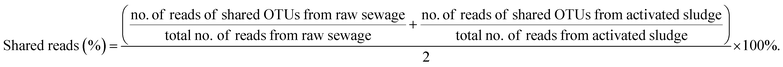Open Access Article
Open Access ArticleThe differential proliferation of AOB and NOB during natural nitrifier cultivation and acclimation with raw sewage as seed sludge†
Lifang Yu *ab,
Yu Wangac,
Ren Li
*ab,
Yu Wangac,
Ren Li ac,
Ru Zhanga,
Xingxiu Zhanga,
Sisi Huaa and
Dangcong Penga
ac,
Ru Zhanga,
Xingxiu Zhanga,
Sisi Huaa and
Dangcong Penga
aSchool of Environmental and Municipal Engineering, Xi'an University of Architecture and Technology, #13, Yanta Road, Xi'an 710055, China. E-mail: yulifang@xauat.edu.cn; wangyu@xauat.edu.cn; Fax: +86 029 82202729; Tel: +86 029 82202729
bShaanxi Key Laboratory of Environmental Engineering, Xi'an University of Architecture and Technology, Xi'an 710055, China
cKey Laboratory of Northwest Water Resource, Environment and Ecology, MOE, Xi'an University of Architecture and Technology, Xi'an 710055, China
First published on 29th July 2020
Abstract
Nitrifier immigration from sewers to wastewater treatment systems is attracting increasing attention for understanding nitrifier community assembly mechanisms, and improving process modeling and operation. In this study, nitrifiers in raw sewage were cultivated and acclimated in a sequencing batch reactor (SBR) for 90 days to investigate the characteristics of the influent nitrifiers after immigration. During the experiment, specific nitrite utilization rate (SNUR) exceeded specific ammonia utilization rate (SAUR) when floc size reached 224 ± 46 μm, and nitrogen loss occurred at the same time. The ratio of nitrite oxidizing bacteria (NOB) to ammonia oxidizing bacteria (AOB) increased from 0.84 to 2.14 after cultivation. The Illumina MiSeq sequencing showed that the dominant AOB was Nitrosomonas sp. Nm84 and unidentified species, and the three most abundant Nitrospira were Nitrospira defluvii, Nitrospira calida, and unidentified Nitrospira spp. in both raw sewage and cultivated activated sludge. The shared reads of raw sewage and activated sludge were 48.76% for AOB and 89.35% for Nitrospira. These indicated that nitrifiers, especially NOB, immigrated from influent can survive and propagate in wastewater systems, which may be a significant hinder to suppress NOB in the application of advanced nitrogen remove process based on partial nitrification in the mainstream.
Introduction
Nitrification is typically the rate-limiting process in activated sludge water resource recovery facilities (WRRFs). Activated sludge systems are common in large-scale municipal wastewater treatment, however, nitrification failure is a too-frequent occurrence in winter due to significant temperature effects and relatively slow growth rate of nitrifiers.1 Besides, how to stably and sustainably eliminate nitrite oxidizing bacteria (NOB) is still the main bottleneck in the application of advanced mainstream biological nitrogen removal (BNR) technologies based on partial nitrification.2,3 Thus, the microorganism community of nitrifiers has attracted more and more attention in the field of biological wastewater treatment design and operation.To maintain a constant amount of biomass in WRRFs, a small fraction of the biomass is removed daily as surplus sludge. Plants in Xi'an, China, which have relatively low temperatures, have a relatively long solids retention time (SRT) of 10–20 days, where 5–10% of the biomass is removed each day. In order to counterbalance this removal in the system, the number of organisms must increase at a rate of 1/SRT per day. This increase is generally considered to be caused by the net growth of bacteria in the bioreactor.
Relatively high abundances of microbes have been confirmed to exist in the influent of WRRFs.4,5 Due to their constant and effective immigration, the incoming cells might be abundant in the activated sludge community, despite possibly being inactive.6 Jauffur et al. investigated three WRRFs located near Montreal during winter and suggested that the nitrifiers in the influent were active and likely seeded activated sludge bioreactors since the most abundant operational taxonomic units (OTUs) in the influent and mixed liquor were the same.4 Saunders et al. investigated three WRRFs in Denmark and showed that the similar relative abundance of Nitrospira and Nitrotoga in the activated sludge and wastewater influent may indicate these organisms are acting as a seed for selection in the plants.6 Moreover, the immigration of influent nitrifiers into activated sludge systems has been shown to enhance the local nitrifier community and function in a lab-scale study.5 These works test the hypothesis that influent organisms are another important source of biomass addition, it also has significant implications regarding the role of influent populations in the construction of activated sludge communities.
There is no consensus on approaches to analyzing immigration, and the assembly of bacterial communities in open biological systems, such as activated sludge systems, has long been considered chaotic and unpredictable.7,8 Therefore, the current best practice for biological wastewater treatment modeling, such as the Activated Sludge Models (ASMs) which are recognized by the International Water Association (IWA), is to assume that there is no active nitrifying biomass in municipal wastewaters at the entrance of treatment facilities.9 However, the neglection of the impact of natural nitrifier immigration may influence the design of biological wastewater treatment facilities, especially the size of aerobic bioreactors performing nitrification. Meanwhile, a continuous supply of nitrifiers, especially NOB, immigrated from raw sewage is very likely to have an adverse effect on achieving partial nitrification in the mainstream application.10 Therefore, immigration from sewers to wastewater treatment systems is attracting increasing attention for understanding community assembly mechanisms and improving process modeling and operation. While so far, almost all of the studies previously discussed were based on the investigation of the similarity of the influent and activated sludge nitrifier communities. Comparatively few experiments have been designed directly to understand how the influent nitrifiers grow and persist in environmental conditions prevalent in WRRFs.
In this study, influent nitrifiers from the 2nd WRRF (without primary settler) in Xi'an were cultivated and acclimated in a sequencing batch reactor (SBR) for 90 days. The floc size, nitrification performance, and nitrifier community were investigated to explore the process of the survival and reproduction of the influent nitrifiers in activated sludge bioreactors and evaluate the impact of nitrifier immigration from influent on nitrifier community assembly. Using this basis, we provided new insight into achieving full nitrification in cold northern regions and meanwhile proposes a great challenge of suppressing NOB in the application of advanced BNR process based on partial nitrification in the mainstream.
Methods
Experimental set-up and operation
A lab-scale SBR with a working volume of 4 L was set-up and operated for 90 days. Throughout, the reactor temperature was maintained at 20 ± 1 °C. The processes of influent, effluent and sludge discharge were controlled by a programmable logic controller (PLC) using the peristaltic pumps. In addition, airflow rate was kept constant and average dissolved oxygen (DO) concentration during an operating cycle was above 2.5 mg L−1. pH was maintained at 7.5–8.0 by manually adding 1.0 M NaHCO3 to maintain the alkalinity required for nitrification.During start-up, SBR was filled with 4 L real influent sewage from the 2nd WRRF in Xi'an and aerated for six days to cultivate biomass (phase I). The characteristics of the 2nd WRRF and influent sewage were described in ESI (Tables S1 and S2†). On the sixth day, once the ammonium was completely degraded, the reactor began operation for 90 days. It was operated in 48 h cycle (phase II, days 7–8, 15.06 g N per m3 per day), 24 h cycle (phase III, days 9–13, 30.11 g N per m3 per day), 12 h cycle (phase IV, days 14–22, 60.23 g N per m3 per day), 8 h cycle (phase V, days 23–27, 90.35 g N per m3 per day), 6 h cycle (phase VI, days 28–33, 120.46 g N per m3 per day), after which the cycle time was gradually reduced to 4 hours (phase VII, days 34–90, 180.69 g N per m3 per day) with a volumetric exchange rate (VER) of 50%. Each cycle consisted of feeding (5 min), aeration (depending on the operating cycle time of different phases), settling (40 min), decanting (5 min) and idle time (10 min). Once the mixed liquor volatile suspended solids (MLVSS) is close to the value of the 2nd WRRF in Xi'an (2.35 g L−1), a specific volume of activated sludge was discharged prior to the end of the aeration phase to keep the SRT at 15–20 days.
Feed medium
The synthetic influent was used as feed to mimic the average composition and quality of domestic wastewater (the effluent of aerated grit chamber, that is, the influent of the bioreactor) in the 2nd WRRF after the sixth day. The composition was as follows: 13 mg L−1 NH4Cl, 17 mg L−1 peptone, 79 mg L−1 NaAc, 122 mg L−1 starch, 116 mg L−1 low fat milk powder, 52 mg L−1 yeast, 92 mg L−1 urea, 23 mg L−1 KH2PO4, and 5.8 mg L−1 FeSO4·7H2O. The feed was autoclaved at 121 °C for 1 h and allowed to cool down prior to feeding the reactors. A mixture of trace elements was then added to the feed and comprised of the following: 0.770 mg L−1 Cr(NO3)3·2H2O, 0.536 mg L−1 CuCl2·2H2O, 0.108 mg L−1 MnSO4·H2O, 0.336 mg L−1 NiSO4·6H2O, 0.100 mg L−1 PbCl2, 0.208 mg L−1 ZnCl2. The total nitrogen (TN) concentration, and COD of the formulated influent recipe were approximately 60.23 mg L−1, and 385 mg L−1, respectively.Analytical methods
Morphology observation of activated sludge in raw sewage and SBR was performed using a microscope (ECLIPSE Ti-S, NIKON, JAPAN). DO and pH were monitored by a DO and pH meter (Mettler Toledo, Switzerland). All other physical and chemical parameters, including sludge volume index (SVI), mixed liquor suspended solids (MLSS), MLVSS, chemical oxygen demand (COD), NH4+–N, NO2−–N, NO3−–N were measured according to the standard methods.11The microscopic digital images and floc size of the sludge samples were obtained by an electron microscope (50i, Nikon, Japan). The values of the equivalent diameter (Deq) were calculated using the eqn (1).
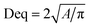 | (1) |
All samples harvested from raw sewage and reactor were investigated with oxygen uptake rates (OUR) for nitrifier activity. The detailed measurement methods of the specific ammonia uptake rate (SAUR) and specific nitrite uptake rate (SNUR) referred to the previous study.5 Fluorescence in situ hybridization (FISH) was performed on day 1 (initial start-up period) and day 70 (stable operation period) with the same primers as recorded previously.5 Concretely, the rRNA-targeted oligonucleotide probes used in FISH were EUBmix (EUB338 + EUB338 II + EUB338 III), AOBmix (Nso1225 + Nsv443 + Nsm156 + NmV), and NOBmix (Ntspa662 + NIT3 + Ntcoc206 + Ntspn693). A confocal laser scanning microscope (CLSM) was used for image acquisition (Leica TCS SP8, Leica Microsystems, Germany).
DNA extraction and PCR amplification
The samples of raw sewage and mixed liquor were collected for community structure analysis on day 1 and day 70, respectively. All samples were pelleted by centrifugation for 5 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. Genomic DNA was extracted from 0.25 g of centrifuged wet solids using the TIAN amp Soil DNA Kit (DP336, TIANGEN Biotech, China) according to the protocol provided by the manufacturer. DNA concentration and purity were determined by analysis with a microvolume spectrophotometer (NanoDrop 2000, USA). And the extracted genomic DNA samples were stored at −20 °C until further analysis.
000 rpm. Genomic DNA was extracted from 0.25 g of centrifuged wet solids using the TIAN amp Soil DNA Kit (DP336, TIANGEN Biotech, China) according to the protocol provided by the manufacturer. DNA concentration and purity were determined by analysis with a microvolume spectrophotometer (NanoDrop 2000, USA). And the extracted genomic DNA samples were stored at −20 °C until further analysis.
The PCR primers used in this study are shown in Table 1. Each 50 μL of PCR mixture containing 1 μL of 10 μmol L−1 forward primer, 1 μL of 10 μmol L−1 reverse primer, 1 μL of 20 ng mL−1 DNA template, 25 μL of 2.5 units per μL 2× Taq MasterMix (CWBIO, China), and 22 μL of UltraPure™ Distilled Water. The thermocycling conditions for PCR amplification included a prior denaturation at 94 °C for 5 min; followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, elongation at 72 °C for 30 s, and extension at 72 °C for 10 min. To reduce the deviation, the PCR reaction per sample was performed in triplicate.
| Target gene | Primera | Sequence (5′–3′) | Reference |
|---|---|---|---|
| a Primer's short name used in the reference. | |||
| Ammonium monooxygenase (amoA) | amoA-1F | GGGGTTTCTACTGGTGGT | 12 |
| amoA-2R | CCCCTCTGCAAAGCCTTCTTC | ||
| nxrB genes of Nitrospira | nxrB-169F | TACATGTGGTGGAACA | 13 |
| nxrB-638R | CGGTTCTGGTCRATCA | ||
| 16S rRNA Nitrobacter sp. | FGPS-1269′ | CTAAAACTCAAAGGAATTGA | 14 |
| FGPS-872 | TTTTTTGAGATTTGCTAG | ||
Quantitative PCR (qPCR)
Real-time PCR was performed to quantify the copy numbers of ammonia oxidizing bacteria (AOB), Nitrobacter spp., Nitrospira spp., and Nitrotoga spp. by ABI 7500 systems (Applied Biosystems, USA). And an SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) (Takara Bio., Dalian, CO., LTD) was used to quantify nitrifiers. Each diluted plasmid, ranging from 101 to 105 copies per microliter, was used for standardization for qPCR assays. Each tube was loaded with 2 μL DNA sample, followed by 10 μL 2× SYBR Green Master Mix reaction solution (Invitrogen, China), 0.5 μL of 10 μmol L−1 forward primer, 0.5 μL of 10 μmol L−1 reverse primer, and 7 μL UltraPure™ Distilled Water. A two-stage amplification protocol was performed as follows: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, and simultaneous annealing and extension at 60 °C for 40 s.Illumina MiSeq sequencing and sequence data analysis
The 16S rRNA gene fragments of nitrifiers were amplified via the PCR with the above primer sets (Table 1). The PCR products were extracted from 2% agarose gels and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) according to the manufacturer's instructions and quantified using QuantiFluor™-ST (Promega, USA). Purified amplicons were pooled in equimolar and paired-end sequenced (2 × 250) on an Illumina MiSeq PE300 platform (Illumina, Inc., CA, USA). The library construction and sequencing were performed by Allwegene Co., Ltd. (Beijing, China).The extraction of high-quality sequences was performed with the Quantitative Insights Into Microbial Ecology (QIIME) package (v1.2.1). Raw sequences were selected based on sequence length, quality, primer, and tag. The raw sequences were selected and low-quality sequences were removed. This included any raw reads shorter than 110 nucleotides, any truncated reads that were shorter than 50 bp, reads containing ambiguous characters were removed. Only sequences with an overlap of longer than 10 bp were assembled according to their overlap sequence. Reads which could not be assembled were discarded.
The unique sequence set was classified into OTUs under the threshold of 97% identity using UCLUST. Chimeric sequences were identified and removed using Usearch (version 8.0.1623). The taxonomy of each 16S rRNA gene sequence was analyzed by UCLUST against then database using a confidence threshold of 90%. All the raw data have been archived at NCBI Sequence Read Archive (SRA) database with accession number of SRR2106467.
Results
Reactor performance
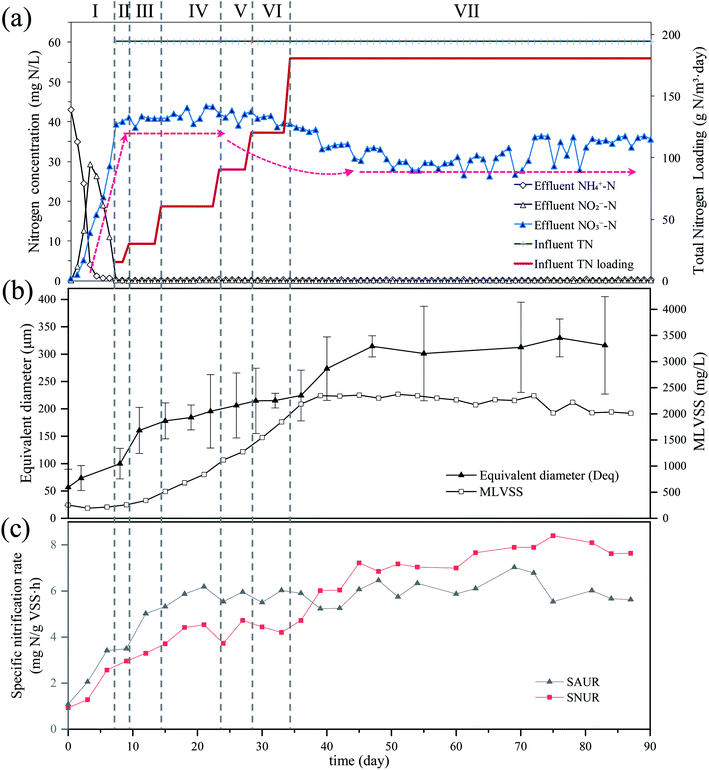
The nitrogen concentrations of the SBR influent and effluent over the 90 day monitoring period are displayed in Fig. 1a. During phase I (days 0–6), the SBR was filled with raw sewage from the 2nd WRRF in Xi'an and aerated without feeding. The effluent NH4+–N concentration gradually decreased from 43.14 mg L−1 (day 0) to 3.97 mg L−1 on day 3 and was less than 1 mg L−1 during days 4–7. Corresponding to the reduction of NH4+–N, the NO2−–N concentration gradually increased from 0 mg L−1 at the beginning of the observation period to 29.38 mg L−1 on day 3, and then gradually decreased to 0.14 mg L−1 by day 7. The NO3−–N concentrations gradually increased from 0 mg L−1 at the beginning of the observation period to 39.39 mg L−1 by day 7. During phase II–VII, the concentration of NH4+–N and NO2−–N in effluent were all lower than the minimum detection limit, while the effluent NO3−–N concentration was 41.36 ± 1.42 mg L−1 during days 7–30. It is, however, worth noting that the effluent NO3−–N concentration decreased gradually during days 30–44 until reaching and maintaining 30.58 ± 2.14 mg L−1 from day 45 until the end of the observation period.As shown in Fig. 1b, the MLVSS in SBR gradually increased with the influent nitrogen loading rate. On day 39, the MLVSS reached 2349 mg L−1, which was close to the MLVSS in the bioreactor of the 2nd WRRF in Xi'an (2.35 g L−1). The excess sludge was then discharged to keep the SRT of the SBR at 15–20 days and to maintain the MLVSS at 2279 ± 96 mg L−1 during days 40–90.
The Deq of flocs in raw sewage was 57 ± 33 μm and then quickly increased to 161 ± 42 μm during days 0–11 (Fig. 1b). During days 11–36, the increasing rate in Deq was slow, with the value increasing from 161 ± 42 μm to 224 ± 46 μm. The floc size increased quickly again from 224 ± 46 μm to 314 ± 19 μm during days 36–47. After day 47, the floc size stabilized at 315 ± 62 μm until the end of the observation period.
Nitrifier activity
Respirometric assays were used to evaluate the activity of nitrifiers. Fig. S1† showed that the AOB and NOB in raw sewage required approximately 0.5 h after ammonia addition and 2 h after nitrite addition, respectively, to acclimatize to the new environment. The maximal OUR of AOB and NOB were 17.06 ± 0.20 mg O2 per L and 8.10 ± 0.21 mg O2 per L, respectively. After the conversion of mg O2 per L to mg N (g VSS h)−1, the maximum SAUR and SNUR of raw sewage in the experiment were 1.09 ± 0.02 mg N (g VSS h)−1 and 0.93 ± 0.07 mg N (g VSS h)−1, respectively. And the SNUR/SAUR of raw sewage was 0.85.The activities of nitrifiers in SBR were analyzed every three days (Fig. 1c). The SAUR increased quickly until day 12 and kept relatively stable at 6.30 ± 0.44 mg N (g VSS h)−1 during the rest of the experiment period. On the contrary, the SNUR increased more slowly than SAUR and reached a relatively stable value of 7.89 ± 0.29 mg N (g VSS h)−1 after day 45; the SNUR/SAUR was 1.25. It was worth noting that the SNUR was lower than the SAUR during days 0–39, while higher than SAUR during days 40–90. Similar results that the average SNUR was higher than SAUR in the investigation of 10 full-scale WRRFs in Xi'an, China, were also reported by Yao and Peng.15
Nitrifier community
| Sample source | AOB (copies per L) | Nitrospira spp. (copies per L) | Nitrobacter spp. (copies per L) | NOB/AOBa (cell/cell) |
|---|---|---|---|---|
| a Cell/cell: the ratio of cell number per liter, cells per L = copies per L ÷ (gene copy number per cell). Assumed gene copy number per cell is 1 for Nitrospira 16S rDNA, 1 for Nitrobacter 16S rRNA, and 2 for amoA gene.16 | ||||
| Raw sewage | 3.70 × 106 | 1.56 × 106 | 1.43 × 103 | 0.84 |
| Activated sludge | 1.06 × 108 | 1.13 × 108 | 2.11 × 104 | 2.14 |
The Chao1 index and Shannon index were used to evaluate the species richness and diversity of nitrifiers, respectively. As shown in Table 3, the Chao1 and Shannon index of raw sewage all were higher than these of cultured activated sludge. This implied that raw sewage had higher species richness and diversity of AOB and NOB.
The relative abundances of the microbial communities at the species level are shown in Fig. 3a. It could be seen that the dominant AOB species in both raw sewage and activated sludge were Nitrosomonas sp. Nm84 and unidentified species. The immense ecological significance of this particular group of bacteria contrasts with our limited knowledge about them because most species that inhabit activated sludge are still uncultured and unidentified.17 Similar to the AOB community profiles, the dominant Nitrospira-related NOB species identified in the activated sludge, are the same as those detected in raw sewage, namely Nitrospira defluvii, Nitrospira calida, and unidentified (Fig. 3b). These have previously been reported to be important NOB species in sewage treatment.18
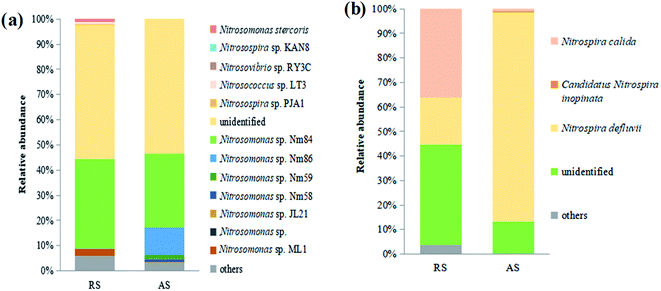 | ||
| Fig. 3 Relative abundances of microbial community of two samples (raw sewage, RS; activated sludge, AS) at species level. (a) AOB; (b) NOB (Nitrospira). | ||
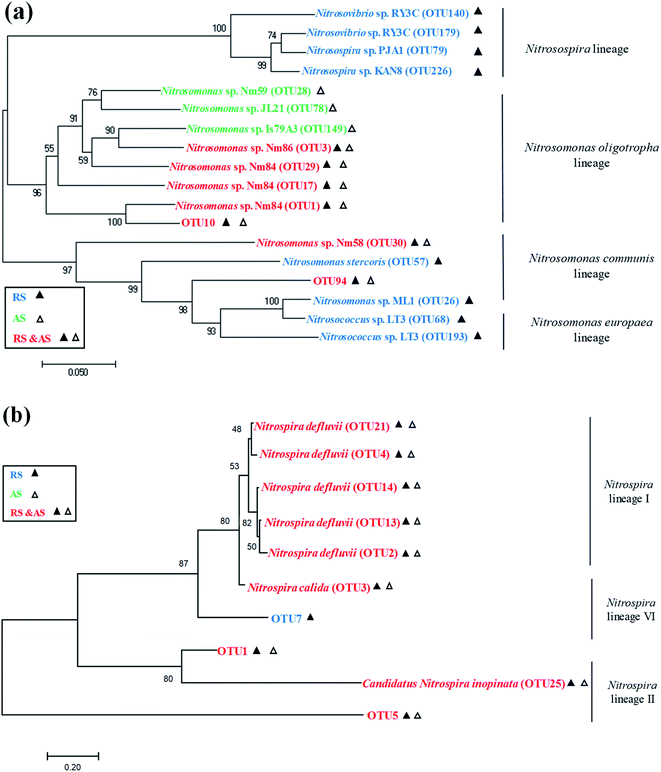
The AOB populations and the NOB (Nitrospira) populations were classified under lineages as shown in Fig. 4. Among the AOB populations, four lineages were found in the raw sewage, namely Nitrosomonas oligotropha, Nitrosomonas communis, Nitrosomonas europaea, and Nitrosospira lineages. But only Nitrosomonas oligotropha lineage and Nitrosomonas communis lineage were found to be dominant in the activated sludge, and these are common AOB present in activated sludge of WRRFs.19 Among the NOB (Nitrospira) populations, Nitrospira lineage I, Nitrospira lineage II, and Nitrospira lineage VI were found in both the raw sewage and activated sludge samples. This is in agreement with Saunders et al. who detected a relatively high abundance of Nitrospira in influent and activated sludge of WRRFs.6
Discussion
Additional nitrite produced by nitrite loop urges the increase of NOB/AOB
Due to mass conservation law, the concentration of NO2−–N cannot be higher than that of NH4+–N in the complete nitrification pathway. Besides, the yield coefficient of AOB was higher than that of NOB,20 thus it seems almost inevitable that SNUR will be lower than SAUR in the sewage treatment systems. This was exactly the case during the initial 39 days of the experiment, however, there was an unexpected turn that SNUR > SAUR after day 40 (Fig. 1c). Furthermore, the floc size gradually increased in natural cultivation and acclimation (Fig. 1b). When the floc size reached 224 ± 46 μm (day 36), the effluent NO3−–N concentration decreased significantly and the nitrogen loss occurred as shown in Fig. 1a. Almost at the same time (day 40–50), the effluent NO3−–N concentration, the Deq of the flocs, and SNUR all tended to be stable.Besides, it is known that in full-nitrification, NOB generates only two electrons from the oxidation of nitrite to nitrate, which is three times lower than the number of electrons generated by AOB during oxidizing of ammonium to nitrite.21,22 Due to ammonium activated by the ammonia monooxygenase (AMO) in the AOB metabolic pathway, these two electrons are not available for energy generation. Thus, it is expected that the biomass yield of NOB is about two times lower than that of AOB per unit of nitrogen. This implies a theoretical NOB/AOB ratio of 0.5.23 However, the FISH and qPCR results both showed the same trend that the ratio of NOB/AOB increased after cultivation and acclimation with raw sewage as seed. Concretely, the NOB/AOB ratio (the ratio of cell number per liter) in the raw sewage was 0.84, relatively close to the theoretical value, however, it increased to 2.14 in the cultured activated sludge (days 90) (Table 2). Winkler et al. also showed higher NOB/AOB ratios (3–4) in aerobic granular sludge samples.23 This has corresponded to the increase of SNUR/SAUR with the floc size as shown in Fig. 1.
Given all that, there is probably another route by which NOB could contact more NO2−–N. In this study, the floc size gradually increased during cultivation. The increasing floc size produced greater mass transport resistance, and the diffusion of oxygen in the inner flocs would be limited. The aerobic region was thus gradually confined to the surface layer, and then an anoxic zone would occur in the inner part of the flocs. Numerous studies have shown experimentally as well as by mathematical modeling that oxygen penetration is restricted to the outer rim of the flocs (<100 μm).24 Therefore, anoxic denitrification should occur along with the increase of the floc size after day 36, which is corresponding to the decrease in effluent NO3−–N concentration during days 30–44. Based on the fact that nitrite is an intermediate compound in both nitrification and denitrification steps,25 Winkler et al. adopted a conceptual “nitrite loop” model to describe the bacterial growth balance in a nitrifying community (Fig. 5).23 Nitrite oxidation is coupled with nitrate reduction. According to the nitrite loop theory, the additional nitrite produced in the denitrification pathway may transfer to the oxic zone and be reoxidized to nitrate by NOB. Therefore, it is possible for NOB to receive a larger amount of NO2−–N than that of NH4+–N received by AOB, with the result that the SNUR will be higher than SAUR after day 36 when the average floc size has increased to 224 ± 46 μm.
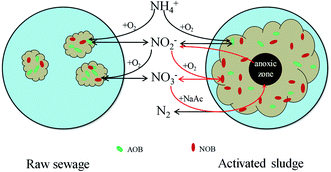 | ||
| Fig. 5 Schematic view of nitrite loop theory (adapted from Winkler et al.23). The additional nitrite produced in the denitrification pathway may transfer to the oxic zone and then be reoxidized to nitrate by NOB. | ||
The effect of nitrifier immigration on the community of activated sludge system
Raw sewage obviously had a certain nitrifier activity that required several hours to recover as shown in Fig. S1.† Meanwhile, the recovery time of AOB in raw sewage significantly is shorter than that of NOB (i.e., AOB activity relatively is easier to recover). Similar results were also obtained by Yu et al. and Jauffur et al.4,5 As the measured recovery time of nitrifier activity is shorter than the conventional SRT of activated sludge systems in WRRFs (15–20 days), nitrifiers immigrated from raw sewage are likely to achieve full induction in biological treatment systems.5 This is confirmed by the qPCR results that the copies number of AOB and NOB in cultured activated sludge were significantly larger than that in raw sewage (Table 2). Therefore, nitrifier immigrated from raw sewage should not be neglected in biological treatment systems.Actually, the nitrifier immigration of raw sewage is accompanied by the variation of species richness and the change of nitrifier community occurred in activated sludge systems. The Illumina MiSeq sequencing showed that raw sewage had higher species richness and diversity of AOB and NOB than cultured activated sludge (Table 3). The reasons for lower diversity of activated sludge may be: (i) the activated sludge process operates some kind of selection,7 (ii) the number of ecological niches in biological treatment process is lower than that in sewer system,26 or (iii) the environmental conditions prevalent in WRRFs may also exert a sort of selective pressure on the species assembly of the nitrifier population.27 The real reasons still need to be further studied. In the community structure of nitrifiers, the dominant AOB species in both raw sewage and activated sludge were Nitrosomonas sp. Nm84 and unidentified species (Fig. 3a). For NOB, the dominant Nitrospira-related NOB species in the activated sludge are Nitrospira defluvii, Nitrospira calida, and unidentified, which are the same as those detected in raw sewage (Fig. 3b). However, Nitrospira defluvii from the raw sewage eventually becomes the most predominant NOB in the acclimated activated sludge. It is worth mentioning that Nitrospira defluvii is always be considered as the most predominant nitrite oxidizer in WRRFs.28 Phylogenetic trees reflected evolutionary relationships among various nitrifiers of raw sewage and activated sludge based upon similarities and differences in their genetic characteristics (Fig. 4). For AOB, only Nitrosomonas oligotropha lineage and Nitrosomonas communis lineage, common AOB present in WRRFs,19 were found to be dominant in the activated sludge. However, for NOB, the Nitrospira lineage I, II, and VI were all found in the raw sewage and activated sludge samples. These results indicated that the nitrifier communities of raw sewage and activated sludge have a certain similarity.
In brief, raw sewage makes a great contribution in supplying valuable AOB and NOB populations to bioreactor by natural continuous seeding, and then plays an important role in the nitrifier community construction of activated sludge systems. Considering that nitrifiers immigrated from raw sewage could survive and propagated in activated sludge systems, theses nitrifiers could partially compensate for the decreasing nitrifier activity of activated sludge systems in cold northern regions. Actually, nitrifiers were reportedly able to form strong microcolonies in flocs, which are more resistant to high shear forces so that it could be more effectively removed by primary settler.29 Based on incubation tests and modelling by Duan et al., the primary settler designed in WRRFs exhibited high efficiencies for AOB removal and NOB removal, at 72.3% and 94.2%, respectively.10 Therefore, primary treatment for raw sewage, e.g. primary settler, may be unnecessary for achieving efficiently full-nitrification in cold northern regions.
The intractable challenge for achieving partial nitrification in the mainstream
However, the nitrifiers immigration from raw sewage also may bring bad consequences to BNR process. It can be observed from Table 3 that the percentages of shared reads in raw sewage and activated sludge were 48.76% for AOB and 89.35% for NOB (Nitrospira), respectively. Similarly, Jauffur et al. reported that the percentage of reads belonging to OTUs that appeared in both influent and mixed liquor of the same WRRF averaged 78% for AOB and 86% for NOB. NOB (Nitrospira), by contrast, is more effective than AOB in seeding the activated sludge systems.4 Thus, it was inferred that the percentage of shared reads could even be used to evaluate the seeding efficiency in the design of biological wastewater treatment modeling (e.g. ASMs).It is known that the key to achieving stable partial nitrification is to sustainably retain AOB while eliminating NOB in the mainstream BNR process.30,31 However, raw sewage that contains a few nitrifiers, especially the more efficient NOB in seeding as shown in Table 3, continuously inoculates the bioreactor, which might have a devastating effect on achieving stable partial nitrification. Duan et al. reported that the continuously seeding of NOB in raw sewage resulted in different extents of ineffective NOB suppression in the mainstream activated sludge systems.10 Meanwhile, the NOB in the raw sewage could stimulate the NOB community shifts under NOB suppression pressure to develop resistance.
As the municipal sewage has too lower carbon-nitrogen ratio to meet the carbon source requirement in the BNR process, canceling primary settling tanks usually are used to increase the concentration of organic carbon source entering the bioreactors in the engineering, like the 2th WRRF in Xi'an, which ultimately leads to the reduced TN concentration of secondary effluent.32 It has been proved that the primary settling tank could remove about 94.2% of NOB contained in the raw sewage.10 Considering that the NOB immigration from raw sewage is definitely a significant hinder to suppress NOB, primary treatment is necessary for ensuring stable mainstream NOB suppression.
To sum up, when the mainstream BNR process in WRRFs is based on traditional nitrification-denitrification process in cold northern regions, if without any special requirements, it may be considered not to set up primary settling tank to achieve efficiently full-nitrification. However, when advanced mainstream BNR process based on partial nitrification was applied in WRRFs, primary settling tanks was necessary for achieving stable partial nitrification process.
Conclusion
(1) In natural cultivation and acclimation with raw sewage as seed sludge, the flocs size gradually increased, which provided valuable habitats for influent nitrifiers to retain and propagate, and then enough nitrifiers could be cultured.(2) Nitrogen loss occurred when the flocs size reached 193 ± 46 μm (day 36), and then SNUR unexpectedly exceeded SAUR and the NOB/AOB ratio increased with the floc sizes due to nitrite loop.
(3) The shared reads were 48.76% for AOB and 89.35% for NOB (Nitrospira) for raw sewage and activated sludge. Thus, nitrifiers, especially NOB, immigrated from influent can survive and propagate in wastewater systems, which should be benefit for achieving full nitrification in cold northern regions and may be a significant hinder to suppress NOB in the application of advanced BNR process based on partial nitrification in the mainstream.
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This research was supported by Natural Science Foundation of Shaanxi Province (2020JM-474).References
- E. B. Szabó, M. Hermansson, O. Modin, F. Persson and B. Wilén, Water, 2016, 8, 172 CrossRef.
- G. Xu, Y. Zhou, Q. Yang, Z. M.-P. Lee, J. Gu, W. Lay, Y. Cao and Y. Liu, Appl. Microbiol. Biotechnol., 2015, 99, 2485–2490 CrossRef CAS PubMed.
- H. Daims, E. V. Lebedeva, P. Pjevac, P. P. Han, C. W. Herbold, M. Albertsen, N. Jehmlich, M. Palatinszky, J. Vierheilig, A. G. Bulaev, R. H. Kirkegaard, M. Von Bergen, T. Rattei, B. Bendinger, P. H. Nielsen and M. Wagner, Nature, 2015, 528, 504–509 CrossRef CAS PubMed.
- S. Jauffur, S. Isazadeh and D. Frigon, Water Sci. Technol., 2014, 70, 1526–1532 CrossRef CAS PubMed.
- L. F. Yu, R. Li, R. Delatolla, R. Zhang, X. L. Yang and D. C. Peng, J. Environ. Sci., 2018, 74, 159–167 CrossRef PubMed.
- A. M. Saunders, M. Albertsen, J. Vollertsen and P. H. Nielsen, ISME J., 2016, 10, 11–20 CrossRef CAS PubMed.
- D. Frigon and G. Wells, Curr. Opin. Biotechnol., 2019, 57, 151–159 CrossRef CAS PubMed.
- A. Ramette and J. M. Tiedje, Microb. Ecol., 2007, 53, 197–207 CrossRef PubMed.
- M. W. Sweeney and J. C. Kabouris, Water Environ. Res., 2014, 87, 1178–1195 CrossRef PubMed.
- H. Duan, L. Ye, Q. Wang, M. Zheng, X. Lu, Z. Wang and Z. Yuan, Water Res., 2019, 162, 331–338 CrossRef CAS PubMed.
- APHA, Standard methods for the examination of water and wastewater, American Public Health Association, Washington, DC, 22nd edn, 2012 Search PubMed.
- J. H. Rotthauwe, K. P. Witzel and W. Liesack, Appl. Environ. Microbiol., 1997, 63, 4704–4712 CrossRef CAS PubMed.
- M. Pester, F. Maixner, D. Berry, T. Rattei, H. Koch, S. Lucker, B. Nowka, A. Richter, E. Spieck and E. N. Lebedeva, Environ. Microbiol., 2014, 16, 3055–3071 CrossRef CAS PubMed.
- J. Geets, M. de Cooman, L. Wittebolle, K. Heylen, B. Vanparys, P. De Vos, W. Verstraete and N. Boon, Appl. Microbiol. Biotechnol., 2007, 75, 211–221 CrossRef CAS PubMed.
- Q. Yao and D. C. Peng, AMB Express, 2017, 7, 25 CrossRef PubMed.
- G. Harms, A. C. Layton, H. M. Dionisi, I. R. Gregory, V. Garrett, S. A. Hawkins, K. G. Robinson and G. S. Sayler, Environ. Sci. Technol., 2003, 37, 343–351 CrossRef CAS PubMed.
- S. Lücker, M. Wagner, F. Maixner, E. Pelletier, H. Koch, B. Vacherie, T. Rattei, J. S. S. Damsté, E. Spieck and D. L. Paslier, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 13479–13484 CrossRef PubMed.
- H. Daims, U. Purkhold, L. Bjerrum, E. Arnold, P. A. Wilderer and M. Wagner, Water Sci. Technol., 2001, 43, 9–18 CrossRef CAS PubMed.
- U. Purkhold, A. Pommereningroser, S. Juretschko, M. Schmid, H. Koops and M. Wagner, Appl. Environ. Microbiol., 2000, 66, 5368–5382 CrossRef CAS PubMed.
- F. Fang, B. Ni, X. Li, G. Sheng and H. Yu, Appl. Microbiol. Biotechnol., 2009, 83, 1159–1169 CrossRef CAS PubMed.
- H. Daims, S. Lücker and M. Wagner, Trends Microbiol., 2016, 24, 699–712 CrossRef CAS PubMed.
- B. B. Ward, in Methods Enzymol., ed. M. G. Klotz, Academic Press, 2011, vol. 486, pp. 307–323 Search PubMed.
- M. K. H. Winkler, J. P. Bassin, R. Kleerebezem, D. Y. Sorokin and M. C. M. Van Loosdrecht, Appl. Microbiol. Biotechnol., 2012, 94, 1657–1666 CrossRef CAS PubMed.
- C. Picioreanu, J. B. Xavier and M. C. M. Van Loosdrecht, Biofilms, 2004, 1, 337–349 CrossRef.
- L. Zhang, G. Zeng, J. Zhang, Y. Chen, M. Yu, L. Lu, H. Li, Y. Zhu, Y. Yuan, A. Huang and L. He, Appl. Microbiol. Biotechnol., 2015, 99, 4059–4070 CrossRef CAS PubMed.
- S. H. Lee, H. J. Kang and H. D. Park, Water Res., 2015, 73, 132–144 CrossRef CAS PubMed.
- L. Wittebolle, M. Marzorati, L. Clement, A. Balloi, D. Daffonchio, K. Heylen, P. De Vos, W. Verstraete and N. Boon, Nature, 2009, 458, 623–626 CrossRef CAS PubMed.
- D. Xu, S. Liu, Q. Chen and J. Ni, AMB Express, 2017, 7, 40 CrossRef PubMed.
- P. Larsen, J. L. Nielsen, T. C. Svendsen and P. H. Nielsen, Water Res., 2008, 42, 2814–2826 CrossRef CAS PubMed.
- X. Liu, S. Q. Ni, W. Guo, Z. Wang, H. A. Ahmad, B. Gao and X. Fang, RSC Adv., 2018, 8, 24305–24311 RSC.
- H. Duan, L. Ye, X. Lu and Z. Yuan, Environ. Sci. Technol., 2019, 53, 1937–1946 CrossRef CAS PubMed.
- X. Zhang, B. Zhao and X. Yue, Ind. Water Treat., 2019, 39, 30–34 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra05252c |
| This journal is © The Royal Society of Chemistry 2020 |