Open Access Article
Open Access ArticleDevelopment and characterization of a novel L-asparaginase/MWCNT nanobioconjugate†
Raquel O. Cristóvão a,
Mafalda R. Almeida
a,
Mafalda R. Almeida b,
Maria A. Barros
b,
Maria A. Barros a,
João C. F. Nunes
a,
João C. F. Nunes ab,
Rui A. R. Boaventura
ab,
Rui A. R. Boaventura a,
José M. Loureiro
a,
José M. Loureiro a,
Joaquim L. Faria
a,
Joaquim L. Faria a,
Márcia C. Neves
a,
Márcia C. Neves b,
Mara G. Freire
b,
Mara G. Freire b,
Valéria C. Ebinuma-Santos
b,
Valéria C. Ebinuma-Santos c,
Ana P. M. Tavares
c,
Ana P. M. Tavares *b and
Cláudia G. Silva
*b and
Cláudia G. Silva *a
*a
aLaboratory of Separation and Reaction Engineering-Laboratory of Catalysis and Materials (LSRE-LCM), Department of Chemical Engineering, Faculty of Engineering, University of Porto, Rua Dr Roberto Frias, 4200-465, Porto, Portugal. E-mail: cgsilva@fe.up.pt
bDepartment of Chemistry, CICECO-Aveiro Institute of Materials, University of Aveiro, 3810-193 Aveiro, Portugal. E-mail: aptavares@ua.pt
cDepartment of Engineering Bioprocess and Biotechnology, School of Pharmaceutical Sciences, UNESP-University Estadual Paulista, Araraquara, Brazil
First published on 24th August 2020
Abstract
The enzyme L-asparaginase (ASNase) presents effective antineoplastic properties used for acute lymphoblastic leukemia treatment besides their potential use in the food sector to decrease the acrylamide formation. Considering their applications, the improvement of this enzyme's properties by efficient immobilization techniques is in high demand. Carbon nanotubes are promising enzyme immobilization supports, since these materials have increased surface area and effective capacity for enzyme loading. Accordingly, in this study, multi-walled carbon nanotubes (MWCNTs) were explored as novel supports for ASNase immobilization by a simple adsorption method. The effect of pH and contact time of immobilization, as well as the ASNase to nanoparticles mass ratio, were optimized according to the enzyme immobilization yield and relative recovered activity. The enzyme–MWCNTs bioconjugation was confirmed by thermogravimetric analysis (TGA), Fourier transform infrared spectroscopy (FTIR), Raman and transmission electron microscopy (TEM) studies. MWCNTs have a high ASNase loading capacity, with a maximum immobilization yield of 90%. The adsorbed ASNase retains 90% of the initial enzyme activity at the optimized conditions (pH 8.0, 60 min, and 1.5 × 10−3 g mL−1 of ASNase). According to these results, ASNase immobilized onto MWCNTs can find improved applications in several areas, namely biosensors, medicine and food industry.
Introduction
L-Asparaginase (L-asparagine amidohydrolase EC 3.5.1.1 – ASNase) presents pharmaceutical relevance in the treatment of acute lymphoblastic leukemia (ALL), a severe disease of the lymphoid system.1 Due to its specificity, ASNase has been a preferred pharmaceutical when compared to chemotherapy or radiation, known to be non-specific therapies.2,3 The catalysis of ASNase consists on the hydrolysis of L-asparagine into ammonia and L-aspartic acid. L-Asparagine is a vital amino acid that is used by both normal and cancer cells for their growth.2 However, unlike normal cells, leukemic lymphoblast cells cannot produce asparagine because the lack of the enzyme asparagine synthetase. Consequently, the depletion of serum L-asparagine by ASNase enzyme leads to tumour cell apoptosis.2,4 ASNase has another essential use in the food industry, the reduction of acrylamide formation. Acrylamide is a potential human carcinogen, being produced by the reaction of reducing sugars with L-asparagine at high-temperature cooking and in certain starchy foods. Thus, ASNase is able to reduce acrylamide formation by converting L-asparagine into L-aspartate.5,6 This enzyme is also used as a biosensor for the L-asparagine detection in both food and pharmaceutical sectors,7 as a more straightforward and economic method compared to different spectroscopy techniques.8 Nevertheless, the ASNase use faces certain hindrances regarding its application. The major limitation when used as a pharmaceutical is due to the adverse reactions caused on patients, like fever, allergic reactions, skin rashes or even anaphylactic shocks.9 In addition, due to its non-human origin, it has a recognized rapid plasma clearance by native proteases.10 Moreover, native ASNases are mostly unstable and thermolabile enzymes, which, together with their short half-life and impossibility of reuse, also makes their application in the food and biosensor industries challenging.11 Taking into account the importance of ASNase applications, the improvement of this enzyme properties by efficient immobilization techniques is in high demand.12ASNase immobilization has been studied using several supports, from organic to inorganic ones. Studied organic supports include polymers from natural sources, such as chitosan,13 silk sericin,14 and dextran,15 and the synthetic ones, like polyacrylamide16 or nylon.17 Activated carbon18 and silica gel19 are some inorganic supports studied for ASNase. During the last decade, nanomaterials appeared as a promising alternative for enzyme immobilization because of their particular characteristics, such as high surface area, efficient capability for enzyme loading and mass transfer resistance.20–23 Nanomaterials include several structures such as nanofibers, nanoparticles and nanotubes. In particular, carbon nanotubes (CNTs) have been subject to extensive research. CNTs are sheets of graphene with cylinder form presenting lengths in the order of μm and diameters up to 100 nm.24,25 Because of their exceptional structural dimensions,26 chemical and mechanical characteristics,27 strong loading capacity28 and biocompatibility,29,30 both forms of CNTs namely single-walled carbon nanotubes (SWCNT) and multi-walled carbon nanotubes (MWCNT), were already employed as carrier materials for the attachment of enzymes by diverse procedures, such as direct physical adsorption31,32 or covalent attachment.33,34 For instance, with laccase, CNTs proved to have a high adsorption capacity35,36 when compared to other carriers like silica,37 spent grain38 or coconut fibre,39 with a 333, 20 and 714 times higher adsorption capacity, respectively. Despite these promising results, to the best of our knowledge, up to now there are no previous reports about ASNase immobilization on pristine MWCNTs. In the current work, we investigated MWCNTs as novel immobilization support for ASNase by physical adsorption. This technique is a simple attachment method, with low associated costs, that allows easy enzyme removal and reload along with facilitated support regeneration when compared to covalent binding methodologies.40 We optimized different adsorption conditions (contact time, media pH, and ASNase/MWCNT ratio) to improve the enzyme loading and enzyme activity while characterizing the ASNase–MWCNT bioconjugate. Fig. 1 presents a schematic diagram that summarizes this research work.
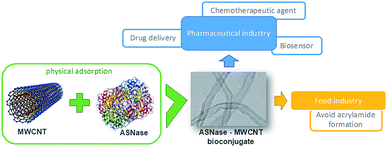 | ||
| Fig. 1 Schematic diagram summarizing the immobilization of L-asparaginase on MWCNTs and respective applications. | ||
Experimental
Materials, enzyme and chemicals
Lyophilized and purified ASNase from Escherichia coli (P1321-10000; 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 IU) was supplied by Deltaclon S.L., Spain. According to the supplier, the enzyme was lyophilized with no additives. MWCNTs were acquired from Nanocyl™ (NC3100). The MWCNTs have the following characteristics: diameter average of 9.5 nm, length average of 1.5 μm, specific surface area (SBET) of 189 m2 g−1 and purity ≥ 95%. L-Asparagine (≥99.0%), tris(hydroxymethyl)aminomethane (TRIS) (≥99.0%) and Na2HPO4 (≥99.0%) were acquired from VWR International, LLC. Trichloroacetic acid (TCA) (≥99.0%) was obtained from J.T. Baker. Citric acid (≥99.5%) and Nessler's reagent were supplied by Merck Chemical Company (Germany).
000 IU) was supplied by Deltaclon S.L., Spain. According to the supplier, the enzyme was lyophilized with no additives. MWCNTs were acquired from Nanocyl™ (NC3100). The MWCNTs have the following characteristics: diameter average of 9.5 nm, length average of 1.5 μm, specific surface area (SBET) of 189 m2 g−1 and purity ≥ 95%. L-Asparagine (≥99.0%), tris(hydroxymethyl)aminomethane (TRIS) (≥99.0%) and Na2HPO4 (≥99.0%) were acquired from VWR International, LLC. Trichloroacetic acid (TCA) (≥99.0%) was obtained from J.T. Baker. Citric acid (≥99.5%) and Nessler's reagent were supplied by Merck Chemical Company (Germany).
L-Asparaginase activity measurement
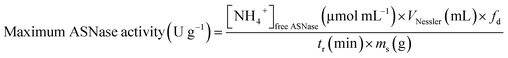
The activity of ASNase was measured based on the Nessler method which quantifies ammonia.41 In this method, the amount of ammonia generated from the L-asparagine hydrolysis by ASNase was monitored. The experimental procedure comprises the addition of 50 μL of free ASNase or 2.0 mg of ASNase–CNTs bioconjugate (immobilized ASNase) to 500 μL of Tris–HCl buffer (pH 8.6, 50 mM), 50 μL of 189 mM L-asparagine solution and 450 μL of deionized water and incubated at 37 °C for 30 min under stirring. Then, the enzymatic reaction was interrupted by adding 250 μL of TCA (1.5 M) to the mixture with the free enzyme. For immobilized enzyme, a centrifugation step was performed to separate the bioconjugate from the supernatant. Then, the ammonia amount released after the hydrolysis of L-asparagine was measured by adding 100 μL of each sample with 250 μL of Nessler's reagent and 2.15 mL of deionized water. After incubation during 30 min, the absorbance was determined spectrophotometrically at 436 nm (JASCO V-560 UV-Vis spectrophotometer). The concentration of ammonia formed was quantified by using a calibration curve with ammonium chloride as standard.One unit of free ASNase activity is described as the quantity of enzyme that produces 1 μmol of ammonia per minute (eqn (1)):
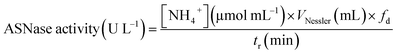 | (1) |
One unit of immobilized ASNase is described as the quantity of enzyme that produces 1 μmol of ammonia per minute and per mass of support at 37 °C (eqn (2)).
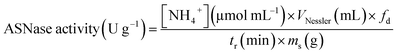 | (2) |
Optimization of L-asparaginase immobilization conditions
ASNase immobilization was performed by adding 2.0 mg of MWCNTs to 200 μL of an ASNase solution (8.6 × 10−5 g mL−1, unless otherwise stated) in an appropriate buffer solution under stirring in a multifunctional tube rotator (Grant Instruments Ltd., model PTR-35). Specific conditions are detailed below.The pH was evaluated by using various buffers: citrate/phosphate buffer (50 mM) for pH value 5.0; and phosphate buffer 50 mM for pH 6.0, 7.0 and 8.0. The contact time was optimized at the selected best pH by performing the immobilization in different intervals of time: from 15 to 120 min at room temperature. The optimal enzyme concentration was performed immobilizing ASNase solution at different concentrations of laccase: from 4.0 × 10−5 to 3.0 × 10−3 g mL−1, pH 8.0, 45 min of contact time.
After all immobilization tests, the mixture was centrifuged at 112g for 10 min and the supernatant was recovered and kept for ASNase activity measurements. MWCNTs were placed in contact with Nessler's reagent or ammonia without adsorbed ASNase in control experiments and no reaction was detected. Each test was carried out in triplicate.
The immobilization yield (IY, %), determined in each condition, is the difference between the enzyme activity (determined spectrophotometrically as previously described) of the free enzyme before ASNase immobilization and the activity of the free enzyme remaining in the supernatant after immobilization, divided by the free enzyme activity before immobilization (eqn (3)).
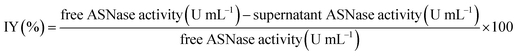 | (3) |
The immobilized enzyme relative recovered activity (RRA, %) was calculated as the ratio between the enzyme activity (determined spectrophotometrically as previously described) of the effectively immobilized enzyme and the maximum theoretical activity that would exist if the free enzyme was totally immobilized (eqn (4)).
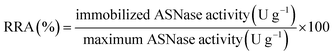 | (4) |
 | (5) |
Adsorption isotherms
The adsorption equilibrium behaviour of the ASNase was determined by Freundlich and Langmuir isotherms. The parameters of Langmuir model were determined by a non-linearized fitting of eqn (6) to the experimental data:
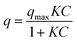 | (6) |
The parameters of the Freundlich model were determined by the non-linear fitting of eqn (7) to the experimental data:
| q = kF × C1/n | (7) |
ASNase concentrations from 4.0 × 10−5 to 3.0 × 10−3 g mL−1 were applied in the equilibrium studies. The equilibrium time was fixed at 45 min and pH 8.0.
Determination of kinetic parameters
The kinetic parameters of the enzymatic reaction were determined according the Michaelis–Menten equation:
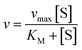 | (8) |
The Michaelis–Menten kinetic parameters KM and vmax of free and immobilized ASNase were determined by measuring the ASNase activity according to the method described before, using L-asparagine as substrate over a wide range of concentrations 1–250 mM for both free and immobilized ASNase. The parameters were obtained by non-linear fitting of eqn (8) to the experimental data (plot of reaction rate versus substrate concentration) using the CurveExpert software. The ASNase efficiency was determined by the ratio of kcat (turnover number) to KM. kcat was determined by dividing vmax by the enzyme total concentration.43
Characterization techniques
The pristine MWCNT textural properties were determined by N2 adsorption–desorption at −196 °C on a Quantachrome NOVA 4200e equipment. Before analysis, each sample (60–80 mg) was degassed at 120 °C for 3 h under vacuum. The specific surface area (SBET) was calculated by multipoint BET analysis of the obtained isotherm in the relative pressure range 0.05–0.15.Transmission electron microscopy (TEM) images were acquired using an analytical electron microscope (JEOL 2010F) equipped with a field-emission gun.
Thermogravimetric (TG) analyses were executed using a STA 490 PC/4/H Luxx Netzsch thermal equipment. For each test, approximately 5 mg of MWCNTs were loaded on the sample crucible and heated at 10 °C min−1 from 50 °C to 900 °C under air flow, while the weight was measured and recorded continuously. The TG profiles are a result of the average of three independent assays, with a maximum deviation of 5%.
Raman spectra were obtained by a Brucker RFS100/S FT-Raman spectrometer (Nd:YAG laser, 1064 nm excitation), with 3000 scans at a resolution of 4 cm−1 at a power of 200 mV.
The Fourier transform infrared (FTIR) spectra (4000–600 cm−1) of ASNase and ASNase–MWCNTs bioconjugate were acquired in a JASCO FT/IR-6800 spectrometer (JASCO Analytical Instruments, USA) equipped with a MIRacle™ Single Reflection ATR (attenuated total reflectance ZnSe crystal plate) accessory (PIKE Technologies, USA). The analyses were performed using 256 scans with a resolution of 4 cm−1.
Results and discussion
L-Asparaginase immobilization
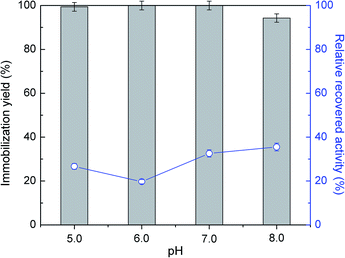
Several studies reported CNTs as an effective support for enzymes immobilization.44,45 They demonstrated to be stable in severe environments and to provide higher enzyme loading with improved catalytic activity, as verified for instance with soybean peroxidase.46 In this work, the immobilization of commercial ASNase was studied through physical adsorption onto MWCNTs. Adsorption is a simple non-covalent mechanism that comprises weak van der Waals forces, hydrogen bonding, electrostatic, hydrophobic and π–π stacking interactions between the biocatalyst and the support.47The immobilization of ASNase onto MWCNTs is expected to be influenced by pH, since it determines the surface charge of the enzyme. In this sense, initial tests were conducted varying the pH between 5.0 to 8.0 during the immobilization of 8.6 × 10−5 g mL−1 of ASNase onto MWCNTs for 60 min. The efficacy of the immobilization was analysed through the balance between the immobilization yield and enzyme relative recovered activity results. According to Fig. 2 (respective detailed data given in Table S1 of the ESI†), the enzyme adsorption onto MWCNTs is total for the pH values 5.0, 6.0 and 7.0. A small decrease in the immobilization yield (94%) is, however, observed for pH 8.0. It is known that the isoelectric point (IP) of ASNase is between 5.0 and 5.7 (ref. 48) and that the point of zero charge of carbon nanotubes used in this work is 7.0.49 Therefore, at alkaline pH values, i.e. pH 8.0, both enzyme and support are negatively charged, suggesting that electrostatic interactions are not expected to be responsible for the enzyme immobilization. On the other hand, the relative enzyme recovery changes between 20 to 40%, being higher at a pH value of 8.0.
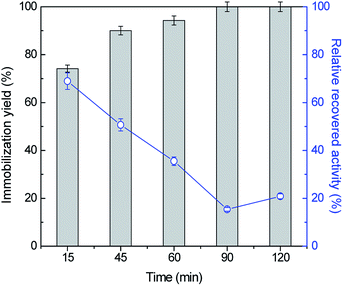
To attain the maximum immobilization yield and relative recovered activity values, the influence of other key conditions, namely ASNase concentration and contact time was then assessed. The adsorption of ASNase onto MWCNTs was evaluated for five contact times, between 15 to 120 min, using 8.6 × 10−5 g mL−1 of ASNase at pH 8.0. Fig. 3 (respective detailed data given in Table S2 in the ESI†) shows the effect of contact time on the immobilization yield and recovery activity. Overall, there is an increase in the immobilization yield and a decrease in the relative recovery ASNase activity with time. For an immobilization time of 15 min, a high recovered enzymatic activity was observed, while a low immobilization yield is detected. In fact, with lower amounts of immobilized enzyme, it will be more able to react by avoiding mass transfer restrictions that may occur with a higher enzyme loading. As more enzyme is immobilized, a multi-layered stacking of adsorbed ASNase or an uncontrolled enzyme packing during the immobilization procedure may occur, blocking the access of substrate molecules to the active site of ASNase. After 45 min of contact between the enzyme and the MWCNTs, an increase in the immobilization yield was obtained, reaching 100% at 90 min, yet with a recovery activity of 15%. Overall, this set of results confirms the fast binding of the enzyme onto MWCNTs without resorting to any surface modification or introduction of crosslinkers. Considering the recovery activity of ≈50% and immobilization yield of ≈90%, 45 min of contact time was selected for further experiments.
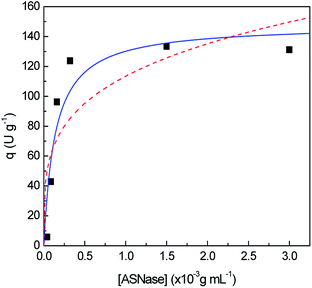
In order to identify improved adsorption capacities of MWCNTs, the enzyme/support mass ratio was also investigated. ASNase was immobilized onto MWCNTs at pH 8.0 during 45 min using six enzyme concentrations, ranging from 4.0 × 10−5 to 3.0 × 10−3 g mL−1, while maintaining the mass of MWCNTs at 2.0 mg, whose results are given in Fig. 4. The respective detailed data are given in Table S3 in the ESI.† There is an almost total adsorption (immobilization yield > 90%) for all the enzyme concentrations studied and an increment in the relative recovered activity with the increase of the ASNase content up to 1.5 × 10−3 g mL−1. A 8.2-fold improvement in the relative recovered activity of the immobilized ASNase, from 11.1 to 90.9%, during the same immobilization time, was observed with the increase of the enzyme concentration from 4.0 × 10−5 to 1.5 × 10−3 g mL−1, respectively. This behavior may be associated to a higher amount of ASNase molecules available to adsorb onto MWCNTs surface, forming an enzyme layer, covering the MWCNT surface. However, the increment in the enzyme concentration up to 3.0 × 10−3 g mL−1 shows a negligible effect on the relative recovered activity when compared with the concentration of 1.5 × 10−3 g mL−1, suggesting that the MWCNTs surface attained its maximum adsorption capacity.
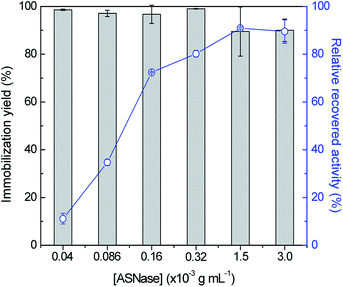 | ||
| Fig. 4 Effect of enzyme concentration on the immobilization yield (columns) and relative recovered activity (symbols, line) with the immobilization of ASNase onto 2 mg of MWCNTs at pH 8.0 for 45 min. | ||
Resorting to the literature (Table 1), the ASNase immobilization on other supports has been reported, for example, the ASNase was encapsulated with poly(lactide-co-glycolide) nanoparticles, preserving 62.8% of the activity.50
| ASNase immobilization method | Support | ASNase source | Results | Ref. |
|---|---|---|---|---|
| Encapsulation | Poly(lactide-co-glycolide) nanoparticles | Recombinant ASNase | 63% RRA | 49 |
| Covalent binding | Natural silk sericin protein microparticles | Escherichia coli | 63% RRA | 14 |
| Entrapment | Ca-ALG/MWCNT-COOH | Escherichia coli | 97% IY | 50 |
| Physical adsorption | MWCNT-COOH | Aspergillus versicolor | 54% IY, 100% RRA | 51 |
| Physical adsorption | Pristine MWCNT | Escherichia coli | >90% IY, >90% RRA | This work |
Zhang et al.14 tested natural silk sericin protein microparticles as support for ASNase by covalent attachment, attaining 62.5% of the enzyme original activity. The immobilization of ASNase on functionalized CNTs was already reported in literature. Ulu et al.51 described the use of a novel calcium-alginate/carboxylated multi-walled carbon nanotube hybrid bead (Ca-ALG/MWCNT-COOH) for the entrapment of ASNase reaching an IY of 97%. Oxidized MWCNTs were also studied by Haroun et al.52 as a support for an Aspergillus versicolor ASNase reporting higher IY values using a physical adsorption technique than with the covalent binding of the enzyme to the support, with a maximum IY attained of 54.4%. On the other hand, the immobilized ASNase retained 100% of the free enzyme activity. In this work, pristine MWCNTs proved to provide a relatively high enzyme loading (higher than 90%) by a simple adsorption method (without any modification over the surface of the MWCNTs), highlighting the easy accessibility of enzyme's active sites by the substrate, which was confirmed by the high relative recovered activity achieved (above 90%). These results prove the potential of using simple pristine MWCNTs as supports for the ASNase immobilization, reaching promising values of both IY and RRA parameters, which together proved to be higher than those reported in the literature (comparison in Table 1), highlighting the use of a simpler and cost-effective immobilization process.
Aiming to better understand how ASNase interacts with the MWCNTs surface leading to the enzyme adsorption, the Langmuir and Freundlich adsorption isotherms were determined, whose results are shown in Fig. 5. The adsorption isotherm parameters were obtained by non-linear fitting of Langmuir and Freundlich models to the experimental data, given by eqn (6) and (7), respectively. The respective detailed data corresponding to the experimental data fitting are given in Table S4 in the ESI.† Taking into account the correlation coefficients (R2) obtained, the Langmuir model better describes the experimental equilibrium data. The Freundlich isotherm is characterized by an exponential distribution of heterogeneous active sites leading to a multilayer adsorption.53 The lowest correlation coefficient observed in the Freundlich isotherm fitting shows that the formation of ASNase multilayers on the MWCNTs probably does not occur. On the other hand, according to the Langmuir isotherm, where a monolayer adsorption on the support surface is predicted, the MWCNTs maximum adsorption capacity (qmax) for ASNase is 148.0 U g−1 and the adsorption equilibrium constant (K), which defines the interactions strength between the enzyme and the surface of MWCNTs is 7.3 mL g−1.
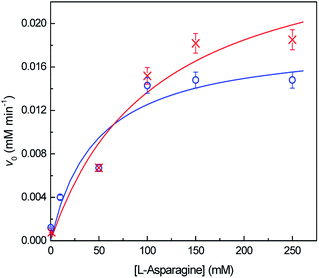
The kinetic parameters were determined by non-linear fitting of Michaelis–Menten equation (eqn (8)) to the experimental data (Fig. 6). Table 2 shows the obtained kinetic constants, vmax and KM, for both free and immobilized ASNase, along with the respective catalytic efficiency (kcat/KM). KM is a measure of the enzyme affinity to the substrate. The lesser the value of KM, the higher is the enzyme affinity to the substrate.39 The higher KM value for immobilized ASNase indicates a lower affinity for L-asparagine. The increase in KM parameter after enzyme immobilization is frequently reported in literature due to the steric hindrances and diffusional limitations promoted by the support.23,54 These factors also lead to a 35% decrease in the ASNase catalytic efficiency (kcat/KM) upon immobilization onto MWCNTs (Table 2). Nevertheless, contrary to what is usually reported, the immobilized ASNase attained a higher vmax value when compared to the free enzyme probably due to the adsorption by the MWCNTs: the local concentration increases, increasing the reaction rate. This property reinforces the great ability of MWCNTs as supports for ASNase immobilization.
| Kinetic parameter | Native ASNase | Immobilized ASNase |
|---|---|---|
| vmax (mM min−1) | 0.019 | 0.029 |
| KM (mM) | 47 | 109 |
| kcat/KM (mM−1 min−1) | 0.008 | 0.006 |
ASNase–MWCNT bioconjugate characterization
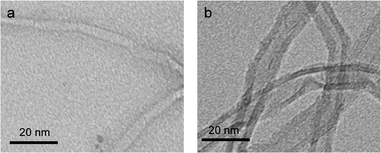
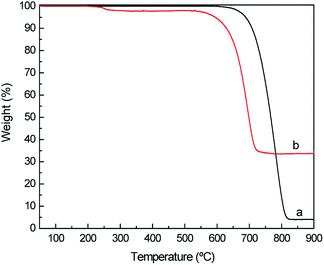
To confirm the immobilization of ASNase onto MWCNTs, the surface morphology of the original MWCNTs and of ASNase–MWCNTs bioconjugates was appraised by TEM. Fig. 7a shows the typical morphology of pristine MWCNTs, exhibiting a tubular shape with a smooth surface. After immobilization of ASNase (Fig. 7b), the MWCNTs limits look more filled/thicker due to the enzyme coating on MWCNTs nanosheets.Thermogravimetric analysis (TGA) was also carried out to confirm ASNase immobilization onto MWCNTs (Fig. 8a). The original MWCNTs (without enzyme) display almost no weight loss up to 700 °C, being practically completely burned above 800 °C. After this temperature a plateau is reached, corresponding to 3.8% of the initial mass, due to the existence of ashes resulting from the pyrolysis of impurities. The TG profile of ASNase has been previously reported by Shakambari et al.55 revealing an accentuated mass loss starting at 250 °C, which is clearly observed in the TG profile of the ASNase–MWCNTs bioconjugate, corresponding to the thermal decomposition of the enzyme (Fig. 8b). The second and higher weight loss begins at 550 °C is attributed to the simultaneous pyrolysis of ASNase and MWCNTs, attaining a plateau at a temperature close to 700 °C. This plateau corresponds to 44.8% of the initial mass of the enzyme–support complex. These results suggest that the enzyme content in the bioconjugate is ca. 41 wt% (difference between the mass loss obtained for the pristine MWCNTs and the one obtained for the bioconjugate).
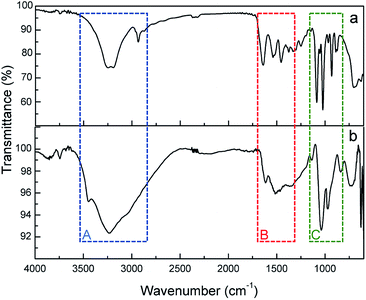
FTIR-ATR spectra of ASNase and ASNase–MWCNTs bioconjugate is shown in Fig. 9. To confirm the presence of ASNase, the spectrum of pristine MWCNTs was subtracted from the spectrum of the ASNase–MWCNTs bioconjugate. The characteristic bands of the stretching vibrations of N–H and O–H bonds appear at ca. 3240 and 3100 cm−1, respectively (Fig. 9, Zone A). In the case of ASNase (Fig. 9a), a peak at 2934 cm−1 assigned to the stretching vibrations of C–H bonds in amino acids is also observed.23 In addition, the broad band observed in the spectrum of the ASNase–MWCNTs bioconjugate (Fig. 9b) between 3500 and 2800 cm−1 is attributed to the stretching vibrations of hydrogen bonded surface water molecules and hydroxyl groups. The characteristic bands of N–H stretching of amide I and N–H bending of amide II groups of asparaginase, can be found in both ASNase and ASNase–MWCNTs spectra at 1638 and 1530 cm−1, respectively (Fig. 9, Zone B).56 The band at ca. 1390 cm−1 corresponds to asymmetric stretching vibrations of carboxyl groups in amino acids. Finally, a number of common bands are observed for ASNase and ASNase–MWCNTs bioconjugate in the range 1200–800 cm−1 (Fig. 9, Zone C), corresponding to the vibrations of C–C, C–O, C–N, N–H and C–H bonds in amino acids structure.57
Raman spectroscopy allows to examine the changes in the MWCNTs surface after ASNase immobilization. The spectra of both pristine MWCNTs and ASNase–MWCNTs bioconjugate (Fig. 10) display 2 main typical bands (D and G peaks) in the 1000–2000 cm−1 spectral region. The first order G mode peaking at ca. 1600 cm−1 corresponds to the regular sp2 graphitic network of MWCNTs which is typical for carbon based materials. The D mode band (ca. 1287 cm−1) reveals the defects and disorder produced by sp3 hybridized carbon in the lattice.58 Moreover, the presence of defect sites and the purity level in the MWCNTs, and the amount of functionalization, are normally assessed by the ratio between D and G bands intensities (ID/IG).59 The ID/IG values obtained for pristine MWCNTs and ASNase–MWCNTs bioconjugate were 0.537 and 0.550, respectively. The slight difference observed for the ID/IG ratio suggests that the enzyme immobilization in the nanomaterials does not cause significant disturbance on the MWCNTs surface, indicating similar degrees of disorder. This behaviour is in accordance to the work described by Monajati et al.60 for the immobilization of ASNase on graphene oxide, suggesting that the ASNase adsorption onto MWCNTs occurs predominantly on the defect sites previously present on MWCNTs surface, not changing the sp2 and sp3 bonds concentrations.
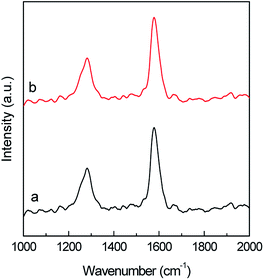 | ||
| Fig. 10 Raman spectra of (a) multi-walled carbon nanotubes (MWCNTs) and (b) ASNase–MWCNTs bioconjugate. | ||
Conclusions
The immobilization of commercial ASNase on MWCNTs was successfully achieved by adsorption, with an immobilization yield and a recovery of free enzyme activity above 90%. The contact time between the ASNase and the MWCNTs through the immobilization procedure influences the results, where short immobilization times may not allow the total enzyme adsorption. An immobilization yield of 100% was achieved after 45 min of immobilization.The pH did not show a considerable effect on the immobilization yield within the range 5.0–8.0, attaining the highest relative recovered activity at pH 8.0. The parameter that revealed the most substantial influence on the immobilization effectiveness was the ASNase/MWCNTs mass ratio. To take full advantage of the use of the ASNase/MWCNT bioconjugate, it is necessary to ensure maximum enzyme adsorption capacity. This occurred with a concentration of ASNase of 1.5 mg mL−1 in 2.0 mg of carbon nanotubes, corresponding to a MWCNT maximum enzyme loading capacity of 148.0 U g−1. Under these conditions, an immobilization efficiency and relative recovered activity of 90% were achieved.
The determined adsorption isotherms allowed to conclude that there is monolayer adsorption of the ASNase on the MWCNTs. The adsorption of the enzyme onto MWCNTs was further confirmed by TEM, FTIR, TGA, and Raman spectroscopic results.
The obtained results show that MWCNTs are efficient supports of ASNase, with no chemical modification or covalent binding required, opening the possibility of using ASNase–MWCNT bioconjugates in several application fields, e.g. as biosensors, in medicine, pharmaceutical and food industry.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by Base Funding – UIDB/EQU/50020/2020 of the Associate Laboratory LSRE-LCM – funded by national funds through FCT/MCTES (PIDDAC), and POCI-01-0145-FEDER-031268 – funded by FEDER, through COMPETE2020 – Programa Operacional Competitividade e Internacionalização (POCI), and by national funds (OE), through FCT/MCTES. This work was developed within the scope of the project CICECO-Aveiro Institute of Materials, UIDB/50011/2020 & UIDP/50011/2020, financed by national funds through the Portuguese Foundation for Science and Technology/MCTES. Ana P. M. Tavares and Cláudia G. Silva acknowledge the FCT Investigator Programme and Exploratory Project (IF/01634/2015 and IF/00514/2014, respectively) with financing from the European Social Fund and the Human Potential Operational Programme. Valéria C. Santos-Ebinuma acknowledges FAPESP (2018/06908-8). Raquel O. Cristóvão acknowledges FCT funding under DL57/2016 Transitory Norm Programme. João C. F. Nunes acknowledges FCT for the PhD fellowship (SFRH/BD/150671/2020).References
- G. Shakambari, B. M. Sumi, B. Ashokkumar, P. Palanivelu and P. Varalakshmi, RSC Adv., 2015, 5, 48729–48738 RSC.
- G. Dorsa and V. Jaleh, Recent Pat. Nanotechnol., 2018, 12, 70–82 CrossRef PubMed.
- M. Shanmugaprakash, C. Jayashree, V. Vinothkumar, S. N. S. Senthilkumar, S. Siddiqui, V. Rawat and M. Arshad, Sep. Purif. Technol., 2015, 142, 258–267 CrossRef CAS.
- G. Shakambari, A. K. Birendranarayan, M. J. Angelaa Lincy, S. K. Rai, Q. T. Ahamed, B. Ashokkumar, M. Saravanan, A. Mahesh and P. Varalakshmi, RSC Adv., 2016, 6, 25943–25951 RSC.
- F. Xu, M.-J. Oruna-Concha and J. S. Elmore, Food Chem., 2016, 210, 163–171 CrossRef CAS PubMed.
- Z. Sun, R. Qin, D. Li, K. Ji, T. Wang, Z. Cui and Y. Huang, Int. J. Biol. Macromol., 2016, 92, 232–239 CrossRef CAS PubMed.
- L. H. R. R. Possarle, J. R. Siqueira Jr and L. Caseli, Colloids Surf., B, 2020, 192, 111032 CrossRef CAS PubMed.
- T. Batool, E. A. Makky, M. Jalal and M. M. Yusoff, Appl. Biochem. Biotechnol., 2016, 178, 900–923 CrossRef CAS PubMed.
- U. Ali, M. Naveed, A. Ullah, K. Ali, S. A. Shah, S. Fahad and A. S. Mumtaz, Eur. J. Pharmacol., 2016, 771, 199–210 CrossRef CAS PubMed.
- M. K. Danks, K. J. Yoon, R. A. Bush, J. S. Remack, M. Wierdl, L. Tsurkan, S. U. Kim, E. Garcia, M. Z. Metz, J. Najbauer, P. M. Potter and K. S. Aboody, Cancer Res., 2007, 67, 22 CrossRef CAS PubMed.
- S. Zuo, T. Zhang, B. Jiang and W. Mu, Appl. Microbiol. Biotechnol., 2015, 99, 1069–1079 CrossRef CAS PubMed.
- B. Ates, A. Ulu, S. Köytepe, S. A. Ali Noma, V. S. Kolat and T. Izgi, RSC Adv., 2018, 8, 36063–36075 RSC.
- E. Bahreini, K. Aghaiypour, R. Abbasalipourkabir, A. R. Mokarram, M. T. Goodarzi and M. Saidijam, Nanoscale Res. Lett., 2014, 9, 340 CrossRef PubMed.
- Y.-Q. Zhang, M.-L. Tao, W.-D. Shen, Y.-Z. Zhou, Y. Ding, Y. Ma and W.-L. Zhou, Biomaterials, 2004, 25, 3751–3759 CrossRef CAS PubMed.
- T. E. Wileman, R. L. Foster and P. N. C. Elliott, J. Pharm. Pharmacol., 1986, 38, 264–271 CrossRef CAS PubMed.
- T. Mori, T. Tosa and I. Chibata, Cancer Res., 1974, 34, 3066 CAS.
- J. P. Allison, L. Davidson, A. Gutierrez-Hartman and G. Barrie Kitto, Biochem. Biophys. Res. Commun., 1972, 47, 66–73 CrossRef CAS PubMed.
- M. Moharam, A. Gamal-Eldeen and S. El-Sayed, J. Am. Sci., 2010, 6, 131–140 Search PubMed.
- I. Al-Kadmy, S. Muslim, N. Hussein, A. Ali and A. Dwaish, International Journal of Advances in Chemical Engineering and Biological Sciences, 2015, 2, 35–39 Search PubMed.
- W. Feng and P. Ji, Biotechnol. Adv., 2011, 29, 889–895 CrossRef CAS PubMed.
- M. N. Gupta, M. Kaloti, M. Kapoor and K. Solanki, Artif. Cells, Blood Substitutes, Biotechnol., 2011, 39, 98–109 CrossRef CAS PubMed.
- H. Jia, G. Zhu and P. Wang, Biotechnol. Bioeng., 2003, 84, 406–414 CrossRef CAS PubMed.
- S. Agrawal and N. Kango, Int. J. Biol. Macromol., 2019, 135, 1142–1150 CrossRef CAS PubMed.
- D. Tasis, N. Tagmatarchis, A. Bianco and M. Prato, Chem. Rev., 2006, 106, 1105–1136 CrossRef CAS PubMed.
- Ihsanullah, Sep. Purif. Technol., 2019, 209, 307–337 CrossRef CAS.
- A. Dettlaff, M. Sawczak, E. Klugmann-Radziemska, D. Czylkowski, R. Miotk and M. Wilamowska-Zawłocka, RSC Adv., 2017, 7, 31940–31949 RSC.
- J. Chen, B. Liu, X. Gao and D. Xu, RSC Adv., 2018, 8, 28048–28085 RSC.
- X. Dong, C. Wei, J. Liang, T. Liu, D. Kong and F. Lv, Colloids Surf., B, 2017, 154, 253–262 CrossRef CAS PubMed.
- S. K. Verma, A. Modi and J. Bellare, Colloids Surf., B, 2019, 181, 890–895 CrossRef CAS PubMed.
- X. Zhao, K. Tian, T. Zhou, X. Jia, J. Li and P. Liu, Colloids Surf., B, 2018, 168, 43–49 CrossRef CAS PubMed.
- D. W. Horn and V. A. Davis, Colloids Surf., B, 2016, 139, 237–243 CrossRef CAS PubMed.
- X. Zhou, J. Su, C. Wang, C. Fang, X. He, W. Lei, C. Zhang and Z. Huang, J. Mater. Sci. Technol., 2020, 46, 74–87 CrossRef.
- M. M. Noor, J. Goswami and V. A. Davis, ACS Omega, 2020, 5, 2254–2259 CrossRef CAS PubMed.
- M. Di Giosia, F. Valle, A. Cantelli, A. Bottoni, F. Zerbetto, E. Fasoli and M. Calvaresi, Carbon, 2019, 147, 70–82 CrossRef CAS.
- C. G. Silva, A. P. M. Tavares, G. Dražić, A. M. T. Silva, J. M. Loureiro and J. L. Faria, ChemPlusChem, 2014, 79, 1116–1122 CrossRef CAS.
- A. P. M. Tavares, C. G. Silva, G. Dražić, A. M. T. Silva, J. M. Loureiro and J. L. Faria, J. Colloid Interface Sci., 2015, 454, 52–60 CrossRef CAS PubMed.
- A. P. M. Tavares, O. Rodríguez, M. Fernández-Fernández, A. Domínguez, D. Moldes, M. A. Sanromán and E. A. Macedo, Bioresour. Technol., 2013, 131, 405–412 CrossRef CAS PubMed.
- A. M. da Silva, A. P. M. Tavares, C. M. R. Rocha, R. O. Cristóvão, J. A. Teixeira and E. A. Macedo, Process Biochem., 2012, 47, 1095–1101 CrossRef CAS.
- R. O. Cristóvão, S. C. Silvério, A. P. M. Tavares, A. I. S. Brígida, J. M. Loureiro, R. A. R. Boaventura, E. A. Macedo and M. A. Z. Coelho, World J. Microbiol. Biotechnol., 2012, 28, 2827–2838 CrossRef PubMed.
- N. R. Mohamad, N. H. C. Marzuki, N. A. Buang, F. Huyop and R. A. Wahab, Biotechnol. Biotechnol. Equip., 2015, 29, 205–220 CrossRef CAS PubMed.
- A. Magri, M. F. Soler, A. M. Lopes, E. M. Cilli, P. S. Barber, A. Pessoa and J. F. B. Pereira, Anal. Bioanal. Chem., 2018, 410, 6985–6990 CrossRef CAS PubMed.
- M. Belhachemi and F. Addoun, Appl. Water Sci., 2011, 1, 111–117 CrossRef CAS.
- R. A. Copeland, in Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis, John Wiley & Sons, Inc., 2nd edn, 2000 Search PubMed.
- L. Wang, L. Wei, Y. Chen and R. Jiang, J. Biotechnol., 2010, 150, 57–63 CrossRef CAS PubMed.
- M. Shim, N. W. Shi Kam, R. J. Chen, Y. Li and H. Dai, Nano Lett., 2002, 2, 285–288 CrossRef CAS.
- P. Asuri, S. S. Bale, S. S. Karajanagi and R. S. Kane, Curr. Opin. Biotechnol., 2006, 17, 562–568 CrossRef CAS PubMed.
- S. Kushagri, M. Abha, S. Deepankar and S. Kavita, Curr. Nanosci., 2019, 15, 234–241 CrossRef.
- S. M. Kumar and K. Selvam, J. Microb. Biochem. Technol., 2011, 3, 73–83 Search PubMed.
- S. Morales-Torres, T. L. S. Silva, L. M. Pastrana-Martínez, A. T. S. C. Brandão, J. L. Figueiredo and A. M. T. Silva, Phys. Chem. Chem. Phys., 2014, 16, 12237–12250 RSC.
- S. Vasudev, S. Ahmad, R. Parveen, F. Ahmad, C. Anish, M. Ali and A. Panda, Pharmazie, 2011, 66, 956–960 CAS.
- A. Ulu, M. Karaman, F. Yapıcı, M. Naz, S. Sayın, E. İ. Saygılı and B. Ateş, Catal. Lett., 2020, 150, 1679–1691 CrossRef CAS.
- A. Haroun, H. Ahmed, A.-T. Mossa, S. Mohafrash and E. Ahmed, Biointerface Res. Appl. Chem., 2020, 10, 5733–5740 Search PubMed.
- F. Eba, E. A.-M. A. Gueu, J. Ondo, B. Yao, J. Nlo and R. Biboutou, Int. J. Eng. Sci. Technol., 2010, 2, 5001–5016 Search PubMed.
- H. A. El-Refai, M. S. Shafei, H. Mostafa, A. M. El-Refai, E. M. Araby, F. M. El-Beih, S. M. Easa and S. K. Gomaa, Pol. J. Microbiol., 2016, 65, 43–50 Search PubMed.
- G. Shakambari, R. Sameer Kumar, B. Ashokkumar, V. Ganesh, V. S. Vasantha and P. Varalakshmi, Sci. Rep., 2018, 8, 18013 CrossRef CAS PubMed.
- E. V. Kudryashova, M. V. Pokrovskaya, S. S. Alexandrova, A. A. Vinogradov and N. N. Sokolov, Anal. Biochem., 2020, 598, 113694 CrossRef CAS PubMed.
- M. Mecozzi and E. Sturchio, Journal of Imaging, 2017, 3, 11 CrossRef.
- P. V. Iyer and L. Ananthanarayan, Process Biochem., 2008, 43, 1019–1032 CrossRef CAS.
- E.-S. Mohamed and E.-C. Ghadir, J. Agrobiol., 2011, 28, 129–137 Search PubMed.
- M. Monajati, S. Borandeh, A. Hesami, D. Mansouri and A. M. Tamaddon, Chem. Eng. J., 2018, 354, 1153–1163 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed information about relative recovered activity (%) and immobilization yield of ASNase; detailed information of the amount of adsorbed active ASNase (U) per gram of MWCNT for Langmuir and Freundlich isotherms. See DOI: 10.1039/d0ra05534d |
| This journal is © The Royal Society of Chemistry 2020 |