissolved in acidified iso-propanol (0.05 HCl in IPA; 100 μL) by constant agitation on a reciprocating orbital shaker. Acidified isopropyl alcohol was used as the blank. Absorbance data was collected at a wavelength of 570 nm using a UV-vis plate reader, and cell viability calculated via the following equation (where A is absorbance intensity)35
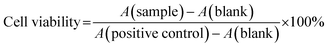 | (1) |
Cytotoxicity study
To determine the cytotoxic activity of the nanocomposites, the growth inhibition of HEK293 cells (Human embryonic kidney cells, ATCC CRL-1573) was measured using the resazurin dye reduction test. Briefly, a final concentration of 25 μg mL−1 resazurin was added into the cell culture, and fluorescence analyzed at ex.: 530/10 nm and em.: 590/10 nm (F560/590) after incubation at 37 °C and 5% CO2 for 3 h. A concentration gradient from 100 μg mL−1 to 0.2 μg mL−1 of each material (ZNPs, ZNP–Cs and pure curcumin) was used in this cytotoxicity assay. The percentage growth inhibition and CC50 (concentration at 50% cytotoxicity) were calculated for each sample in triplicate. A negative control of pure culture media, and a positive control in the form of tamoxifen were also utilized.36Results and discussion
Morphological analysis
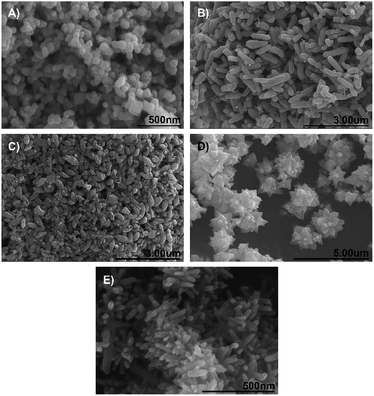
The successful synthesis of a variety of different morphologies of ZnO nanoparticles was confirmed by the scanning electron microscope (SEM) images given in Fig. 2. Spherical particles (Fig. 2A) were observed to have a diameter within the 40–100 nm range, with some slight ovoid features.Rod-shaped nanoparticles with a length of between 600 and 900 nm and a width of <300 nm are shown in Fig. 1B. Javelin-shaped nanoparticles (Fig. 2C) with a length of between 300 and 600 nm and widths of <300 nm were also successfully synthesized, with the main difference between these and the rod-shaped nanoparticles being the more ovoid, pellet-like shape of the former. Fig. 2D and E show the two nanoflowers with short- and long ‘petals’ respectively. The short petal nanoflowers are, as particles, larger than the long petal nanoflowers, with the former having particle diameters (petal tip to petal tip) of between 2 and 4 μm, while the latter having particle diameters of approx. 500 nm. Nevertheless, both types of nanoflowers have protrusions (here referred to as ‘petals’) that emanate from a central point, which have dimensions in the nanoscale.
Various methods have been used to prepare zinc oxide nanoparticles such as hydrothermal,37–39 solvothermal,40,41 microemulsion,42 sol–gel43,44 and thermal decomposition of precursors.45,46 The shape and size of ZNPs are dependent on various factors affecting kinetics, including concentration of reactant, type of base, pH, temperature, stir speed and surfactant. The particles formed may be nanorods,47 nanoplates,48 nanospheres,49 nanoboxes, hexagonal, tripods,50 tetrapods,51 nanowires,52 nanotubes, nanorings,53 nanocages, and nanoflowers54 ranging from a few nanometres to around 900 nm. This study employed both solvothermal and sol–gel methods in a simpler, one-pot reaction attempted by others such as Tong et al. (2013),53 using only NaOH at different concentrations as a precipitator, and CTAC, PEG and starch as structure-directing surfactants. PEG and starch likely played the role of organic templates for the formation of the two ZNP nanoflowers.55,56 In general, the OH groups provided by a base such as NaOH leads to the formation of the [Zn(OH)4]2− complex, the crucial growth unit of ZNPs. This can then be induced to grow preferentially along one or more crystalline planes through the modulation of the kinetic factors noted above, particularly the Zn-base ratio, and the type and concentration of surfactant, which affect the rate of both nucleation and subsequent growth of ZnO nanostructures.56
Chemical characterization
FTIR spectra of pure ZnO, pure curcumin and ZNP–C nanocomposites (RZNP–C, SZNP–C, LPZNP–C, SPZNP–C and JZNP–C) are shown in Fig. 3. FTIR of all ZNP samples clearly showed a broad peak between 3600 and 3050 cm−1. The FTIR spectrum of the ZnO nanoparticles, curcumin, and ZNP–Cs nanocomposites were scanned in the range of 4000–500 cm−1. The nanocomposite showed the expected Zn–O stretching vibrations at 677 cm−1, as well as the bands caused by interatomic vibrations of metal oxides at 802, 833, and 881 cm−1. The benzoate trans-C–H vibration was also observed at 962 cm−1. The peaks at 1197 and 3346 cm−1 are likely caused by O–H deformation and stretching, respectively, on account of the moisture adsorbed on the NP surface.29 The peak at 1274 cm−1 was assigned to the enol C–O, and that at 1508 cm−1 was likely due to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O. Symmetric stretching vibrations of the aromatic C
O. Symmetric stretching vibrations of the aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds produces a band at 1600 cm−1, while the peak at 3014 cm−1 pertains to C–H stretching, and the broad peak at 3512 cm−1 is the characteristic O–H stretch.
C bonds produces a band at 1600 cm−1, while the peak at 3014 cm−1 pertains to C–H stretching, and the broad peak at 3512 cm−1 is the characteristic O–H stretch.
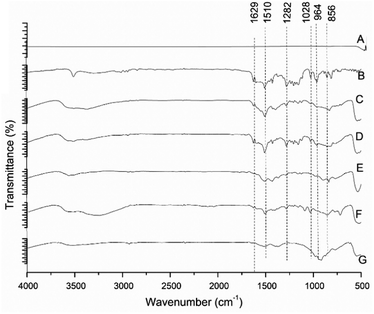 | ||
| Fig. 3 FT-IR spectra of different morphologies of ZnO nanoparticles. (A) Pure ZnO, (B) pure curcumin, (C) JZNP–C, (D) LPZNP–C, (E) RZNP–C, (F) SPZNP–C and (G) SZNP–C. | ||
Relative to curcumin, the reduction in intensity of the band at 962 cm−1, and shifting of the ZnO band at 881 to 871 cm−1 in the nanocomposite could indicate chelation of carbonyl or hydroxyl groups of curcumin with zinc.57 Hence, the FTIR spectra serve to confirm the formation of the metal–curcumin complex.
Physical characterization
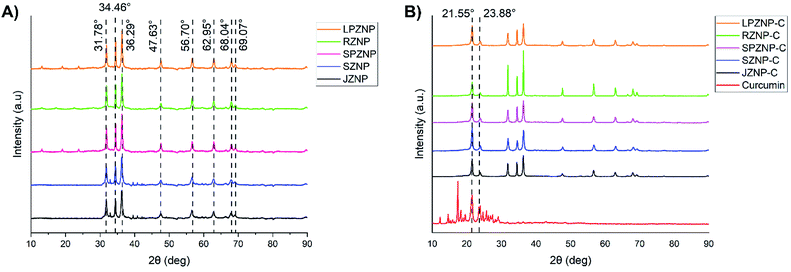
The XRD patterns of bare ZNPs are shown in Fig. 4A, with the results matching JCPDS card no. 00-0361451, showing the presence of crystalline ZnO in its customary wurtzite phase. The patterns exhibit characteristic 2θ/basal plane pairs of 31.78°/(100), 34.46°/(002), 36.29°/(101), 47.63°/(102), 56.70°/(110), 62.95°/(103), 68.04°/(200) and 69.07°/(201). No great differences are apparent between the various ZNP morphologies, but the lack of peaks other than those regarded as characteristic points to a high degree of sample purity. Fig. 4B shows XRD patterns obtained for the loaded ZNP–Cs, compared with that of pure curcumin. The latter shows good crystallinity by the presence of peaks between 2θ values of 10 and 30°,58 two of which are present in the patterns of the nanocomposites (21.55° and 23.88°)59,60 albeit at reduced intensities. This reduction in intensity and the absence of other characteristic curcumin peaks (such as that at 17.44°) may point to the formation of inclusion complexes of curcumin within the surface of their host ZNPs, exposing only a select number of basal planes.Thermogravimetric analysis (TGA) of ZNP–C nanocomposites (RZNP–C, SZNP–C, LPZNP–C, SPZNP–C and JZNP–C) was carried out to approximate curcumin loading (Fig. 5). Loss of water occurred at approximately 110 °C, and all thermograms showed characteristic curcumin degradation (to varying degrees) between 280 and 420 °C. Loading of curcumin onto ZNPs was calculated approximately by subtracting the mass loss of the bare nanoparticle from that of the loaded one. These results are presented in Table 1 with the result of post-digestion amount of curcumin. The mean of both studies was taken as the loaded percentage of curcumin.
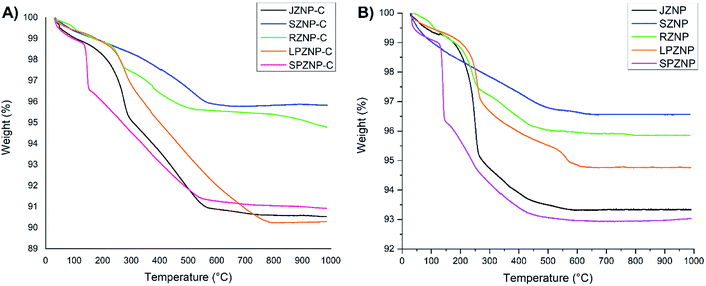 | ||
| Fig. 5 Thermograms of different morphologies of ZnO nanocomposites. (A) With no loaded curcumin, (B) with loaded curcumin. | ||
| Nanoparticle shape | % Curcumin loaded (w/w) | |
|---|---|---|
| TGA (mass loss) | Post-digestion (UV-vis) | |
| Rod | 4.36 | 3.93 |
| Sphere | 4.20 | 5.72 |
| Long petal | 9.03 | 10.25 |
| Short petal | 4.58 | 3.14 |
| Javelin | 9.00 | 8.37 |
Results of both the TGA and post-digestion of nanoparticles give a similar pattern of curcumin loading on the nanoparticles. The greatest percentage of curcumin-containing nanocomposites was observed in LPZNP–C, interpreted as being indicative of the highest curcumin loading of all synthesized nanocomposites. This was to be expected due to the comparatively high surface area lent to this particle by the presence of long needle-like ‘petals’, and in particular the deep crevices between them. Though the sphere has the highest surface area-to-volume ratio, the percentage of loaded curcumin is comparatively modest, likely due to the leaking of trapped curcumin during washing. The percentage mass loss of curcumin was observed to be the lowest in SPZNP–C, while lowest post-digestion curcumin percentage was observed in SZNP–C. This is most likely due to the comparatively low surface area-to-volume ratio.
Comparison of Fig. 5A and B also provides an interesting insight: whereas the order of mass loss amongst bare ZNPs were in the order SPZNP > JZNP > LPZNP > RZNP > SZNP, the.mass loss amongst loaded nanocomposites is changed to LPZNP–C > JZNP–C > SPZNP–C > RZNP–C > SZNP–C. This is most likely due to the higher curcumin loading (see Table 1) in the long petal nanoflowers compared to the lower loading in javelin- and small petal forms, resulting in greater mass loss in the composite containing the highest amount of loaded curcumin, and the other two following in order of loaded curcumin.
Antibacterial assay
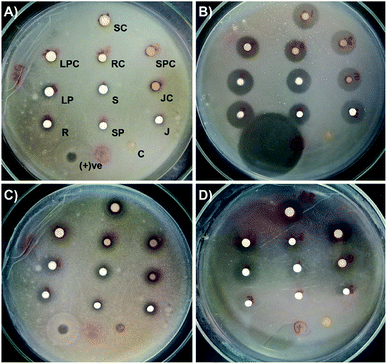
The degree of antibacterial activity of nanoparticles is usually size-dependent, being an inversely proportional relationship. Therefore, nanoscale ZnO was expected to show better antibacterial activity than bulk ZnO.61,62 Apart from the size, activity also depends on shape. In this work, oblong particles showed higher activity than spherical ones.The antibacterial activities of ZNP- and ZNP–C nanocomposites (RZNP–C, SZNP–C, LPZNP–C, SPZNP–C and JZNP–C) were tested by the disc- and well diffusion agar methods on Gram-positive strains (S. aureus, B. cereus and S. epidermidis) as well as Gram-negative strains (E. coli) accordingly. The size of the inhibition zones (Fig. 6) appeared to vary dependent upon the type of bacteria and the type of ZnO/ZnO–curcumin nanocomposites; E. coli appeared to be the least susceptible to the antibacterial activity of all nanocomposites tested, most likely due to the thicker cell membrane. Tyagi et al. 2015,64 in their work with curcumin I observed E. coli to be among the bacteria least susceptible to it.6 Nevertheless, all ZNPs and ZNP–Cs showed some antibacterial activity against both the Gram-positive and Gram-negative strains. In the drop diffusion assay, SZNP–C showed the most promising activity against S. epidermidis (22.2 ± 0.3 mm), while RZNP–C had the most potency against B. cereus (20.2 ± 0.8 mm), S. aureus (17.7 ± 0.1), and E. coli (10.7 ± 0.4 mm). The results for S. aureus are consistent with those obtained by others, wherein it is one of the most susceptible to curcumin's antibacterial activity.63 The inhibition zones for these curcumin-loaded ZnO nanocomposites were consistently larger than their bare counterparts or pure curcumin, revealing an additive effect between the ZnO and curcumin components. This pattern of ZNP–Cs having a higher antibacterial effect than the correspondingly shaped ZNPs holds true across all shapes of the nanocomposites. These general trends hold true for the well diffusion assay as well, albeit at a lower magnitude, likely due to the generally slower diffusion of nanoparticles through the agar bulk as opposed to lateral diffusion on its surface.
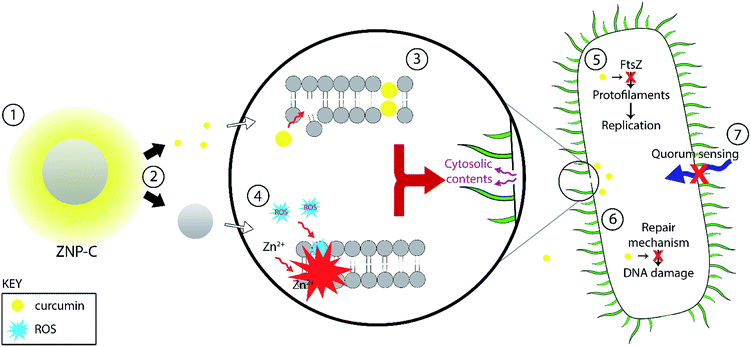
The antibacterial action of curcumin has been shown to be exerted through a number of different mechanisms. It has been shown to be capable of permeabilizing the bacterial membrane regardless of Gram status,64 leading to lysis. This may take place due to curcumin's demonstrated ability to bind to peptidoglycans.63 Curcumin has also been demonstrated to interact with the prokaryotic FtsZ protein in vitro to inhibit assembly of its protofillaments, interfering with the formation of the z ring, thereby destabilizing the bacteria replication process.65 Other antibacterial mechanisms include: inhibition of bacterial quorum sensing (affecting both biofilm formation and virulence; possibly through inhibition of sortaseA) and the inhibition of bacterial DNA damage repair process (contributing to bacteriostatic effects) (Table 2).66
| Antimicrobial activity (5 mg mL−1) (mean inhibition zone diameter ± standard error − mm) | ||||||||
|---|---|---|---|---|---|---|---|---|
| E. coli | S. epidermidis | S. aureus | B. cereus | |||||
| Well dif. | Drop dif. | Well dif. | Drop dif. | Well dif. | Drop dif. | Well dif. | Drop dif. | |
| SZNP | 6.2 ± 0.4 | 8.6 ± 0.5 | 14.1 ± 0.3 | 18.6 ± 0.5 | 13.1 ± 0.7 | 14.6 ± 0.6 | 13.2 ± 0.8 | 14.1 ± 1.0 |
| SZNP–C | 8.4 ± 0.8 | 10.2 ± 0.3 | 19.1 ± 0.1 | 22.2 ± 0.3 | 15.4 ± 1.1 | 17.0 ± 0.4 | 17.4 ± 0.7 | 19.6 ± 1.5 |
| LPZNP | 8.2 ± 0.2 | 8.5 ± 0.6 | 15.7 ± 1.2 | 16.0 ± 0.7 | 13.1 ± 0.4 | 13.4 ± 0.7 | 13.0 ± 0.5 | 14.6 ± 0.2 |
| LPZNP–C | 9.8 ± 0.3 | 10.0 ± 0.4 | 20.1 ± 0.2 | 20.8 ± 0.4 | 17.0 ± 0.5 | 16.8 ± 0.2 | 14.2 ± 0.5 | 17.8 ± 0.4 |
| RZNP | 8.4 ± 0.1 | 9.7 ± 0.2 | 14.4 ± 0.4 | 17.4 ± 0.2 | 12.5 ± 0.3 | 14.3 ± 0.6 | 15.4 ± 0.8 | 18.9 ± 0.5 |
| RZNP–C | 10.1 ± 0.5 | 10.7 ± 0.4 | 17.2 ± 0.4 | 20.0 ± 0.8 | 16.6 ± 0.7 | 17.7 ± 0.1 | 18.7 ± 0.2 | 20.2 ± 0.6 |
| SPZNP | 7.8 ± 0.6 | 8.2 ± 0.4 | 14.1 ± 1.1 | 14.3 ± 0.1 | 13.0 ± 0.6 | 13.1 ± 0.7 | 11.4 ± 0.6 | 11.3 ± 0.1 |
| SPZNP–C | 8.1 ± 0.1 | 8.9 ± 1.2 | 16.4 ± 0.4 | 17.7 ± 0.2 | 15.2 ± 0.8 | 15.9 ± 0.4 | 14.2 ± 0.4 | 13.5 ± 0.6 |
| JZNP | 9.0 ± 0.5 | 10.3 ± 0.7 | 16.0 ± 0.6 | 16.7 ± 0.5 | 12.4 ± 0.1 | 15.0 ± 0.8 | 15.3 ± 0.4 | 16.0 ± 0.5 |
| JZNP–C | 11.1 ± 0.2 | 10.5 ± 0.6 | 19.4 ± 0.1 | 20.9 ± 0.8 | 13.4 ± 0.4 | 15.5 ± 0.8 | 18.7 ± 0.3 | 19.3 ± 0.2 |
| Curcumin (C) | 7.4 ± 1.3 | 7.8 ± 0.4 | 8.2 ± 0.3 | 9.0 ± 0.3 | 8.1 ± 0.4 | 8.8 ± 0.2 | 8.4 ± 1.1 | 7.9 ± 0.3 |
| +ve control | 11.3 ± 0.8 | 14.1 ± 1.0 | 24.0 ± 0.4 | 28.1 ± 1.1 | 18.8 ± 1.1 | 22.1 ± 1.7 | 21.2 ± 2.0 | 24.0 ± 0.9 |
| −ve control | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Several reports suggest that the action of ZnO on bacterial species is due to creation of reactive oxygen species (ROS) and release of zinc ions. Generated ROS (i.e. hydrogen peroxide (H2O2), OH− (hydroxyl radicals), O22− (peroxide)) and zinc ions from ZnO nanoparticles bind to the negative surface of the cell membrane, leading to disruption of the cells, followed by leakage of cellular material to cause cell death.4,21,67 Additive effects of curcumin with different antibiotics such as ampicillin, oxacillin, and norfloxacin against methicillin-resistant S. aureus have been previously reported.68 The MIC of ZNPs against S. aureus was reported to be 1 mg mL−1.69 ZnO combined with ciprofloxacin or gentamicin has also been demonstrated to be effective against S. aureus.70,71 A recent study using nanocurcumin, ZNPs, and curcumin/ZNP nanocomposites against S. aureus, B. subtilis, E. coli, and P. aeruginosa showed the nanocomposite to have higher efficacy than the pristine ZNPs and nanocurcumin on their own.57 The antibacterial activity of curcumin-loaded ZnO nanocomposites were significantly higher compare to the pure curcumin and ZnO NPs. From this, it is reasonable to conclude that there exists additive effect between the ZnO and curcumin components in the composites studied herein as well, and that this combination can be used as a potent antibacterial agent for various purposes including medical.68,70 The combined effects of ZNP–Cs on bacteria are outlined in Fig. 7.
Anticancer assay
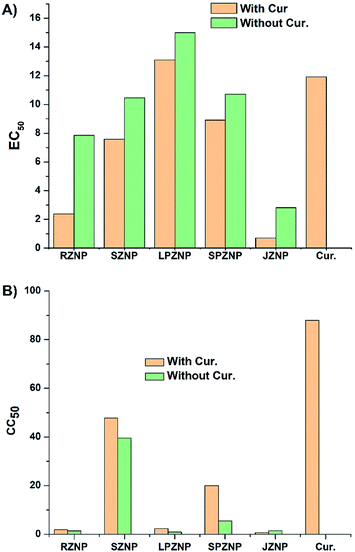
The anticancer activity of ZnO–curcumin nanocomposites was assessed against rhabdomyosarcoma (RD) cells, a cancer that affects children and adolescents in particular,72,73 and has previously been used in similar studies.74,75The results of this assessment (performed by use of the MTT assay) are summarized in Fig. 8A, 9 and S2 in the ESI.† Pure curcumin resulted in 50% cell viability in rhabdomyosarcoma (RD) cells after treatment with a 12 μg mL−1 solution. RZNP, SZNP, LPZNP, SPZNP and JZNP showed 50% cell death at concentrations of 7.8, 9.6, 2.9, 15.0 and 11.0 μg mL−1 respectively. In contrast, ZnO–curcumin nanocomposites displayed a general tendency to have lower EC50 values. As such, RZNP–C, SZNP–C, LPZNP–C, SPZNP–C and JZNP–C produced 50% cell death at concentrations of 2.4, 7.6, 13.0, 8.9 and 0.8 μg mL−1 respectively (Fig. 8).
During this assay, the same mass of bare nanoparticles and curcumin-loaded nanoparticles were used in all cases. Thus, the weight percentage of ZnO in the assay of curcumin-loaded particles is lower than in the case of the bare particles, since a proportion of the curcumin-containing sample consists of curcumin itself. The reduction of ZnO should logically lead to a concomitant decrease in anticancer toxicity had the composites had the same anticancer activity as the bare nanoparticles. However, the results show that the addition of even a small quantity of curcumin to this reduced ZnO mass leads to a sharp increase in anticancer activity, reflected by the decrease in EC50. The anticancer activity of pure curcumin lends support to this observation and previous literature that note the compound's activity against cancer cell lines.55,56
Various in vitro studies have shown that curcumin induces apoptosis in oncogenic cells by inhibiting various intracellular transcription factors and secondary messengers such as NF-kB, AP-1, c-Jun, the JAK-STAT pathway, and various others.3,4,7
Curcumin is well-known for its potential to inhibit carcinogenesis induced by chemical carcinogens, at both initiation and progression stages in various preclinical studies.76 It is known to inhibit cytochrome P450 (CYP) enzyme mediated bioactivation of environmental carcinogens like benzo[a]pyrene (B[a]P).77 Curcumin is a competitive inhibitor of B[a]P metabolism and thus depresses its bioactivation via CYP1A1.78 Curcumin also increases the levels of other endogenous antioxidants via the Nrf2 pathway to strengthen the body's defences against reactive oxygen species.4,7
According to the results obtained herein, the ZnO nanoparticle itself showed some anticancer properties. Several studies have demonstrated that ROS generation is a key cytotoxic mechanism of ZNPs. ROS (mainly superoxide, hydroxyl radicals and hydrogen peroxide) are constantly generated and eliminated in biological systems. It is well-known that ROS play important roles in a variety of normal biochemical functions and aberrant pathological processes. Elevated ROS accumulation that exceeds the level of the cellular antioxidant defence system causes oxidative stress in cells, which leads to the damage of cellular components, including lipids, proteins, and DNA.4,20,21,79 In addition, Zn2+ ions have also been shown to induce a cytotoxic effect in cancer cells, also by inducing oxidative stress.80 In the present study, ZNP–Cs showed the additive anticancer effect of both ZnO NP and curcumin, and therefore, it can be considered as a potential candidate or supplement for cancer chemotherapy.
Cytotoxicity study
In the cytotoxicity assay against the designated healthy Human Embryonic Kidney (HEK) cells, pure curcumin caused a 50% drop in cell viability at a concentration of 88 μg mL−1. RZNP, SZNP, LPZNP, SPZNP and JZNP showed 50% cell death at concentrations of 1.42, 39, 0.96, 5.5 and 1.5 μg mL−1 respectively. In contrast, ZnO–curcumin nanocomposites, displayed a general tendency to have higher CC50 values in this case. RZNP–C, SZNP–C, LPZNP–C, SPZNP–C and JZNP–C produced 50% cell death at concentrations of 1.9, 48, 2.4, 20 and 0.66 μg mL−1 respectively. This decrease in toxicity cannot be wholly attributed to the presence of curcumin, since as noted before the amount of ZnO in the sample is lower than in the case of the assay with bare nanoparticles. Nevertheless, it is interesting to note that the pure curcumin has a high CC50 against this healthy cell line (see Fig. 8B and S3 in the ESI†).The highest cytotoxicity against HEK cells was shown by JZNP–C, which rules it out as a potential anticancer agent in the form tested in this work. RZNP–C has the next-highest toxicity against the healthy cell line; its anticancer activity against RD cells is considerable, but due to its nonspecific toxicity, it must also be ruled out. The same holds true for LPZNP–C, whose CC50 against HEK cells is almost a six-fold increase over its EC50 against the cancer cells-a situation that is highly undesirable. In these cases, the high degree of toxicity can be attributed to the high aspect ratio of the ZnO structures.
The lowest cytotoxicity against HEK cells was exhibited by SPZNP–C and SZNP–C. Both of these have higher CC50 values against HEK cells than EC50 values against RD cells, with SPZNP–C having an approximately 2-fold decrease, and SZNP–C having an approximately 6-fold decrease in cytotoxicity against healthy cells compared to toxicity towards the cancer cell line. Given this, SZNP–C can be clearly identified as having the best/optimal balance between toxicity against healthy cells and anticancer activity against the RD cell line.
Conclusions
In this study, we investigated the potential of curcumin-loaded ZnO nanocomposites/nanoparticles (ZNP–Cs) as efficient antibacterial and anticancer agents. Upon successful loading of curcumin onto ZnO nanoparticles, an additive activity between the two components resulted in markedly improved antibacterial- and anticancer activities. Moreover, the spherical (SZNP–C) nanocomposite showed good antibacterial/anticancer activity with considerably less cytotoxic activity than other ZNP–Cs. SZNP–C nanocomposites could therefore be a promising nanoformulation for anticancer therapy and antibacterial applications. Of particular note is the potential of this platform to act as an antibiotic-free formulation for use against infections caused by a range of different bacterial pathogens. Given the anticancer activity of the platform, it may prove to be of great use as an oncotherapy supplement, helping manage both the disease condition and opportunistic bacterial infections.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by the Research Fund of the Sri Lanka Institute of Nanotechnology (PRO0088). The authors would like to thank Dr Malini Damayanthi and Samantha Weerasinghe for their support in SEM imaging. Anticancer studies were supported by Prof. Preethi de Soysa, Department of Biochemistry, Faculty of Medicine, University of Colombo.References
- B. Chauhan, G. Kumar, N. Kalam and S. H. Ansari, J. Adv. Pharm. Technol. Res., 2013, 4, 4–8 CrossRef PubMed.
- A. G. Atanasov, B. Waltenberger, E. M. Pferschy-Wenzig, T. Linder, C. Wawrosch, P. Uhrin, V. Temml, L. Wang, S. Schwaiger, E. H. Heiss, J. M. Rollinger, D. Schuster, J. M. Breuss, V. Bochkov, M. D. Mihovilovic, B. Kopp, R. Bauer, V. M. Dirsch and H. Stuppner, Biotechnol. Adv., 2015, 33, 1582–1614 CrossRef CAS PubMed.
- T. Kurita and Y. Makino, Anticancer Res., 2013, 33, 2807–2822 CAS.
- M. Kundu, P. Sadhukhan, N. Ghosh, S. Chatterjee, P. Manna, J. Das and P. C. Sil, J. Adv. Res., 2019, 18, 161–172 CrossRef CAS PubMed.
- Y. Manolova, V. Deneva, L. Antonov, E. Drakalska, D. Momekova and N. Lambov, Spectrochim. Acta, Part A, 2014, 132, 815–820 CrossRef CAS PubMed.
- W.-H. Lee, C.-Y. Loo, M. Bebawy, F. Luk, R. Mason and R. Rohanizadeh, Curr. Neuropharmacol., 2013, 11, 338–378 CrossRef CAS PubMed.
- S. S. Bansal, M. Goel, F. Aqil, M. V. Vadhanam and R. C. Gupta, Cancer Prev. Res., 2011, 4, 1158–1171 CrossRef CAS PubMed.
- P. Somu and S. Paul, New J. Chem., 2019, 43, 11934–11948 RSC.
- J. Jiang, J. Pi and J. Cai, Bioinorg. Chem. Appl., 2018, 2018, 1–18 CrossRef PubMed.
- P. Anand, H. B. Nair, B. Sung, A. B. Kunnumakkara, V. R. Yadav, R. R. Tekmal and B. B. Aggarwal, Biochem. Pharmacol., 2010, 79, 330–338 CrossRef CAS PubMed.
- S. Bisht, G. Feldmann, S. Soni, R. Ravi, C. Karikar, A. Maitra and A. Maitra, J. Nanobiotechnol., 2007, 5, 1–18 CrossRef PubMed.
- M. R. Gasco, Adv. Drug Delivery Rev., 2007, 59, 377–378 CrossRef CAS PubMed.
- M. Üner and G. Yener, Int. J. Nanomed., 2007, 2, 289–300 Search PubMed.
- P. Dahlman, Diss. - Chalmers Tek. Hoegsk., 2005, 50, 1–63 Search PubMed.
- A. Kunwar, A. Barik, R. Pandey and K. I. Priyadarsini, Biochim. Biophys. Acta, Gen. Subj., 2006, 1760, 1513–1520 CrossRef CAS PubMed.
- P. Santos, A. C. Watkinson, J. Hadgraft and M. E. Lane, Skin Pharmacol. Physiol., 2008, 21, 246–259 CrossRef CAS PubMed.
- A. K. Dash and G. C. Cudworth, J. Pharmacol. Toxicol. Methods, 1998, 40, 1–12 CrossRef CAS PubMed.
- H. J. Anthony, J. Pharm. Sci., 2013, 102, 1165–1172 CrossRef PubMed.
- Y. Li, Z. Lin, G. Gong, M. Guo, T. Xu, C. Wang, M. Zhao, Y. Xia, Y. Tang, J. Zhong, Y. Chen, L. Hua, Y. Huang, F. Zeng and B. Zhu, J. Mater. Chem. B, 2019, 7, 4252–4262 RSC.
- A. Sirelkhatim, S. Mahmud, A. Seeni, N. H. M. Kaus, L. C. Ann, S. K. M. Bakhori, H. Hasan and D. Mohamad, Nano-Micro Lett., 2015, 7, 219–242 CrossRef CAS PubMed.
- X. Cai, Y. Luo, W. Zhang, D. Du and Y. Lin, ACS Appl. Mater. Interfaces, 2016, 8, 22442–22450 CrossRef CAS PubMed.
- X. Xu, D. Chen, Z. Yi, M. Jiang, L. Wang, Z. Zhou, X. Fan, Y. Wang and D. Hui, Langmuir, 2013, 29, 5573–5580 CrossRef CAS PubMed.
- A. Lipovsky, Z. Tzitrinovich, H. Friedmann, G. Applerot, A. Gedanken and R. Lubart, J. Phys. Chem. C, 2009, 113, 15997–16001 CrossRef CAS.
- V. Lakshmi Prasanna and R. Vijayaraghavan, Langmuir, 2015, 31, 9155–9162 CrossRef CAS PubMed.
- P. Rai, S. K. Tripathy, N. H. Park, K. J. O, I. H. Lee and Y. T. Yu, J. Mater. Sci.: Mater. Electron., 2009, 20, 967–971 CrossRef CAS.
- A. Umar, S. H. Kim, E. Suh and Y. B. Hahn, Chem. Phys. Lett., 2007, 440, 110–115 CrossRef CAS.
- R. Herrera-Rivera, M. D. L. L. Olvera and A. Maldonado, J. Nanomater., 2017, 2017, 16–20 Search PubMed.
- M. F. Khan, M. Hameedullah, A. H. Ansari, E. Ahmad, M. B. Lohani, R. H. Khan, M. M. Alam, W. Khan, F. M. Husain and I. Ahmad, Int. J. Nanomed., 2014, 9, 853–864 CrossRef CAS PubMed.
- H. Kumar and R. Rani, Int. Lett. Chem., Phys. Astron., 2013, 19, 26–36 Search PubMed.
- R. T. De Silva, R. K. Dissanayake, M. M. M. G. P. G. Mantilaka, W. P. S. L. Wijesinghe, S. S. Kaleel, T. N. Premachandra, L. Weerasinghe, G. A. J. Amaratunga and K. M. N. De Silva, ACS Appl. Mater. Interfaces, 2018, 10, 33913–33922 CrossRef CAS PubMed.
- R. K. Dissanayake, P. B. Ratnaweera, D. E. Williams, C. D. Wijayarathne, R. L. C. Wijesundera, R. J. Andersen and E. D. de Silva, J. Appl. Pharm. Sci., 2016, 6, 001–006 CAS.
- B. Budiarto, A. Mustopa and K. Tarman, J. Coastal Life Med., 2015, 3, 56–63 CAS.
- W. D. Ratnasooriya, S. G. Ratnasooriya and R. Dissanayake, J. Coastal Life Med., 2016, 4, 623–627 CrossRef CAS.
- A. R. N. Silva, D. M. R. K. Dissanayake, C. B. Ranaweera, R. Pathirana and W. D. Ratnasooriya, Int. J. Pharm. Res. Allied Sci., 2015, 4, 54–57 Search PubMed.
- P. Soysa, I. S. De Silva and J. Wijayabandara, BMC Complementary Altern. Med., 2014, 14, 1–8 CrossRef PubMed.
- J. O'Brien, I. Wilson, T. Orton and F. Pognan, Eur. J. Biochem., 2000, 5426, 5421–5426 CrossRef PubMed.
- B. Liu and H. C. Zeng, J. Am. Chem. Soc., 2003, 125, 4430–4431 CrossRef CAS PubMed.
- X. M. Sun, X. Chen, Z. X. Deng and Y. D. Li, Mater. Chem. Phys., 2003, 78, 99–104 CrossRef.
- M. Bitenc, P. Podbršček, Z. C. Orel, M. A. Cleveland, J. A. Paramo, R. M. Peters and Y. M. Strzhemechny, Cryst. Growth Des., 2009, 9, 997–1001 CrossRef CAS.
- S. H. Yu, J. Yang, Y. T. Qian and M. Yoshimura, Chem. Phys. Lett., 2002, 361, 362–366 CrossRef CAS.
- L. Guo, Y. L. Ji, H. Xu, P. Simon and Z. Wu, J. Am. Chem. Soc., 2002, 124, 14864–14865 CrossRef CAS PubMed.
- D. Mondelaers, G. Vanhoyland, H. Van Den Rul, J. D'Haen, M. K. Van Bael, J. Mullens and L. C. Van Poucke, Mater. Res. Bull., 2002, 37, 901–914 CrossRef CAS.
- S. Lee, S. Jeong, D. Kim, S. Hwang, M. Jeon and J. Moon, Superlattices Microstruct., 2008, 43, 330–339 CrossRef CAS.
- Z. Wang, H. Zhang, Z. Wang, L. Zhang, J. Yuan, S. Yan and C. Wang, J. Mater. Res., 2003, 18, 151–155 CrossRef CAS.
- S. C. Pillai, J. M. Kelly, D. E. McCormack, P. O'Brien and R. Ramesh, J. Mater. Chem., 2003, 13, 2586–2590 RSC.
- R. A. Bohara, N. D. Thorat and S. H. Pawar, RSC Adv., 2016, 6, 43989–44012 RSC.
- T. X. Wang and T. J. Lou, Mater. Lett., 2008, 62, 2329–2331 CrossRef CAS.
- J. S. Jang, C. J. Yu, S. H. Choi, S. M. Ji, E. S. Kim and J. S. Lee, J. Catal., 2008, 254, 144–155 CrossRef CAS.
- K. Kakiuchi, E. Hosono, T. Kimura, H. Imai and S. Fujihara, J. Sol-Gel Sci. Technol., 2006, 39, 63–72 CrossRef CAS.
- S. Mahmud and M. J. Abdullah, in Nanotripods of Zinc Oxide, IEEE, 2006, vol. 2006, pp. 442–446 Search PubMed.
- L. Shen, H. Zhang and S. Guo, Mater. Chem. Phys., 2009, 114, 580–583 CrossRef CAS.
- Y. Ding and Z. L. Wang, Micron, 2009, 40, 335–342 CrossRef CAS PubMed.
- Z. L. Wang, J. Phys.: Condens. Matter, 2004, 16, 829–858 CrossRef.
- A. Moezzi, M. Cortie and A. McDonagh, Dalton Trans., 2011, 40, 4871–4878 RSC.
- J. Duan, X. Huang and E. Wang, Mater. Lett., 2006, 60, 1918–1921 CrossRef CAS.
- G. X. Tong, F. F. Du, Y. Liang, Q. Hu, R. N. Wu, J. G. Guan and X. Hu, J. Mater. Chem. B, 2013, 1, 454–463 RSC.
- K. R. Soumya, S. Snigdha, S. Sugathan, J. Mathew and E. K. Radhakrishnan, 3 Biotech, 2017, 7, 1–10 Search PubMed.
- S. Ji, X. Lin, E. Yu, C. Dian, X. Yan, L. Li, M. Zhang, W. Zhao and L. Dian, J. Nanotechnol., 2018, 2018, 1–9 CrossRef.
- R. S. Pandit, S. C. Gaikwad, G. A. Agarkar, A. K. Gade and M. Rai, 3 Biotech, 2015, 5, 991–997 CrossRef PubMed.
- I. M. Krishnakumar, A. Ravi, D. Kumar, R. Kuttan and B. Maliakel, J. Funct. Foods, 2012, 4, 348–357 CrossRef.
- Z. Emami-Karvani, Afr. J. Microbiol. Res., 2012, 5, 1368–1373 Search PubMed.
- P. J. P. Espitia, N. d. F. F. Soares, J. S. d. R. Coimbra, N. J. de Andrade, R. S. Cruz and E. A. A. Medeiros, Food Bioprocess Technol., 2012, 5, 1447–1464 CrossRef CAS.
- S.-Y. Teow, K. Liew, S. A. Ali, A. S.-B. Khoo and S.-C. Peh, J. Trop. Med. Hyg., 2016, 2016, 1–10 Search PubMed.
- P. Tyagi, M. Singh, H. Kumari, A. Kumari and K. Mukhopadhyay, PLoS One, 2015, 10, 1–15 Search PubMed.
- L. G. Morão, C. R. Polaquini, M. Kopacz, G. S. Torrezan, G. M. Ayusso, G. Dilarri, L. B. Cavalca, A. Zielińska, D. J. Scheffers, L. O. Regasini and H. Ferreira, Microbiologyopen, 2019, 8, 1–12 CrossRef PubMed.
- B. Li, X. Li, H. Lin and Y. Zhou, Biomed Res. Int., 2018, 2018, 1–11 Search PubMed.
- P. P. Patil, J. V. Meshram, R. A. Bohara, S. G. Nanaware and S. H. Pawar, New J. Chem., 2018, 42, 14620–14629 RSC.
- S. H. Mun, D. K. Joung, Y. S. Kim, O. H. Kang, S. B. Kim, Y. S. Seo, Y. C. Kim, D. S. Lee, D. W. Shin, K. T. Kweon and D. Y. Kwon, Phytomedicine, 2013, 20, 714–718 CrossRef CAS PubMed.
- K. M. Reddy, K. Feris, J. Bell, D. G. Wingett, C. Hanley and A. Punnoose, Appl. Phys. Lett., 2007, 90, 10–13 Search PubMed.
- P. Patra, S. Mitra, N. Debnath, P. Pramanik and A. Goswami, Bull. Mater. Sci., 2014, 37, 199–206 CrossRef CAS.
- G. Voicu, O. Oprea, B. S. Vasile and E. Andronescu, Dig. J. Nanomater. Biostruct., 2013, 8, 1191–1203 Search PubMed.
- E. El Nadi, E. A. H. Moussa, W. Zekri, H. Taha, A. Yones, M. S. Zaghloul, M. El Wakeel and R. M. Labib, Sarcoma, 2015, 2015, 1 CrossRef PubMed.
- W. Huh and D. Egas Bejar, Adolesc. Health, Med. Ther., 2014, 5, 115–125 CrossRef PubMed.
- A. J. Jasim Makkawi, N. H. Aysa and F.-A. G. Gassim, Asian J. Pharm. Clin. Res., 2019, 12, 535–539 CrossRef.
- G. Chadalapaka, I. Jutooru, S. Sreevalsan, S. Pathi, K. Kim, C. Chen, L. Crose, C. Linardic and S. Safe, Int. J. Cancer, 2013, 132, 795–806 CrossRef CAS PubMed.
- P. Anand, S. G. Thomas, A. B. Kunnumakkara, C. Sundaram, K. B. Harikumar, B. Sung, S. T. Tharakan, K. Misra, I. K. Priyadarsini, K. N. Rajasekharan and B. B. Aggarwal, Biochem. Pharmacol., 2008, 76, 1590–1611 CrossRef CAS PubMed.
- R. L. Thangapazham, A. Sharma and R. K. Maheshwari, AAPS J., 2006, 8, 443–449 CrossRef PubMed.
- S. V. Singh, X. Hu, S. K. Srivastava, M. Singh, H. Xia, J. L. Orchard and H. A. Zaren, Carcinogenesis, 1998, 19, 1357–1360 CrossRef CAS PubMed.
- Y. Li, Z. Lin, M. Zhao, T. Xu, C. Wang, L. Hua, H. Wang, H. Xia and B. Zhu, ACS Appl. Mater. Interfaces, 2016, 8, 24385–24393 CrossRef CAS PubMed.
- S.-H. Moon, W. J. Choi, S.-W. Choi, E. H. Kim, J. Kim, J.-O. Lee and S. H. Kim, Toxicol. Rep., 2016, 3, 430–438 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra05755j |
| This journal is © The Royal Society of Chemistry 2020 |