Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Mass spectrometric analysis of lipid A obtained from the lipopolysaccharide of Pasteurella multocida
Abdul Tawab ab,
Noor Akbara,
Mujtaba Hasssana,
Fazale Habiba,
Aamir Alia,
Moazur Rahmanac,
Abdul Jabbarad,
Waqar Rauf
ab,
Noor Akbara,
Mujtaba Hasssana,
Fazale Habiba,
Aamir Alia,
Moazur Rahmanac,
Abdul Jabbarad,
Waqar Rauf a and
Mazhar Iqbal
a and
Mazhar Iqbal *ab
*ab
aHealth Biotechnology Division, National Institute for Biotechnology and Genetic Engineering (NIBGE), P.O. Box 577, Jhang Road, Faisalabad-38000, Pakistan. E-mail: hamzamgondal@gmail.com; Tel: +92 419201403
bDepartment of Biotechnology NIBGE, Pakistan Institute of Engineering and Applied Sciences (PIEAS), Lehtrar Road, Nilore, Islamabad-45650, Pakistan
cSchool of Biological Sciences, University of the Punjab, Lahore, Pakistan
dDepartment of Biotechnology, Mirpur University of Science and Technology (MUST), Mirpur-10250, Azad Jammu and Kashmir, Pakistan
First published on 20th August 2020
Abstract
Haemorrhagic septicaemia is mainly caused by an opportunistic pathogen, Pasteurella multocida, a major threat to the livestock dependent economies. The main endotoxins are lipopolysaccharides. The lipid A, a key pathogenic part of lipopolysaccharides, anchors it into the bacterial cell membrane. Hence, profiling of the lipid A is important to understand toxicity of this pathogen. Despite a significant progress made on glycan analyses of core regions of lipopolysaccharides from various P. multocida strains, the structure of lipid A has not been reported yet. The lipid A of P. multocida type B:2 was analyzed using ESI-MS/MS to identify the acylation patterns, number and length of various acyl fatty acids, phosphorylation level and lipid A modifications. The MSn data revealed the existence of multiple lipid A variants, i.e. mono and bisphosphorylated hepta-, hexa-, penta- and tetra-acylated structures, decorated with varied levels of 4-amino-4-deoxy-L-arabinose (Ara4N) on C-1 and/or C-4′ phosphate groups of proximal and distal glucosamine lipid A backbone. The detailed mass spectrometric analyses revealed that even within the same class, lipid A exhibits several sub-variant structures. A primary and secondary myristoylation at C-2, C-3, C-2′ and C-3′ was observed in all variants except hepta-acylated lipid A that carried a secondary palmitate at C-2 position. The lipid A profiling described herein, may contribute in exploring the mechanisms involved in endotoxicity of P. multocida type B:2 in haemorrhagic septicaemia disease.
Introduction
Pasteurella multocida is an opportunistic Gram-negative bacterial pathogen associated with various septic diseases of wild as well as domestic animals.1 Based on capsular chemistry, P. multocida strains have been classified into 5 sub-groups i.e. A, B, D, E and F; whereas, on the basis of the variations in their lipopolysaccharides (LPS), there are around 16 Heddleston serovars.2,3 Haemorrhagic septicaemia (HS) disease is mostly associated with serotypes B:2 and E:2 of P. multocida subsp. multocida (Carter and Heddleston system), corresponding to the 6:B and 6:E serotypes in Namioka–Carter system.4 HS is endemic in ungulates, predominantly water buffalo (Bubalus bubalis) and cattle in most parts of tropical Asia and Africa.5,6 In fact, HS is classified among list B cattle diseases notifiable to the Office International des Epizooties (OIE) with 100% mortality rate in infected animals if not treated at early stage.7 The diseased animals usually collapse and die within a few hours to a few days after the onset of the illness. Sudden death with few or no clinical signs can also be seen while the symptomatic animals, especially buffalo, rarely recover.4,8 This makes HS, the most important disease of bovines in terms of economic impact in various tropical and sub-tropical parts of the world.9 In Pakistan, having cattle and buffalo populations of 47.8 and 40 million respectively,8 young buffalo, being the most affected group,10 has been reported as an important reservoir of P. multocida.4The pathogenesis of the P. multocida is poorly understood as compared to other Gram-negative bacteria. The haemagglutinin surface adhesin,11 capsular polysaccharides and LPS are critical virulence factors.12 The LPS plays a significant role in adhesion of P. multocida to the respiratory epithelium of host.13 The mutants attenuated to acapsular as well as truncated LPS, showed no or significantly reduced virulence and found to be more prone to the host antimicrobial peptides.14–16 Indeed, intravenous administration of the purified LPS induced the clinical signs resembling haemorrhagic septicaemia in buffalo i.e. endotoxic shock.17 Notably, in LPS, the lipid A part is considered as the active endotoxic part leading to inflammation, sepsis and septic shock in Gram-negative bacterial infections.18,19 Lipid A is primary pathogen-associated molecular pattern (PAMP) that binds to the pattern recognition receptors (PRR) e.g. toll-like-receptors (TLRs) of various eukaryotic cells. The first encounter of bacterial LPS occurs with a host cell plasma protein called lipopolysaccharide binding protein (LBP). LBP-bound LPS is recognized by a protruding CD14 receptor on the phagocyte cell membrane for onward presentation to TLR4. An intracellular signalling cascade, causing the induction of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), is triggered by this complex oligomerization. The downwards signalling leads to the NF-κB activation, pro-inflammatory cytokines production and induction of more and more host immune cells.20–22 Consequently, excessive inflammatory response of the innate immune system leads to sepsis and septic shock.23
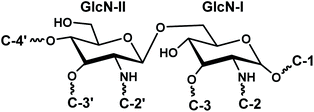
The disaccharide backbone of lipid A is highly conserved among most of the bacterial species24,25 with few exceptions.26,27 The overall chemical structure of Gram-negative bacterial lipid A consists of disaccharide backbone of two covalently bonded pyranosidic hexaosamine residues (β-D-GlcpN4P-(1→6)-α-D-GlcpN1P). The skeletal disaccharide moiety is bonded to fatty acid chains at C-2/C-2′, C-3/C-3′, and mono or bisphosphorylation modifications at C-1, C-4′. The length of the fatty acid chains varies from C10 to C16 carbon atoms; however, some higher length chain may also exist.28 The endotoxicity of lipid A is associated with its structure i.e. number and length of fatty acyl chains, nature and phosphorylation of its glycan disaccharide backbone.29–31 The bisphosphorylated lipid A, substituted by six or seven asymmetrically distributed fatty acids linked via ester and amide linkages, is found to be highly endotoxic to the mammalian host.32 Such asymmetric (4/2) hexa-acylated and (4/3) hepta-acylated lipid A variants are reported in diverse pathogenic Gram-negative bacteria e.g. Salmonella Typhimurium,28 E. coli,33 Klebsiella pneumoniae,34 Hafnia alvei.35 For intracellular survival, the Gram-negative bacteria modify their lipid A structures by adding phosphoethanolamine (PEtn),36 4-amino-4-deoxy-L-arabinose (Ara4N), and palmitate residues.37 These modifications promote resistance against host antimicrobial peptides that are mobilized at the time of infection to destroy the invading microbes. Moreover, such changes help the pathogen to evade recognition by the innate immune receptor, TLR4.38
A significant progress has been made towards characterization of the polysaccharide components of LPS i.e. inner and outer core regions, produced by many P. multocida isolates belonging to serovars 1, 2, 3, 5, 9 and 14.12,66–68 Moreover, the outer glycan part of LPS obtained from P. multocida genome strain Pm70 has also been structurally characterised.39 However, despite its strong correlation with endotoxicity, the structure of lipid A part of the LPS of P. multocida has not yet been determined. Thus, in order to fill this knowledge gap, lipid A structure of pathogenic P. multocida type B:2 isolate (termed as PM2) from Pakistan was investigated using mass spectrometric technique that will help in better understanding of pathogenicity of this bacterium.
Results
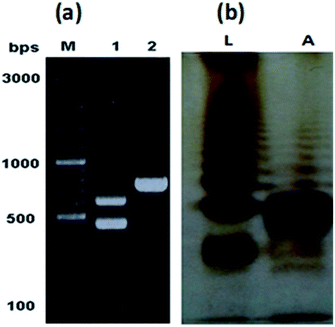
The characteristic colonies of P. multocida: grayish, translucent and mucoid having around 1 mm diameter, were obtained on CSY agar plates. The molecular identification of the isolate resulted in amplification of 460 bp fragment of KMT1 gene, 590 bp fragment of 6b gene and 758 bp fragment of bcbD gene (Table 2), confirming the isolate identity as P. multocida belonging to B:2 type (Fig. 1a). No amplification was observed in case of negative controls. Large scale fermentation, resulted in 5 g L−1 yield of wet cell pellet and the LPS extraction yielded 55 mg of purified LPS per gram of bacterial cell pellet. The DOC-PAGE analysis showed the typical heterogenic patterns of the purified rough type LPS (Fig. 1b).Characterization of lipid A structure
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and injected to ESI-MS using direct syringe pump. The full scan data at negative ion mode is shown in Fig. 2, which revealed that lipid A structure of P. multocida exhibited a high-level of heterogeneity, comprising of different fatty acyl chains and covalent modifications. Apparently, lipid A consists of hepta-, hexa-, penta- and a minor variant of tetra-acylation having 7, 6, 5 and 4 fatty acid acyl chains, respectively, attached to the glucosamine disaccharide backbone, which is mono and/or bisphosphorylated at C-1 and C-4′ positions with extended substitution(s) of one or two 4-amino-4-deoxy-L-arabinopyranose (Ara4N) molecules (Fig. 2).
1) and injected to ESI-MS using direct syringe pump. The full scan data at negative ion mode is shown in Fig. 2, which revealed that lipid A structure of P. multocida exhibited a high-level of heterogeneity, comprising of different fatty acyl chains and covalent modifications. Apparently, lipid A consists of hepta-, hexa-, penta- and a minor variant of tetra-acylation having 7, 6, 5 and 4 fatty acid acyl chains, respectively, attached to the glucosamine disaccharide backbone, which is mono and/or bisphosphorylated at C-1 and C-4′ positions with extended substitution(s) of one or two 4-amino-4-deoxy-L-arabinopyranose (Ara4N) molecules (Fig. 2).
 | ||
| Fig. 2 ESI-MS full scan of lipid A from PM2 in negative ionization mode exhibiting various structures. | ||
Notably, these lipid A variants show highly capricious endotoxicity. Multiple lipid A structures complicate interpretation and can have variable effects on innate host immune response. The immunogenic properties of lipid A are directly correlated with the number of acyl chains, phosphate groups and covalent modifications, i.e., the incorporation of palmitate, the addition of PEtN and/or Ara4N.38 Usually, bisphosphorylated asymmetric (4/2) hexa-acylated lipid A induces the strongest proinflammatory immune reactions after binding to TLR4, whereas, penta-acylated lipid A exhibits lower binding activity. Overall, bisphosphorylated versions of hepta-acylated (4/3), asymmetric hexa-acylated (4/2) and penta-acylated (3/2) lipid A activate NF-κB pathway and are considered as TLR4 expression agonists. Conversely, bisphosphorylated tetra-acylated (2/2) lipid A, monophosphorylated penta-acylated (3/2) and bisphosphorylated symmetric (3/3) hexa-acylated lipid A antagonize proinflammatory NF-κB pathway.40,41 Overall, endotoxicity of the pathogens i.e. P. multocida, is the collective impression of all these lipid A variants. Therefore, the determination of precise structure of lipid A species is important to visualize the endotoxicity as well as pathogenicity of any Gram-negative bacteria. Hence, the accurate structure of these lipid A variants of the tested P. multocida PM2 strain have been illustrated below in descending order, on the basis of their mass/charge (m/z) values and is summarized in Table 1. The conjugation sites were confirmed by correlating the acyl group fragmentations with the cross-ring fragmentations that are denoted in the manuscript according to the nomenclature proposed by Domon and Costello42 and Morrison et al.43
| S. # | m/z | RAa | C-4′ | C-3′ | C-2′ | C-3 | C-2 | C-1 |
|---|---|---|---|---|---|---|---|---|
| a RA = Relative abundance in full scan MS.b Ara4N–P = 4-amino-4-deoxy-L-arabinose bonded with phosphate group.c P = phosphate group. | ||||||||
| Hepta-acylated | ||||||||
| 1 | 2325 | 05 | Ara4N–Pb | 3-OH–C14 (C14) | 3-OH–C14 (C14) | 3-OH–C14 | 3-OH–C14 (C16) | P–Ara4N |
| 2 | 2194 | 25 | Ara4N–P | 3-OH–C14 (C14) | 3-OH–C14 (C14) | 3-OH–C14 | 3-OH–C14 (C16) | Pc |
| 3 | 2166 | 06 | Ara4N–P | 3-OH–C14 (C14) | 3-OH–C14 (C12) | 3-OH–C14 | 3-OH–C14 (C16) | P |
| 4 | 2114 | 08 | Ara4N–P | 3-OH–C14 (C14) | 3-OH–C14 (C12) | 3-OH–C14 | 3-OH–C14 (C16) | H |
| 5 | 2063 | 22 | P | 3-OH–C14 (C14) | 3-OH–C14 (C14) | 3-OH–C14 | 3-OH–C14 (C16) | P |
| 6 | 2035 | 07 | P | 3-OH–C14 (C14) | 3-OH–C14 (C12) | 3-OH–C14 | 3-OH–C14 (C16) | P |
| 7 | 1983 | 13 | P | 3-OH–C14 (C14) | 3-OH–C14 (C12) | 3-OH–C14 | 3-OH–C14 (C16) | H |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||||
| Hexa-acylated | ||||||||
| 8 | 2087 | 15 | Ara4N–P | 3-OH–C14 (C14) | 3-OH–C14 (C14) | 3-OH–C14 | 3-OH–C14 | P–Ara4N |
| 9 | 1967 | 10 | Ara4N–P | 3-OH–C14 (C14) | 3-OH–C14 (C14) | H | 3-OH–C14 (C16) | P |
| 10 | 1956 | 42 | Ara4N–P | 3-OH–C14 (C14) | 3-OH–C14 (C14) | 3-OH–C14 | 3-OH–C14 | P |
| 11 | 1927 | 05 | Ara4N–P | 3-OH–C14 (C14) | 3-OH–C14 (C12) | 3-OH–C14 | 3-OH–C14 | P |
| 12 | 1888 | 09 | Ara4N–P | 3-OH–C14 (C14) | 3-OH–C14 (C14) | H | 3-OH–C14 (C16) | H |
| 13 | 1875 | 05 | Ara4N–P | 3-OH–C14 (C14) | 3-OH–C14 (C14) | 3-OH–C14 | 3-OH–C14 | H |
| 14 | 1853 | 05 | P | 3-OH–C14 | 3-OH–C14 (C14) | 3-OH–C14 | 3-OH–C14 (C16) | P |
| 15 | 1837 | 10 | P | 3-OH–C14 (C14) | 3-OH–C14 (C14) | H | 3-OH–C14 (C16) | P |
| 16 | 1824 | 31 | P | 3-OH–C14 (C14) | 3-OH–C14 (C14) | 3-OH–C14 | 3-OH–C14 | P |
| 17 | 1797 | 15 | P | 3-OH–C14 (C14) | 3-OH–C14 (C12) | 3-OH–C14 | 3-OH–C14 | P |
| 18 | 1773 | 08 | P | 3-OH–C14 | 3-OH–C14 (C14) | 3-OH–C14 | 3-OH–C14 (C16) | H |
| 19 | 1756 | 21 | P | 3-OH–C14 (C14) | 3-OH–C14 (C14) | H | 3-OH–C14 (C16) | H |
| 20 | 1744 | 20 | P | 3-OH–C14 (C14) | 3-OH–C14 (C14) | 3-OH–C14 | 3-OH–C14 | H |
| 21 | 1739 | 18 | P | 3-OH–C14 (C14) | 3-OH–C14 (C14) | H | 3-OH–C14 (C16) | –H2O |
| 22 | 1726 | 20 | P | 3-OH–C14 (C14) | 3-OH–C14 (C14) | 3-OH–C14 | 3-OH–C14 | –H2O |
| 23 | 1717 | 09 | P | 3-OH–C14 (C14) | 3-OH–C14 (C12) | 3-OH–C14 | 3-OH–C14 | H |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||||
| Penta-acylated | ||||||||
| 24 | 1888 | 09 | Ara4N–P | 3-OH–C14 | 3-OH–C14 (C14) | H | 3-OH–C14 (C16) | P–Ara4N |
| 25 | 1876 | 12 | Ara4N–P | 3-OH–C14 | 3-OH–C14 (C14) | 3-OH C14 | 3-OH–C14 | P–Ara4N |
| 26 | 1860 | 04 | Ara4N–P | 3-OH–C14 (C14) | 3-OH–C14 (C14) | H | 3-OH–C14 | P–Ara4N |
| 27 | 1757 | 20 | Ara4N–P | 3-OH–C14 | 3-OH–C14 (C14) | H | 3-OH–C14 (C16) | P |
| 28 | 1745 | 27 | Ara4N–P | 3-OH–C14 | 3-OH–C14 (C14) | 3-OH–C14 | 3-OH–C14 | P |
| 29 | 1729 | 20 | Ara4N–P | 3-OH–C14 (C14) | 3-OH–C14 (C14) | H | 3-OH–C14 | P |
| 30 | 1649 | 28 | Ara4N–P | 3-OH–C14 (C14) | 3-OH–C14 (C14) | H | 3-OH–C14 | H |
| 31 | 1614 | 12 | P | 3-OH–C14 | 3-OH–C14 (C14) | 3-OH–C14 | 3-OH–C14 | P |
| 32 | 1598 | 11 | P | 3-OH–C14 (C14) | 3-OH–C14 (C14) | H | 3-OH–C14 | P |
| 33 | 1546 | 06 | P | 3-OH–C14 | 3-OH–C14 (C14) | H | 3-OH–C14 (C16) | H |
| 34 | 1534 | 10 | P | 3-OH–C14 | 3-OH–C14 (C14) | 3-OH–C14 | 3-OH–C14 | H |
| 35 | 1518 | 100 | P | 3-OH–C14 (C14) | 3-OH–C14 (C14) | H | 3-OH–C14 | H |
| 36 | 1500 | 30 | P | 3-OH–C14 (C14) | 3-OH–C14 (C14) | H | 3-OH–C14 | –H2O |
| 37 | 1490 | 10 | P | 3-OH–C14 (C14) | 3-OH–C14 (C12) | H | 3-OH–C14 | H |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||||
| Tetra-acylated | ||||||||
| 38 | 1439 | 05 | Ara4N–P | 3-OH–C14 | 3-OH–C14 (C14) | H | 3-OH–C14 | H |
| 39 | 1308 | 18 | P | 3-OH–C14 | 3-OH–C14 (C14) | H | 3-OH–C14 | H |
| 40 | 1290 | 14 | P | 3-OH–C14 (–H2O) | 3-OH–C14 (C14) | H | 3-OH–C14 | H |
| 41 | 1262 | 05 | P | 3-OH–C14 (–H2O) | 3-OH–C14 (C12) | H | 3-OH–C14 | H |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GlcN-I disaccharide backbone (Fig. 3A).
GlcN-I disaccharide backbone (Fig. 3A).
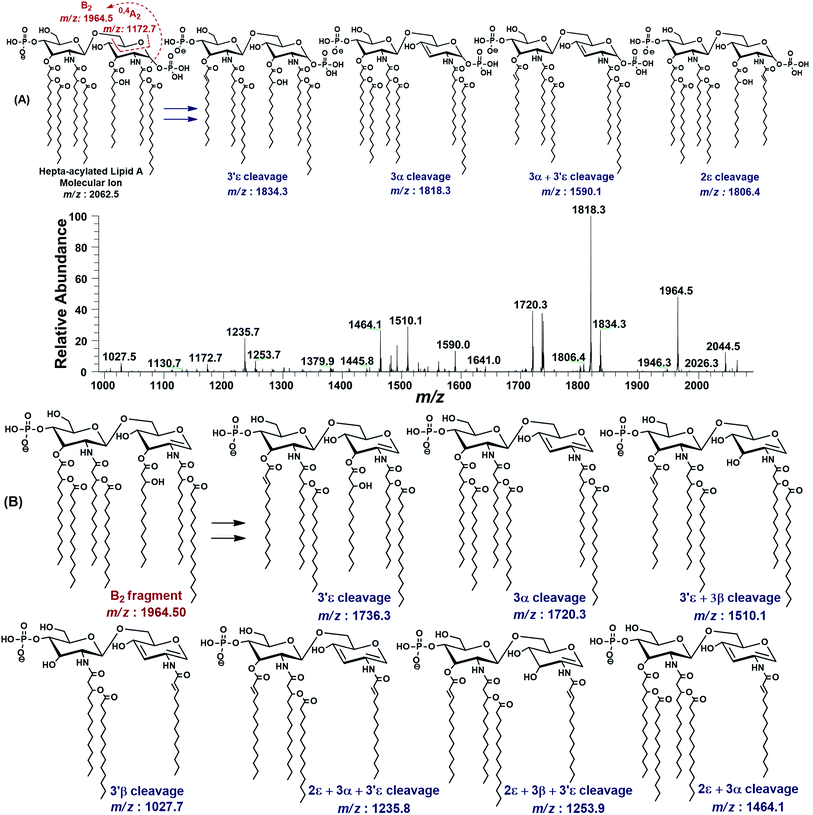 | ||
| Fig. 3 The MS2 of representative hepta-acylated lipid A molecular ion at m/z 2062.5 @CID 24, showing its fragmentation patterns. Red labels denote standard Domon and Costello fragments42 and blue labels denote the fragments arising from the loss of acyl chains that have been symbolized by their cleavage sites adopting the terminology proposed by Morrison et al.43 (A) Fragmentation (deacylation) while retaining H3PO4 at 1-position. (B) Simultaneous dephosphorylation and de-acylation. | ||
The base peak at m/z 1818.3 in MS2 spectrum of m/z 2062.5 was attributed to apparently the most stable daughter ion as a result of 3α elimination of 3-hydroxymyristate (244 Da) at GlcN-I leading to a bisphosphorylated fragment with fatty acyl chain distribution of 4/2 on GlcN-II![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GlcN-I disaccharides, respectively. The fragment ion at m/z 1818.3 further lost the secondary acyl chain (C14:0) from the ester-linked 3-OH–C14:0 fatty acid at 3′ε position of GlcN-II, generating the subsequent fragment at m/z 1590.1, corresponding to a bisphosphorylated species with 3/2 fatty acyl chains on GlcN-II
GlcN-I disaccharides, respectively. The fragment ion at m/z 1818.3 further lost the secondary acyl chain (C14:0) from the ester-linked 3-OH–C14:0 fatty acid at 3′ε position of GlcN-II, generating the subsequent fragment at m/z 1590.1, corresponding to a bisphosphorylated species with 3/2 fatty acyl chains on GlcN-II![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GlcN-I disaccharides (Fig. 3A).
GlcN-I disaccharides (Fig. 3A).
The minor peak at m/z 1806.5 was correlated with the concomitant 2ε loss of palmitate [C16:0] at the secondary acyl of GlcN-I. The low abundance of m/z 1806.5 is due to the fact that C16:0 is attached to the amide linked 3-OH–C14:0 at C-2 position. Secondary acyl fatty acids are strongly bonded with the amide linked 3-OH–C14:0 at C-2 and C-2′ positions on GlcN-I and GlcN-II, as compared to the ester linked primary acyl chains at C-3 and C-3′ positions.
The MS2 of m/z 2062.5 resulted in several additional peaks due to the formation of B2 fragment at m/z 1964.5 by the neutral loss of one phosphoric acid (H3PO4), followed by fatty acyl chains (Fig. 3B). The fragments at m/z 1736.3 and m/z 1720.3 were ascribed to 3′ε cleavage of myristic acid C14:0 and 3α cleavage of 3-OH–C14:0, respectively, from B2 fragment. The concurrent fragmentations of the B2 ion also exhibited the tendency for multiple cleavages of fatty acids. The fragments at m/z 1510.1 and m/z 1464.1 were produced by the loss of C14:0 at 3′ε site in conjunction with 3β cleavage of 3-OH–C14:0 and cleavage of 3-OH–C14:0 at 3α combined with that of C16:0 at 2ε position, respectively. Deacylation of three fatty acids yielded the ion peaks at m/z 1253.7 and m/z 1235.7. Similarly, removal of four fatty acids generated the ion peak at m/z 1027.7.
The MS2 fragmentation pattern of the precursor ion (m/z 2062.5) and its analysis suggested its structure as bisphosphorylated hepta-acylated disaccharide with 4/3 stoichiometric distribution of acyl chains. GlcN-I![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GlcN-II disaccharide carries a 3-hydroxymyristate appendage at C-2 (primary position) attached via an amide linkage that further has an ester linkage with a palmitate (secondary acyl chain). The C-3 position also has a 3-hydroxymyristate moiety but through ester linkage. Another 3-hydroxymyristate is anchored at C-2′ (primary position) utilizing its hydroxyl group for ester linkage with a myristate (secondary). The C-3′ exhibits yet another 3-hydroxymyristate (primary position) through ester linkage with O-myristoylation (secondary) (Table 1, S. # 5). The cross-ring fragmentation represented as 0,4A2 at m/z 1172.7 further supported the distribution of different fatty acids (Fig. 3).
GlcN-II disaccharide carries a 3-hydroxymyristate appendage at C-2 (primary position) attached via an amide linkage that further has an ester linkage with a palmitate (secondary acyl chain). The C-3 position also has a 3-hydroxymyristate moiety but through ester linkage. Another 3-hydroxymyristate is anchored at C-2′ (primary position) utilizing its hydroxyl group for ester linkage with a myristate (secondary). The C-3′ exhibits yet another 3-hydroxymyristate (primary position) through ester linkage with O-myristoylation (secondary) (Table 1, S. # 5). The cross-ring fragmentation represented as 0,4A2 at m/z 1172.7 further supported the distribution of different fatty acids (Fig. 3).
After confirming the structure of m/z 2062.5, the other hepta-acylated lipid A structures i.e. m/z 1982.8, 2034.8, 2114.0, 2165.9, 2194.0 and 2325.0 were successfully correlated as illustrated in Fig. 4. The ion peak at m/z 1982.8 was the monophosphorylated version of the structure proposed for m/z 2062.5 (Δ = 80 Da). The ion peak at m/z 2034.8 emerged due to loss of 28 Da (variation of ethylene group C2H4) from m/z 2063 indicating that one of the secondary myristoyl chains is replaced with laurate [C12:0]. Notably, almost all of the lipid A variants i.e. with hepta-, hexa-, penta- and tetra acylation, both in their monophosphorylated and bisphosphorylated forms and having covalent modification with Ara4N, demonstrated the substitution of myristate with laurate (reduction of m/z by 28 Da), suggesting a minor alternative lipid A variant. The exact position of laurate fatty acyl chain was determined to be 2′ through fragmentation of hexa-acylated precursor ion at m/z 1716.4 (Fig. 7).
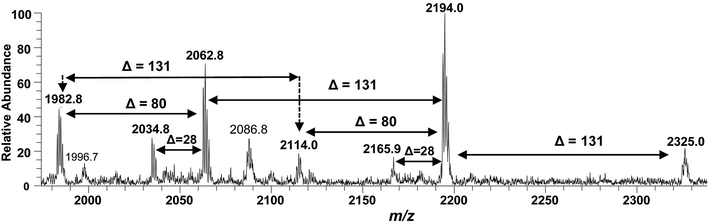 | ||
| Fig. 4 ESI-MS full scan of lipid A from PM2, in negative ionization mode, showing the correlation among hepta-acylated lipid A variants. | ||
The ion peak at m/z 2194 (Fig. 4) represented the addition of 131 Da to the bisphosphorylated hepta-acylated lipid A structure corresponding to the peak at m/z 2063, indicating that one of the phosphate group (more likely at 4′-position of GlcN-II) is decorated with Ara4N (Δ = 131 Da). The presence and position of mono and di-Ara4N on phosphate groups have been further confirmed in the following data analysis of hexa- and penta-acylated lipid A species and is summarized in Table 1. The ion peak at m/z 2194.0 exhibited the highest abundance among all of the hepta-acylated variants. The ion peaks at m/z 2165.9 and m/z 2114.0 demonstrated the reduction of 28 and 80 mass units from m/z 2194.0, indicating the subtraction of methylene and phosphate groups, respectively. The peak at m/z 2114.0 can be further interrelated with the peak at m/z 1982.8 by difference of 131 mass units (Ara4N).
Finally, the ion peak at m/z 2325 can be correlated by adding another 131 Da to m/z 2194.0 (2063 + 131 + 131 = 2325). The absence of any other peak between m/z 2194 and m/z 2325, especially the dephosphorylation possibility (2325 − 80 = 2245), indicated that both phosphate groups (1-position of GlcN-I and 4′-position of GlcN-II) of the entity corresponding to m/z 2325 are decked with Ara4N (Fig. 4 and Table 1, S. # 1). In short, the proposed structure for m/z 2325.0 comprises of a di-phosphorylated di-glucosamine backbone primarily acylated with 3-OH–C14:0 at the 2, 3, 2′, and 3′ positions and secondarily acylated with C16:0 at position 2, and with C14:0 at the 2′ as well as 3′ positions while both phosphate groups decorated with Ara4N.
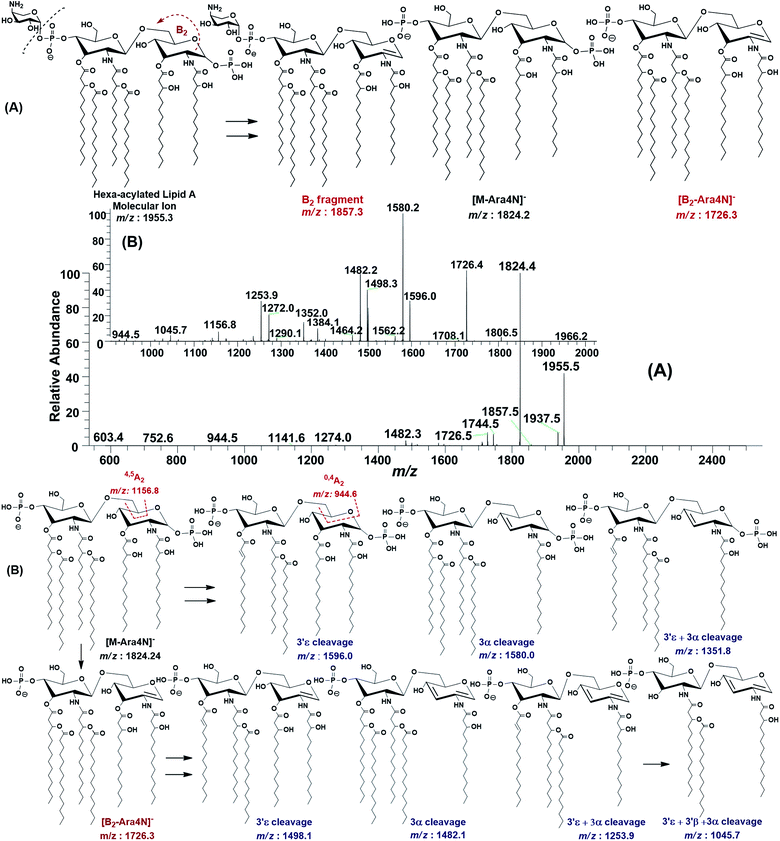
The ESI-MS2 fragmentation of the ion at m/z 1955.3 (@CID 20) generated the daughter ions at m/z 1937.5, 1857.5, 1824.4, 1744.5, 1726.5 and 1482 along with a few minor peaks (Fig. 5A). The fragment at m/z 1937.5 [M − 18] conforms to the loss of H2O, while that at m/z 1857.3 to the B2 fragment formed after the removal of H3PO4 (Δ = 98 Da) from the C-1 position, causing unsaturation between C-1 and C-2 carbon atoms. Although the ion abundance of m/z 1857.3 is very low but its appearance gives an important hint about the presence of a stripped phosphate group at C-1 position without any covalent modification. The most abundant daughter ion at m/z 1824.4 corresponded to the elimination of Ara4N (Δ = 131 Da). This reflects that C-4′ phosphate group holds an Ara4N sugar as the covalent bond between the phosphate group and Ara4N sugar is among the most susceptible bonds. The removal of two moieties i.e. Ara4N and H2PO3 (Δ = 131 + 80) from m/z 1955.3, generated the ion peak at m/z 1744. Similarly, m/z 1726.4 was produced by the loss of Ara4N and H3PO4 (Δ = 131 + 98) (Fig. 5A). The removal of fatty acyl chains along with Ara4N were spotted in minor peaks. Due to highly labile nature of covalent bond between Ara4N and phosphate group (C-4′ position), the possible loss of the fatty acyl chains cannot be associated with the base peak at m/z 1824.4. This made us to pursue the MS3 of m/z 1824.4 to pinpoint the positions and number of fatty acyl chains.
After isolating m/z 1824.4 in ion trap, it was subjected to the MS3 fragmentation (@CID 22.0) that yielded multiple sub-fragments arising from the simultaneous loss of one or both of the phosphate groups (m/z 1726.4) and one or more of the fatty acyl chains (Fig. 5B). The ion peaks at m/z 1596.0 and m/z 1580.2 can be correlated to the loss of one C14:0 at 3′ε cleavage site and one 3-OH–C14:0 at 3α site, respectively, while losing both fatty acyl chains generated the peak at m/z 1352. The fragments at m/z 1498.1 and m/z 1482.1 were attributed to the loss of phosphate group along with the 3′ε cleavage of C14:0 and 3α cleavage of 3-OH–C14:0, respectively (Fig. 5B). The m/z 1272.0 and m/z 1253.9 were produced by the simultaneous loss of phosphate with one of the two pairs of fatty acids i.e. (i) C14:0 by 3′ε cleavage paired with 3-OH–C14:0 by 3α cleavage and (ii) C14:0 by 3′ε cleavage paired with C14:0 by 3β cleavage, respectively. The removal of phosphate together with cleavages at 3′ε, 3′β and 3α gave rise to the peak at m/z 1045.7. Similarly, the removal of four fatty acids produced the ion peak at m/z 1027.7.
The data analysis of MS3 fragmentation of the ion peak at m/z 1824.5 suggested its structure as bisphosphorylated hexa-acylated GlcN-I![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GlcN-II disaccharide with 4/2 acyl chain stoichiometric distribution as 3-OH–C14:0 (C-2), 3-OH–C14:0 (C-3), 3-OH–C14:0 + C14:0 (C-2′) and 3-OH–C14:0 + C14:0 (C-3′) (Fig. 5B). The cross-ring fragments represented as 0,4A2 at m/z 944.5 and 4,5A2 at m/z 1156.8 further supported the distribution of different fatty acids. Out of the four primary chains, two have amide linkages at C-2 and at C-2′ positions, while the remaining two have ester linkages at C-3 and C-3′ positions of the core disaccharide skeleton as described in Table 1 (S. # 16).
GlcN-II disaccharide with 4/2 acyl chain stoichiometric distribution as 3-OH–C14:0 (C-2), 3-OH–C14:0 (C-3), 3-OH–C14:0 + C14:0 (C-2′) and 3-OH–C14:0 + C14:0 (C-3′) (Fig. 5B). The cross-ring fragments represented as 0,4A2 at m/z 944.5 and 4,5A2 at m/z 1156.8 further supported the distribution of different fatty acids. Out of the four primary chains, two have amide linkages at C-2 and at C-2′ positions, while the remaining two have ester linkages at C-3 and C-3′ positions of the core disaccharide skeleton as described in Table 1 (S. # 16).
Consequently, by combining the fragmentation data analysis of m/z 1955.3 (MS2) and m/z 1824.4 (MS3), the structure of the precursor at m/z 1955.3 was proposed as a bisphosphorylated, hexa-acylated lipid A variant of the P. multocida LPS. The C-1 and C-4′ positions are phosphorylated with an additional decoration of one covalently bonded Ara4N moiety at the phosphate group of C-4′ position. The acylation pattern looks to be 4/2 with four primary acyl chains. The two primary 3-OH–C14:0 chains are amide linked at C-2 and C-2′, while the rest are ester linked at C-3 and C-3′ positions of the GlcN disaccharide backbone moiety. The secondary acylation can be docked at the 3-hydroxy positions of primary fatty acid moieties attached at C-2′ and C-3′ locations (Table 1, S. # 10).
The structure elucidation of the precursor ion corresponding to m/z 1955.3 significantly contributed in assigning the other hexa-acylated ion peaks (Fig. 6). The parent ion at m/z 2086.8 represented the addition of a 131 Da unit, indicating that both of phosphate groups of bisphosphorylated hexa-acylated lipid A were decorated with Ara4N (Table 1, S # 8). The ion peak m/z 1926.5 emerged due to the loss of 28 Da (variation of ethylene group C2H4) from m/z 2063 suggesting that one of the C14:0 is replaced with C12:0 fatty acid possibly at C-2′ secondary position. Whereas, m/z 1874.5 was assigned to the mono dephosphorylated (Δ = 80 Da) version of m/z 1955.5. The precursor ions at m/z 1824.5 and m/z 1744.5 can be further interrelated with m/z 1955.5 by removing Ara4N (Δ = 131 Da) from C-4′ phosphate group and HPO3 (Δ = 80 Da) from C-1 positions, respectively (Fig. 6 and Table 1). Similarly, another hexa-acylated lipid A variant can be spotted at m/z 1726.5 by retreating Ara4N (Δ = 131) along with H3PO4 (Δ = 98).
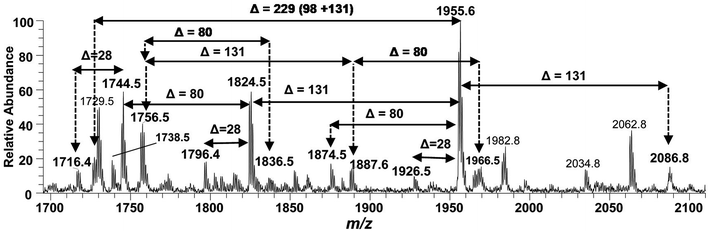 | ||
| Fig. 6 Negative ion mode ESI-MS full scan of lipid A from PM2, showing the correlation among hexa-acylated lipid A variants. | ||
In the ion peak at m/z 1716.4, one of the secondary myristate was found to have been replaced with laurate (Δ = 28 Da). The exact position of C12:0 was determined by MS2 of m/z 1716.4, which yielded daughter ions by losing one of the fatty acyl chains at m/z 1516 (removal of laurate at 2′ε cleavage site), m/z 1488 (3′ε cleavage of myristate) and m/z 1472 (3α cleavage of 3-hydroxymyristate) (Fig. 7). The product ions arising from the elimination of two acyl chains were also detected at m/z 1288 [elimination of C12:0 (2′ε cleavage) together with C14:0 (3′ε cleavage)], m/z 1272 [elimination of C12:0 (2′ε cleavage) and β-OH–C14:0 (3α cleavage) as a ketene derivative], m/z 1261.8 [elimination of C14:0 (3′ε cleavage) and β-OH–C14:0 (3β cleavage)], and at m/z 1243.8 [elimination of C14:0 (3′ε cleavage) and 3-OH–C14:0 (3α cleavage)]. The plausible elimination of C14:1 both by charge-remote (loss of a free fatty acid) and charge-driven processes (loss of a ketene derivative) suggested its location at O-3′ as well as its secondary substitution by C14:0 (ion peak of m/z 1243.8).44
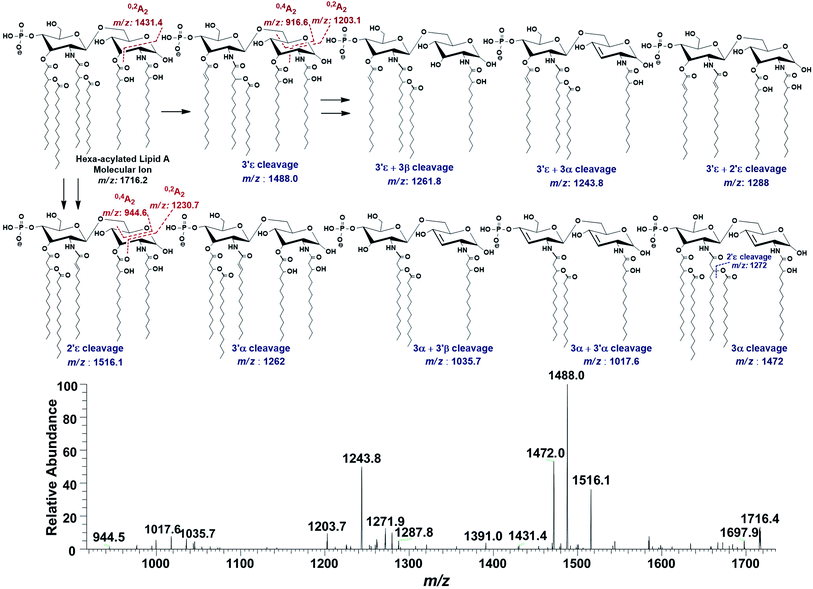 | ||
| Fig. 7 ESI-MS2 of hexa-acylated lipid A molecular ion at m/z 1716.4 @CID 20, showing its fragmentation patterns. | ||
The product ions at m/z 1431.4, 1230.7 and m/z 1203.1 represented 0,2A2 fragments formed from the ions at m/z 1716.2, 1516.1 and 1488.0, respectively. Whereas, cross-ring fragments 0,4A2 at m/z 944.6 and m/z 916.6 were produced by m/z 1516 and 1488, respectively. These ions yielded by the in-source cross-ring fragmentation further established the distribution of the identified fatty acids on specific positions. After interpreting the types of fragment ions, it was possible to define fatty acid distribution on both GlcN residues. From this data analysis, it is suggested that the ion peak at m/z 1716.4 can be attributed to mono-phosphorylated, hexa-acylated lipid A form consisting of two GlcN, four primary acyl chains of β-OH–C14:0 at 2, 3, 2′ & 3′ positions and two secondary acyl chains C12:0 and C14:0 at 2′ and 3′ positions, respectively.
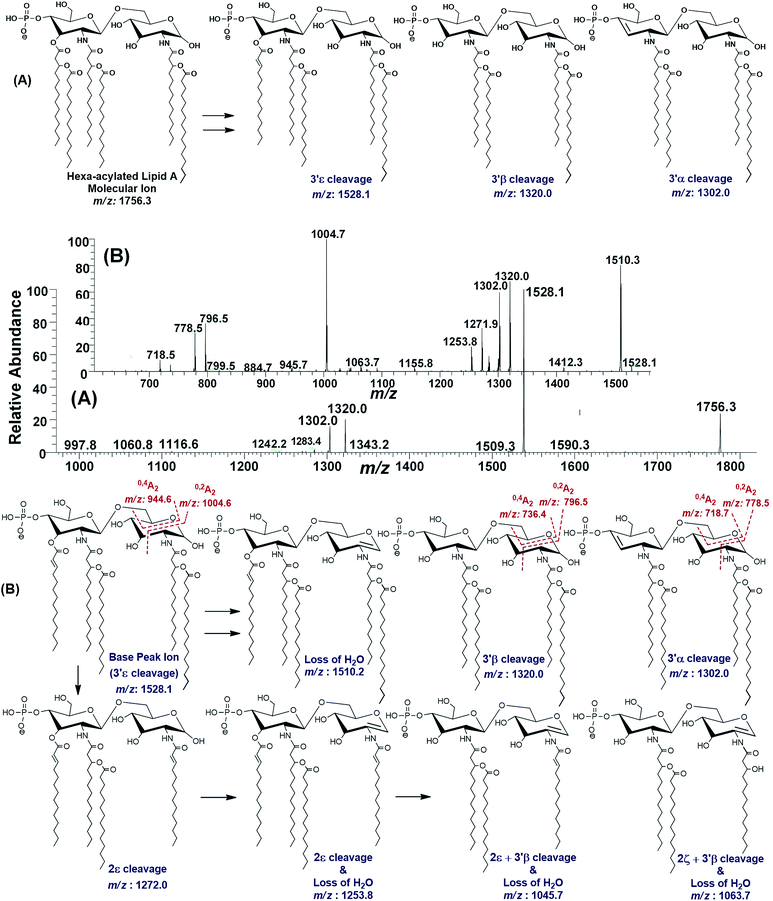 | ||
| Fig. 8 (A) ESI-MS2 of hexa-acylated lipid A molecular ion at m/z 1756.3 @CID 47, showing its fragmentation patterns, (B) MS3 of base peak at m/z 1528.5 @CID 22, obtained from MS2 of m/z 1756.3. | ||
The MS3 of the base peak m/z 1528.1, obtained from m/z 1756.3, generated product ions at m/z 1510, 1320, 1302, 1272, 1254, 1064, 1046, 1005, 946, 796, 778 and 718 (Fig. 8B). The ion at m/z 1510 was generated due to the water loss presumably C-1 on GlcN-I, resulting in an unsaturation between C-1 and C-2 atoms. The notion can be justified by the presence of m/z 1004.6, and absence of m/z 986.6 in case of water loss from the C-3 position of GlcN-I, due to cross-ring 0,2A2 fragmentation. The presence of both m/z 1320.0 and 1302.0 are indicative of the charge-driven as well as charge-remote type 3′α and 3′β cleavages entailing C-3′/C-4′ positions. The explanation is also supported by the presence of other ions at m/z 796.5, 778.5 (0,2A2) as well as at 736.4 and 718.7 (0,4A2) as a result of cross-ring fragmentations. The presence of m/z 1272.0 indicated the loss of a unit with Δ = 256 Da reflecting the elimination of C16:0 from 2ε position. Similarly, the generation of a peak at m/z 1253.8 can be spotted due to the loss of water from C-1 position along with the secondary deacylation from C-2 (2ε cleavage) (Fig. 8B). A less frequent 2ζ cleavage was also observed in combination with 3′β cleavage and water loss from C-1 leading to a product ion at m/z 1063.7.
The ESI-MS2 spectrum of m/z 1756.3 followed by MS3 of its daughter ion at m/z 1528.1 revealed its structure as hexa-acylated mono-phosphorylated variant with three primary acylations of 3-OH–C14:0 at C-2, C-2′ and C-3′ positions. The first two of these primary acyl chains are amide linked while the remaining one is ester linked. The secondary acylation of C14:0 was also observed at C-2′ and C-3′ while a C16:0 at C-2 position. The successful profiling of m/z 1756.3 contributed in correlating the peaks at m/z 1836 and 1887.6 in full scan data (Fig. 6): the addition of 80 and 131 mass units suggested the substitution of HPO3 at C-1 and Ara4N decoration on phosphate at C-4′ positions, respectively.
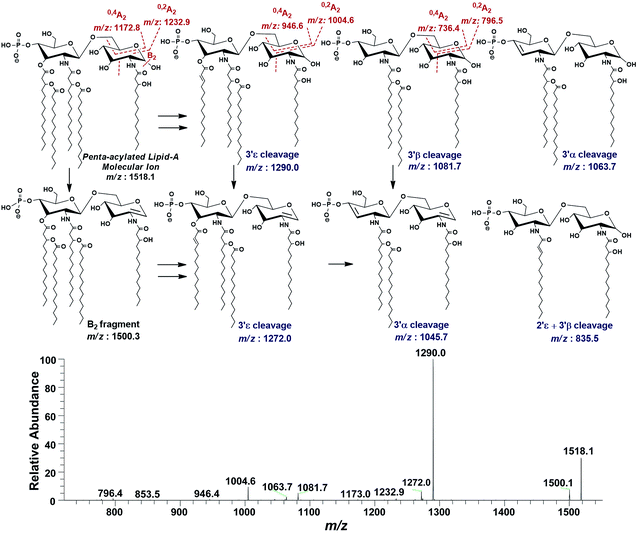 | ||
| Fig. 9 ESI-MS2 of penta-acylated lipid A molecular ion at m/z 1518.1 @CID 20, showing its fragmentation patterns. | ||
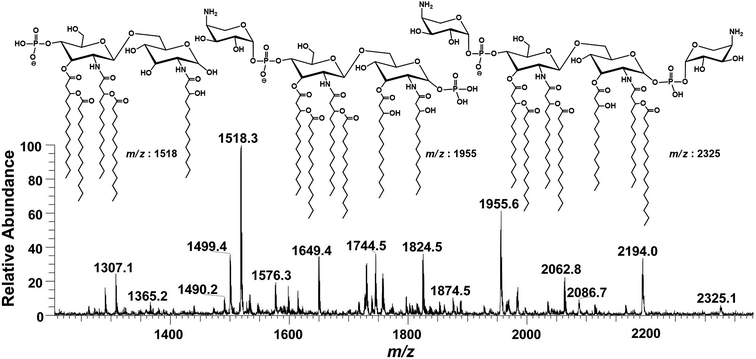
Successful profiling of m/z 1518.1 contributed in correlation of other penta-acylated lipid A variants (Fig. 10). The ion at m/z 1490.2 (Δ = 28 Da) indicated a variation in fatty acid ruler such that a laurate (C12:0) is present at secondary acyl position of C-2′ with 4/1 fatty acyl distribution as described in the analysis of m/z 1518.1 (Table 1, S. # 36). There could also be a dehydrated moiety at m/z 1499.4 owing to the water loss from the anomeric C-1 position of the ion corresponding to the base peak at m/z 1518.0 (Table 1, S. # 35). The addition of HPO3 and Ara4N (Δ = 80 and 131 Da) to the base peak at m/z 1518.1 gave rise to the ions at m/z 1649.4 and m/z 1598.3, respectively (Table 1, S. # 30 & 32), which further proceeded to m/z 1729.5 after the addition of another set of HPO3 (1649 + 80 = 1729 Da) and Ara4N (1598 + 131 = 1729 Da) (Table 1, S. # 29).
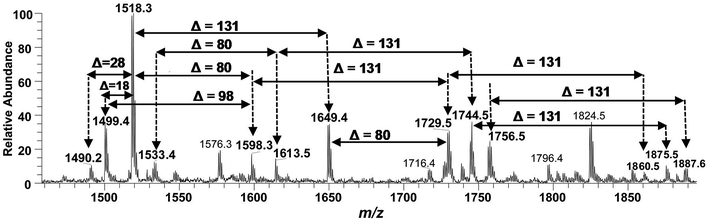 | ||
| Fig. 10 Negative ion mode ESI-MS full scan of lipid A from PM2, showing the correlation among penta-acylated lipid A variants. | ||
The ion peak at m/z 1533.4 corresponded to another set of mono-phosphorylated penta-acylated lipid A molecules having 3-OH–C14:0 appendages at C-2, C-3, C-2′ and C-3′ positions while only one secondary C14:0 acyl chain at C-2′. Its bisphosphorylated (Δ = 80 Da) and Ara4N (Δ = 80 + 131 Da) analogues were spotted at m/z 1614 and 1745, respectively (Table 1, S. # 31 & 28). Similarly, there was another ion peak at m/z 1757 (Table 1, S. # 27), which indicated penta-acylation with 3/2 pattern and an Ara4N moiety on C-4′ phosphate group and a palmitate at secondary position of C-2. By the addition of 131 Da, a second Ara4N on C-1 phosphate group was also observed at m/z 1887.6 (Table 1, S. # 24). Another 3/2 penta-acylated lipid A appeared at m/z 1546 having only one phosphate group at C-4′. The ESI-MS/MS data also depicted a small proportion of tetra-acylated lipid A variants at m/z 1439, 1308, 1290 and 1262 (Table 1, S. # 38–41).
Discussion
Haemorrhagic septicaemia (HS) is associated with P. multocida (serotype B:2 and E:2) infections in cloven-hoofed ungulates.45 The disease occurs in acute, sub-acute and chronic forms. Its initial phase includes temperature elevation, a stage of respiratory involvement and the terminal phase involves septicaemia and recumbence leading to death.9 Although the HS disease etiology is strongly related to the endotoxicity of lipid A component of the bacterial LPS, the structure elucidation of the lipid A has not been reported yet.ESI-MS analysis of lipid A obtained from P. multocida isolate PM2 in current study, showed a high degree of complexity and heterogeneity. The overall heterogeneity of lipid A varies from hepta-acylated to tetra-acylated variants decorated with varied level of Ara4N moiety on C-1 and/or C-4′ phosphate groups of proximal and distal glucosamine lipid A backbone. The full scan mass spectrometric data revealed that even within the same class of lipid A, several sub-variant structures exist, which have been summarized in Table 1.
The hepta-acylated lipid A consists of seven structures with 4/3 acyl distribution on GlcN-II![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GlcN-I sugar moieties of lipid A backbone, respectively (Table 1, S. # 1–7). All of these structures have four β-hydroxy C14:0 primary acyl chain, palmitate secondary acyl chain at C-2 position and myristate at C-3′ position. The secondary acyl chain at C-2′ position in majority of the hepta-acylated variants is C14:0, whereas, C12:0 is also found as a minor alternative (Table 1, S. # 3 and 6). Majority of hepta-acylated lipid A structures are bisphosphorylated (Table 1, S. # 1–3, 5 & 6). Ara4N was mostly found to decorate only C-4′ phosphate group (Table 1, S. # 2–4) except that the entity at m/z 2325 exhibits two Ara4N moieties at both phosphate groups. Similar to other Gram-negative bacteria, the palmitoylation as well as regulated addition of Ara4N in PM2 isolate may be environment-dependent and can contribute in protecting the bacterium from certain host immune defenses.38
GlcN-I sugar moieties of lipid A backbone, respectively (Table 1, S. # 1–7). All of these structures have four β-hydroxy C14:0 primary acyl chain, palmitate secondary acyl chain at C-2 position and myristate at C-3′ position. The secondary acyl chain at C-2′ position in majority of the hepta-acylated variants is C14:0, whereas, C12:0 is also found as a minor alternative (Table 1, S. # 3 and 6). Majority of hepta-acylated lipid A structures are bisphosphorylated (Table 1, S. # 1–3, 5 & 6). Ara4N was mostly found to decorate only C-4′ phosphate group (Table 1, S. # 2–4) except that the entity at m/z 2325 exhibits two Ara4N moieties at both phosphate groups. Similar to other Gram-negative bacteria, the palmitoylation as well as regulated addition of Ara4N in PM2 isolate may be environment-dependent and can contribute in protecting the bacterium from certain host immune defenses.38
Addition of palmitate fatty acyl chain to lipid A is catalyzed by PagP, a palmitoyl transferase enzyme, which is the component of PhoP–PhoQ regulatory system.46 The pagP gene and its functional homologs are distributed among a narrow group of primary pathogenic bacteria (Salmonella species,47,48 E. coli,33 Proteus mirabilis,49 Acinetobacter baumannii,50 Pseudomonas aeruginosa,51 L. pneumophila, B. bronchiseptica, Y. enterocolitica, and Y. pseudotuberculosis52) and its regulation is often correlated with their pathogenic lifestyle.53 The addition of palmitate not only strengthens the outer membrane of the bacterial cell wall through the hydrophobic and van der Waals interactions, which prevents translocation of the cationic antimicrobial peptides across the bilayer, but also contributes in evading the host immune response by lowering TLR4 related signal transduction pathway as compared to hexa-acylated lipid A variants.53 Similarly, insertion of Ara4N is catalyzed by ArnT through activation of PmrA transcription factor. Being positively charged at neutral pH, Ara4N can neutralize the phosphate groups, consequently, reducing bacterial susceptibility to the cationic host antimicrobial peptides and polymyxin antibiotics.38
The hexa-acylated lipid A of the PM2 isolate exhibited a high degree of variation with most of the structures bisphosphorylated at C-1 and C-4′ positions. In five structures, phosphates were decorated with Ara4N at C-4′ positions and only one structure demonstrated Ara4N sugars on both of its phosphate groups (Table 1: S. # 8–23). There was a significant heterogeneity of fatty acyl patterns with respect to their relative positions and variation in chain length of fatty acids. Most of the hexa-acylated structures exhibited 4/2 fatty acyl stoichiometry with C14 carbon ruler at primary and secondary acyl chains. The presence of laurate (C12:0) at secondary acyl chain at C-2′ position was also spotted in minor proportion (Table 1: S. # 11, 17 and 23).
The bisphosphorylated 4/2 structures have been reported to warrant maximal immunostimulatory activities of LPS.54,55 In P. multocida infections these structures presumably induce high inflammatory response of the innate immune system, thus playing a pivotal role in causing HS. However, hexa-acylated structures having 4/2 fatty acyls also exist in which palmitate occupy secondary position at C-2 with absence of 3-OH–C14:0 at C-3 positions (Table 1: S. # 9, 12, 15, 19 and 21). The outer membrane-bound lipid A 3-O-deacylase, encoded by the pagL gene removed the fatty acyl chain from C-3 position of hepta-acylated lipid A. The PagL enzyme was initially reported in Salmonella, but later its homologs were found to be widely distributed among pathogenic and non-pathogenic Gram-negative bacterial species.33 The presence of hexa-acylated lipid A variants without fatty acyl chains at C-2 position hints the presence of PagL in P. multocida as well. The addition of palmitate at C-2 by PagP and the removal of primary acyl chain at C-3 by PagL have been reported to get activated by cationic antimicrobial peptides induced through the PhoP transcription factor.46 Hence, these structures are likely to contribute in evading the host immune defense in P. multocida infections as well. Moreover, hexa-acylated lipid A structures with 3/3 stoichiometry (Table 1: S. # 14 and 18), were also spotted in a minor proportion. Rather than acting as agonist, the 3/3 stoichiometric structures have been found to antagonize TLR4 receptor.40
Like hexa-acylated lipid A, penta-acylated class also exhibited several variants and most of those were bisphosphorylated with Ara4N placement only at C-4′ position in four structures, while three structures displayed Ara4N sugars on both of its phosphate groups (Table 1: S. # 24–37). Significant heterogeneity was found in the positions and types of fatty acyl chains with predominant 4/1 stoichiometry (Table 1: S. # 26, 29, 30, 32, 35–37) as demonstrated by a base peak at m/z 1518. Fatty acid distribution with 3/2 stoichiometry demonstrated two sub-categories, one with secondary palmitate at C-2 (Table 1: S. # 24, 27 and 33), while the second contained two primary acyl chains at C-2 and C-3 positions (Table 1: S. # 25, 28, 31 and 34). In small proportion, mono-phosphorylated tetra-acylated variants having 3/1 stoichiometry were also spotted (Table 1: S. # 38–41) in the lipid A of the PM2 isolate. TLR4 activation has been markedly decreased by tetra- and penta-acylated lipid A structures as compared to hexa-acylated variants. Even antagonistic trend to the TLR4 activity has been found in penta-acylated lipid A having 3/2 fatty acyl distribution stoichiometry lacking phosphate at C-1 position, as well as in bisphosphorylated tetra-acylated lipid A structures.40,56 Gram-negative bacteria modulate lipid A structures to evade host immune response e.g. H. pylori synthesizes a hexa-acylated lipid A intrinsically, but trims it down to tetra-acylated by the action of deacylase enzymes and also removes phosphates (by LpxE & LpxF) to switch the biological activity from TLR4 agonistic to antagonistic, which can turn its infection to a chronic one for the duration of host life.57
The lipid A of PM2 depicts a variety of acylation, and other modes of modifications e.g. phosphorylation and Ara4N decoration. The presence of both phosphate and Ara4N moieties can be located on C-1 and/or C-4′ positions of the basic glucosamine backbone. The ESI-MSn further reveals the C-2, C-3, C-2′ and C-3′ bonded with primary and secondary myristoylation except few lipid A variants, where there is palmitoylation at C-2 position (Table 1, S # 1–7, 9, 12, 14, 15, 18, 19, 21, 24, 27 and 33) and lauric acid at secondary positions of C-2′ carbon (Table 1, S # 3, 4, 6, 7, 11, 17, 23, 37 and 41). There could be a strong correlation between the heterogeneous lipid A modification capabilities of P. multocida and its sporadic HS epidemics, which needs further investigations.
In addition to the number and length of the acyl groups, the stereochemistry and degree of their unsaturation may also have important biological implications. In our study, all the acyl groups bonded to the disaccharide backbone of lipid A happened to be saturated (Table 1), but in many other microbial strains, the unsaturated fatty acids may also exist.58 The carbon–carbon double bond on these unsaturated acyl groups can be located via a recently developed technique using visible-light activated [2 + 2] cycloaddition reaction and tandem mass spectrometry.59
Experimental
Material and method
| Primers | Oligonucleotide sequence (5′-3′) | Targeted gene | Amplicon size (bp) |
|---|---|---|---|
| PMPCR-F | ATCCGCTATTTACCCAGTGG | KMT1 | 460 |
| PMPCR-R | GCTGTAAACGAACTCGCCAC | ||
| KTT72 | AGGCTCGTTTGGATTATGAAG | 6b | 590 |
| KTSP61 | ATCCGCTAACACACTCTC | ||
| CAPB-F | CATTTATCCAAGCTCCACC | bcbD | 758 |
| CAPB-R | GCCCGAGAGTTTCAATCC |
LPS extraction and purification
The PCR confirmed PM2 isolate was grown in sterile CSY broth using 6 × 2 L flasks at 37 °C and 180 rpm. After overnight growth, the cells were treated with formalin (0.5% v/v) for 2 hours and then harvested by 7000 rpm centrifugation at 10 °C. The wet cell biomass was subjected to LPS extraction by hot-phenol method.62 The extracted LPS were dialyzed for 3 consecutive days against deionized water using ZelluTrans/Roth dialysis membrane (T2 MWCO 6000–8000 cat # E665.1), with 2 water changes daily and lyophilized. The LPS were analyzed by deoxycholate-polyacrylamide gel electrophoresis (DOC-PAGE) followed by silver staining.63,64Acid hydrolysis and extraction of lipid A
For lipid A extraction, the purified LPS was subjected to mild acid hydrolysis.65 Briefly, 10 mg L−1 of the purified LPS was dissolved in 1% glacial acetic acid and heated at 95 °C using water bath for 90 minutes. The solution was cooled and ultracentrifuged at 96![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g at 4 °C for 5 hours with a drop of 1 M CaCl2. The pellet was dissolved in chloroform
000g at 4 °C for 5 hours with a drop of 1 M CaCl2. The pellet was dissolved in chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol
methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water solution as 2
water solution as 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v/v) ratio and kept overnight. The chloroform extract was dried under nitrogen and redissolved in chloroform
1 (v/v/v) ratio and kept overnight. The chloroform extract was dried under nitrogen and redissolved in chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol
methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetonitrile solution as 3
acetonitrile solution as 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v/v) ratio and used for Electron Spray Ionization (ESI) mass spectrometric analysis.
1 (v/v/v) ratio and used for Electron Spray Ionization (ESI) mass spectrometric analysis.
Mass spectrometry of lipid A
The ESI-MS/MS was conducted using LTQ XL mass spectrometer (Thermo Electron Corporation, USA). The sample was analyzed through direct injection to the ESI probe set at negative ionization mode by syringe pump at a flow rate of 0.3 mL min−1. The temperature of capillary was kept at 198 °C. After getting the full scan mass spectrum data in the range of m/z 100 to m/z 3000, the generated ions were isolated in the ion trap and fragmented by collision induced dissociation (CID) energies ranging from 10 to 40 according to the stability of the target precursor ions selected for tandem mass spectrometry.Data analysis
The data was analysed using Xcalibur 2.2 and Chemdraw softwares. The identification was conducted using MS/MS fragmentation patterns of various precursor ions. The details for each specific fragmentation can be seen in the footnotes of all the corresponding figures.Conclusion
The pathogenic P. multocida PM2 isolate was found to exhibit a variety of lipid A variants, i.e. mono and bisphosphorylated hepta-, hexa-, penta- and tetra-acylated versions with the 4/3, 4/2, 3/3, 3/2, 4/1 and 3/1 patterns on the Glc-II and Glc-I backbone, respectively. It can also decorate its lipid A with the Ara4N sugar moiety, either at one or both of the phosphate moieties at C-1 and C-4′ positions. Majority of the lipid A variants are hexa-acylated with a total of 16 observed variants, followed by 14 penta, 7 hepta and 4 tetra-acylated variants. Based on the relative abundance, the penta-acylated version at m/z 1518.1 is the most abundant, followed by hexa-acylated lipid A variant having an m/z 1955.5. The presence of highly endotoxic hexa-acylated lipid A variants may have a leading role in inducing the disease, the haemorrhagic septicaemia. While covalently modified hexa-acylated lipid A variants (palmitoylation at C-2 position and Ara4N placement at C-4′ and/or C-1 phosphate positions) can contribute in evading the host immune responses.List of abbreviations
| LPS | Lipopolysaccharides |
| ESI-MS | Electron spray ionization mass spectrometry |
| Ara4N | 4-Amino-4-deoxy-L-arabinose |
| HS | Haemorrhagic septicaemia |
| OIE | Office International des Epizooties |
| PAMP | Pathogen-associated molecular pattern |
| PRR | Pattern recognition receptors |
| TLRs | Toll-like-receptors |
| LBP | Lipopolysaccharide binding protein |
| NFκB | Nuclear factor-κB |
| PEtn | Phosphoethanolamine |
| CSY | Casein sucrose yeast |
| PCR | Polymerase chain reaction |
| DOC-PAGE | Deoxycholate-polyacrylamide gel electrophoresis |
| CID | Collision induced dissociation |
| RA | Relative abundance |
Conflicts of interest
The authors declare no conflict of interests and no permission is required for publication.Acknowledgements
We acknowledge the funding support of Higher Education Commission (HEC) of Pakistan, for the academic visit of Abdul Tawab. We are also thankful to Dr Ijaz Ali, Veterinary Research Institute (VRI), Peshawar, Pakistan, for providing P. multocida (PM2) isolate, Dr Parastoo Azadi, Complex Carbohydrate Research Center, UGA, USA for assisting in DOC-PAGE gel chromatography.Notes and references
- I. W. Wilkie, M. Harper, J. D. Boyce and B. Adler, Pasteurella multocida: siseases and pathogenesis, Microbiol. Immunol., 2012, 361, 1–22 CAS.
- G. R. Carter, The type specific capsular antigen of Pasteurella multocida, Can. J. Med. Sci., 1952, 30(1), 48–53 CAS.
- K. L. Heddleston, J. E. Gallagher and P. A. Rebers, Fowl cholera: gel diffusion precipitin test for serotyping Pasteruella multocida from avian species, Avian Dis., 1972, 16, 925–936 CrossRef CAS PubMed.
- A. R. Spickler, Hemorrhagic septicemia, in Factsheet, 2019, pp. 1–6, available from http://www.cfsph.iastate.edu/Factsheets/pdfs/hemorrhagic_septicemia.pdf Search PubMed.
- M. P. Dagleish, J. C. Hodgson, S. Ataei, A. Finucane, J. Finlayson and J. Sales, et al., Safety and protective efficacy of intramuscular vaccination with a live aroA derivative of Pasteurella multocida B:2 against experimental hemorrhagic septicemia in calves., Infect. Immun., 2007, 75(12), 5837–5844 CrossRef CAS PubMed , available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2168370%26tool=pmcentrez%26rendertype=abstract, cited 2014 Mar 27.
- M. A. Wilson, R. B. Rimler and L. J. Hoffman, Comparison of DNA fingerprints and somatic serotypes of serogroup B and E Pasteurella multocida isolates, J Clin Microbiol, 1992, 30(6), 1518–1524 CrossRef CAS PubMed.
- S. K. Srivastava, A. A. Kumar, P. Chaudhuri and M. P. Yadav, Haemorrhagic septicaemia; manual of diagnostic tests and vaccines for terrestrial animals: mammals, birds and bees, in Terrestrial Manual, Office International Des Epizooties (OIE), Paris, France, 2008, vol. 2, pp. 739–751, http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.04.12_HS.pdf Search PubMed.
- S. E. Wasti, S. Hanif, M. Asif, M. S. Malik, Z. Husnain and S. Khatoon, et al., Pakistan Economic Survey 2018-19, in Pakistan Economic Survey 2018-19, Economic Adviser's Wing, Finance Division, Government of Pakistan, Islamabad, 2019, p. 27, available from http://finance.gov.pk/survey/chapters_19/Economic_Survey_2018_19.pdf Search PubMed.
- S. B. Shivachandra, K. N. Viswas and A. A. Kumar, A review of hemorrhagic septicemia in cattle and buffalo, Anim. Health Res. Rev., 2011, 12(1), 67–82 CrossRef CAS PubMed.
- M. Ahmad, M. Aziz, M. T. Tunio, R. u. Rehman, A. Rehman and K. u. Rehman, et al., Seroprevalence of hemorrhagic septicemia in buffalo and cattle in flood, irrigated and sandy areas of Punjab, Pakistan, Pure Appl. Biol., 2018, 7(4), 1234–1243 Search PubMed.
- T. E. Fuller, M. J. Kennedy and D. E. Lowery, Identification of Pasteurella multocida virulence genes in a septicemic mouse model using signature-tagged mutagenesis, Microb. Pathog., 2000, 29, 25–38 CrossRef CAS PubMed , available from http://www.ncbi.nlm.nih.gov/pubmed/10873488.
- M. Harper and J. D. Boyce, The myriad properties of Pasteurella multocida lipopolysaccharide, Toxins, 2017, 9(8), 1–21 Search PubMed.
- C. Gallego, S. Romero, P. Esquinas, P. Patiño, N. Martínez and C. Iregui, Assessment of Pasteurella multocida a lipopolysaccharide, as an Adhesin in an in vitro model of rabbit respiratory epithelium, Vet. Med. Int., 2017, 2017, 1–13 CrossRef PubMed.
- J. D. Boyce and B. Adler, The capsule is a virulence determinant in the pathogenesis of Pasteurella multocida M1404 (B:2), Infect. Immun., 2000, 68(6), 3463–3468 CrossRef CAS PubMed , available from http://iai.asm.org/content/68/6/3463.
- M. Harper, A. D. Cox, F. St Michael, I. W. Wilkie, J. D. Boyce and B. Adler, A heptosyltransferase mutant of Pasteurella multocida produces a truncated lipopolysaccharide structure and is attenuated in virulence, Infect. Immun., 2004, 72(6), 3436–3443 CrossRef CAS PubMed.
- M. Harper, A. Cox, F. St Michael, H. Parnas, I. Wilkie and P. J. Blackall, et al., Decoration of Pasteurella multocida lipopolysaccharide with phosphocholine is important for virulence, J. Bacteriol., 2007, 189(20), 7384–7391 CrossRef CAS PubMed.
- N. U. Horadagoda, J. C. Hodgson, G. M. Moon, T. G. Wijewardana and P. D. Eckersall, Development of a clinical syndrome resembling haemorrhagic septicaemia in the buffalo following intravenous inoculation of Pasteurella multocida serotype B:2 endotoxin and the role of tumour necrosis factor-α, Res. Vet. Sci., 2002, 72(3), 194–200 CrossRef CAS PubMed.
- Z. Wang, J. Li and E. Altman, Structural characterization of the lipid A region of Aeromonas salmonicida subsp. salmonicida lipopolysaccharide, Carbohydr. Res., 2006, 341(17), 2816–2825 CrossRef CAS PubMed.
- C. Erridge, E. Bennett-Guerrero and I. R. Poxton, Structure and function of lipopolysaccharides, Microbes Infect., 2002, 4(8), 837–851 CrossRef CAS PubMed.
- J. Da Silva Correia, K. Soldau, U. Christen, P. S. Tobias and R. J. Ulevitch, Lipopolysaccharide is in close proximity to each of the proteins in its membrane receptor complex, J. Biol. Chem., 2001, 276(24), 21129–21135 CrossRef CAS PubMed.
- K. Murphy and C. Weaver, The induced responses of innate immunity, Garland Science/Taylor and Francis Group, LLC, New York, NY, USA, 9th edn, Janeway's Immunobiology, 2017, p. 924 Search PubMed.
- R. P. Darveau, T.-T. T. Pham, K. Lemley, R. A. Reife, B. W. Bainbridge and S. R. Coats, et al., Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both toll-like receptors 2 and 4, Infect. Immun., 2004, 72(9), 5041–5051 CrossRef CAS PubMed , available from http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=517442%26tool=pmcentrez%26rendertype=abstract.
- A. Molinaro, O. Holst, F. D. Lorenzo, M. Callaghan, A. Nurisso and G. D'Errico, et al., Chemistry of lipid A: At the heart of innate immunity, Chem.–Eur. J., 2015, 21(2), 500–519 CrossRef CAS PubMed.
- C. M. Crittenden, L. D. Akin, L. J. Morrison, M. S. Trent and J. S. Brodbelt, Characterization of lipid A variants by energy-resolved mass spectrometry: Impact of acyl chains, J. Am. Soc. Mass Spectrom., 2016, 28(6), 1–9, DOI:10.1007/s13361-016-1542-6.
- D. S. Kabanov and I. R. Prokhorenko, Structural analysis of lipopolysaccharides from gram-negative bacteria, Biochemistry, 2010, 75(4), 383–404 CAS.
- B. Jeyaretnam, J. Glushka, V. S. K. Kolli and R. W. Carlson, Characterization of a Novel Lipid-A from Rhizobium Species Sin-1 a Unique Lipid-A Structure That is Devoid of Phosphate and has a Glycosyl Backbone Consisting of Glucosamine and 2-Aminogluconic Acid, J. Biol. Chem., 2002, 277(44), 41802–41810 CrossRef CAS PubMed , available from http://www.jbc.org/content/suppl/2008/05/20/M803225200.DC1.htmlhttp://www.jbc.org/content/283/29/20361.full.html#ref-list-1.
- N. L. S. Que, S. Lin, R. J. Cotter and C. R. H. Raetz, Purification and mass spectrometry of six Lipid A Species from the bacterial endosymbiont Rhizobium etli, J. Biol. Chem., 2008, 275(36), 28006–28016 Search PubMed.
- Z. Zhou, S. Lin, R. J. Cotter and C. R. H. Raetz, Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4 VO3 in Escherichia coli K12, J. Biol. Chem., 1999, 274(26), 18503–18514 CrossRef CAS PubMed.
- X. Wang and P. J. Quinn, Lipopolysaccharide: diosynthetic pathway and structure modification, Prog. Lipid Res., 2010, 49(2), 97–107, DOI:10.1016/j.plipres.2009.06.002.
- M. Caroff, D. Karibian, J. M. Cavaillon and N. Haeffner-Cavaillon, Structural and functional analyses of bacterial lipopolysaccharides, Microbes Infect., 2002, 4(9), 915–926 CrossRef CAS PubMed.
- J. Lodowska, D. Wolny, L. Weglarz and Z. Dzierzewicz, The structural diversity of lipid A from Gram-negative bacteria, Postepy Hig. Med. Dosw., 2007, 61, 106–121 Search PubMed.
- C. Alexander and U. Zähringer, Chemical structure of lipid A - the primary immunomodulatory center of bacterial lipopolysaccharides, Trends Glycosci. Glycotechnol., 2002, 14(76), 69–86 CrossRef.
- J. Geurtsen, L. Steeghs, J. t. Hove, P. V. D. Ley and J. Tommassen, Dissemination of Lipid A Deacylases (PagL) among Gram-negative bacteria, J. Biol. Chem., 2005, 280(9), 8248–8259 CrossRef CAS PubMed.
- G. Mills, A. Dumigan, T. Kidd, L. Hobley and J. A. Bengoecheaa, Identification and characterization of two Klebsiella pneumoniae lpxL lipid A late Acyltransferases and their role in virulence, Infect. Immun., 2017, 85(9), 1–19 CrossRef PubMed.
- J. Lukasiewicz, W. Jachymek, T. Niedziela, L. Kenne and C. Lugowski, Structural analysis of the lipid A isolated from Hafnia alvei 32 and PCM 1192 lipopolysaccharides, J. Lipid Res., 2010, 51(3), 564–574 CrossRef CAS PubMed.
- S.-H. Jo, H.-G. Park, W.-S. Song, S.-M. Kim, E.-J. Kim and Y.-H. Yang, et al., Structural characterization of phosphoethanolamine-modified lipid A from probiotic: Escherichia coli strain Nissle 1917, RSC Adv., 2019, 9(34), 19762–19771 RSC.
- B. D. Needham and M. S. Trent, Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis, Nat. Rev. Microbiol., 2013, 11(7), 467–481, DOI:10.1038/nrmicro3047.
- C. R. H. Raetz, C. M. Reynolds, M. S. Trent and R. E. Bishop, Lipid A modificaton system in Gram-negative bacteria, Annu. Rev. Biochem., 2007, 76(3), 295–329 CrossRef CAS PubMed.
- F. St Michael, E. Vinogradov, J. Li and A. D. Cox, Structural analysis of the lipopolysaccharide from Pasteurella multocida genome strain Pm70 and identification of the putative lipopolysaccharide glycosyltransferases, Glycobiology, 2005, 15(4), 323–333 CrossRef CAS PubMed.
- A. Steimle, I. B. Autenrieth and J. S. Frick, Structure and function: Lipid A modifications in commensals and pathogens, Int. J. Med. Microbiol., 2016, 306(5), 290–301, DOI:10.1016/j.ijmm.2016.03.001.
- R. A. Reife, S. R. Coats, M. Al-Qutub, D. M. Dixon, P. A. Braham and R. J. Billharz, et al., Porphyromonas gingivalis lipopolysaccharide lipid A heterogeneity: differential activities of tetra- and penta-acylated lipid A structures on E-selectin expression and TLR4 recognition, Cell. Microbiol., 2006, 8(5), 857–868 CrossRef CAS PubMed.
- B. Domon and C. E. Costello, A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates, Glycoconjugate J., 1988, 5(4), 397–409 CrossRef CAS.
- L. J. Morrison, W. R. Parker, D. D. Holden, J. C. Henderson, J. M. Boll and M. S. Trent, et al., UVliPiD: A UVPD-based hierarchical approach for de novo characterization of lipid A structures, Anal. Chem., 2016, 88(3), 1812–1820 CrossRef CAS PubMed.
- A. Kussak and A. Weintraub, Quadrupole ion-trap mass spectrometry to locate fatty acids on lipid A from Gram-negative bacteria, Anal. Biochem., 2002, 307(1), 131–137 CrossRef CAS PubMed.
- CABI, Detailed coverage of invasive species threatening livelihoods and the environment worldwide, data sheet on Pasteurella multocida infections, CABI Invasive Species Compendium, 2020, available from https://www.cabi.org/isc/datasheet/70916#todistribution, last updated on 10th Jan, 2020 Search PubMed.
- M. W. Bader, S. Sanowar, M. E. Daley, A. R. Schneider, U. Cho and W. Xu, et al., Recognition of antimicrobial peptides by a bacterial sensor kinase, Cell, 2005, 122, 461–472 CrossRef CAS PubMed.
- M. S. Trent, A. A. Ribeiro, S. Lin, R. J. Cotter and C. R. H. Raetz, An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-L-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor, J. Biol. Chem., 2001, 276(46), 43122–43131 CrossRef CAS PubMed , available from http://www.ncbi.nlm.nih.gov/pubmed/11535604, cited 2015 Dec 29.
- Z. Zhou, A. A. Ribeiro, S. Lin, R. J. Cotter, S. I. Miller and C. R. H. Raetz, Lipid A modifications in polymyxin-resistant Salmonella typhimurium: PmrA-dependent 4-amino-4-deoxy-L-arabinose, and phosphoethanolamine incorporation, J. Biol. Chem., 2001, 276(46), 43111–43121 CrossRef CAS PubMed.
- A. J. McCoy, H. Liu, T. J. Falla and J. S. Gunn, Identification of Proteus mirabilis mutants with increased sensitivity to antimicrobial peptides, Antimicrob. Agents Chemother., 2001, 45(7), 2030–2037 CrossRef CAS PubMed.
- A. Beceiro, E. Llobet, J. Aranda, J. A. Bengoechea, M. Doumith and M. Hornsey, et al., Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system, Antimicrob. Agents Chemother., 2011, 55(7), 3370–3379 CrossRef CAS PubMed , available from http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3122444%26tool=pmcentrez%26rendertype=abstract, cited 2015 Dec 29.
- R. K. Ernst, E. C. Yi, L. Guo, K. B. Lim, J. L. Burns, M. Hackett and S. I. Miller, Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa, Science, 1999, 286(5444), 1561–1565 CrossRef CAS PubMed.
- R. E. Bishop, The lipid A palmitoyltransferase PagP: molecular mechanisms and role in bacterial pathogenesis, Mol. Microbiol., 2005, 57(4), 900–912 CrossRef CAS PubMed.
- H. Loppnow, L. Brade, H. Brade, E. T. Rietschel, S. Kusumoto and T. Shiba, et al., Induction of human interleukin 1 by bacterial and synthetic lipid A, Eur. J. Immunol., 1986, 16(10), 1263–1267 CrossRef CAS PubMed.
- N. Qureshis, K. Takayamas, D. Hellerq and C. Fenselauq, Position of ester groups in the lipid A backbone of lipopolysaccharides obtained from Salmonella typhimurium, J. Biol. Chem., 1983, 258(21), 12947–12951 Search PubMed.
- J. V. Hankins, J. A. Madsen, D. K. Giles, J. S. Brodbelt and M. S. Trent, Amino acid addition to Vibrio cholerae LPS establishes a link between surface remodeling in Gram-positive and Gram-negative bacteria, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(22), 8722–8727 CrossRef CAS PubMed.
- S. R. Coats, A. B. Berezow, T. T. To, S. Jain, B. W. Bainbridge and K. P. Banani, et al., The lipid A phosphate position determines differential host toll-like receptor 4 responses to phylogenetically related symbiotic and pathogenic bacteria, Infect. Immun., 2011, 79(1), 203–210 CrossRef CAS PubMed.
- J. A. Gaddy, J. N. Radin, T. W. Cullen, W. J. Chazin, E. P. Skaar and M. S. Trent, et al., Helicobacter pylori resists the antimicrobial activity of calprotectin via lipid A modification and associated biofilm formation, mBio, 2015, 6(6), 1–14 CrossRef PubMed.
- N. J. Phillips, D. M. Adin, E. V. Stabb, M. J. McFall-Ngai, M. A. Apicella and B. W. Gibson, The lipid A from Vibrio fischeri lipopolysaccharide: a unique structure bearing a phosphoglycerol moiety, J. Biol. Chem., 2011, 286(24), 21203–21219 CrossRef CAS PubMed.
- G. Feng, Y. Hao, L. Wu and S. Chen, A visible-light activated [2 + 2] cycloaddition reaction enables pinpointing carbon-carbon double bonds in lipids, Chem. Sci., 2020, 11(27), 7244–7251 RSC.
- K. M. Townsend, A. J. Frost, C. W. Lee, J. M. Papadimitriou and H. J. S. Dawkins, Development of PCR assays for species- and type-specific identification of Pasteurella multocida isolates, J. Clin. Microbiol., 1998, 36(4), 1096–1100 CrossRef CAS PubMed.
- H. Masoud, A. Martin, P. Thibault, E. Richard Moxon and J. C. Richards, Structure of extended lipopolysaccharide glycoforms containing two globotriose units in Haemophilus influenzae serotype b strain RM7004, Biochemistry, 2003, 42(15), 4463–4475 CrossRef CAS PubMed.
- O. Westphal and K. Jann, Bacterial lipopolysaccharides: extraction with phenol–water and further applications of procedure, ed. R. L. Whisler, Methods Carbohydr Chem, Acad Press Inc, New York, 1965, vol. 5, pp. 83–91 Search PubMed.
- J. Guard-Petter, B. Lakshmi, R. Carlson and K. Ingram, Characterization of lipopolysaccharide heterogeneity in Salmonella enteritidis by an improved gel electrophoresis method, Appl. Environ. Microbiol., 1995, 61(8), 2845–2851 CrossRef CAS PubMed.
- R. Mahat, C. Seebart, F. Basile and N. L. Ward, Global and targeted lipid analysis of Gemmata obscuriglobus reveals the presence of lipopolysaccharide, a signature of the classical Gram-negative outer membrane, J. Bacteriol., 2016, 198(2), 221–236 CrossRef CAS PubMed.
- C. Chu, B. Liu, D. Watson, S. Szu, D. Bryla and J. Shiloach, et al., Preparation, characterization, and immunogenicity of conjugates composed of the O-specific polysaccharide of Shigella dysenteriae type 1 (Shiga's bacillus) bound to tetanus toxoid, Infect. Immun., 1991, 59(12), 4450–4458 CrossRef CAS PubMed.
- F. St Michael, J. Li and A. D. Cox, Structural analysis of the core oligosaccharide from Pasteurella multocida strain X73, Carbohydr. Res., 2005, 340(6), 1253–1257 CrossRef PubMed.
- F. St Michael, J. Li, E. Vinogradov, S. Larocque, M. Harper and A. D. Cox, Structural analysis of the lipopolysaccharide of Pasteurella multocida strain VP161: identification of both Kdo-P and Kdo-Kdo species in the lipopolysaccharide, Carbohydr. Res., 2005, 340(1), 59–68 CrossRef PubMed.
- M. Harper, F. St Michael, E. Vinogradov, M. John, J. D. Boyce, B. Adler and A. D. Cox, Characterization of the lipopolysaccharide from Pasteurella multocida Heddleston serovar 9: Identification of a proposed bi-functional dTDP-3-acetamido-3,6-dideoxy-α-D-glucose biosynthesis enzyme, Glycobiology, 2012, 22(3), 332–344 CrossRef CAS PubMed.
- L. N. Sarangi, P. Thomas, S. K. Gupta, A. Priyadarshini, S. Kumar, V. K. Nagaleekar, A. Kumar and V. P. Singh, Virulence gene profiling and antibiotic resistance pattern of Indian isolates of Pasteurella multocida of small ruminant origin, Comp. Immunol. Microbiol. Infect. Dis., 2015, 38, 33–39 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2020 |