 Open Access Article
Open Access ArticleOrgan-on-a-chip: the next generation platform for risk assessment of radiobiology
Yi Quan†
a,
Miao Sun†*b,
Zhaoyi Tan*a,
Jan C. T. Eijkelc,
Albert van den Berg c,
Andries van der Meer
c,
Andries van der Meer d and
Yanbo Xie
d and
Yanbo Xie b
b
aInstitute of Nuclear Physics and Chemistry, China Academy of Engineering Physics (CAEP), Mianyang, Sichuan 621000, China. E-mail: tanzhaoyi@caep.cn
bJoint Laboratory of Nanofluidics and Interfaces, School of Physical and Technology, Northwestern Polytechnical University, Xi'an, Shaanxi 710072, China. E-mail: 615sunmiao@163.com
cBIOS, Lab on a Chip Group, MESA+ Institution for Nanotechnology, University of Twente, 7522 NB Enschede, The Netherlands
dDepartment of Applied Stem Cell Technologies, University of Twente, 7522 NB Enschede, The Netherlands
First published on 28th October 2020
Abstract
Organ-on-a-chip devices have been widely used in biomedical science and technology, for example for experimental regenerative medicine and precision healthcare. The main advantage of organ-on-a-chip technology is the facility to build a specific human model that has functional responses on the level of organs or tissues, thereby avoiding the use of animal models, as well as greatly improving new drug discovery processes for personal healthcare. An emerging application domain for organs-on-chips is the study of internal irradiation for humans, which faces the challenges of the lack of a clear model for risk estimation of internal irradiation. We believe that radiobiology studies will benefit from organ-on-a-chip technology by building specific human organ/tissues in vitro. In this paper, we briefly reviewed the state-of-the-art in organ-on-a-chip research in different domains, and conclude with the challenges of radiobiology studies at internal low-dose irradiation. Organ-on-a-chip technology has the potential to significantly improve the radiobiology study as it can mimic the function of human organs or tissues, and here we summarize its potential benefits and possible breakthrough areas, as well as its limitations in internal low-dose radiation studies.
Introduction
Organ-on-a-chip systems are in vitro cell culture models in which the microenvironment of human organs or tissues is mimicked on artificial microstructures built on a microfluidic chip.1 Organ-on-a-chip technology integrates concepts from microfabrication, chemical science, cell biology, medical science and biotechnologies, and is expected to have major applications in biomedical science,2 therapy development,3 drug discovery,4 and regenerative medicine.5 An increasing number of studies have demonstrated that organ-on-a-chip systems can be successfully applied in the aforementioned research scenarios. We believe organ-on-a-chip also has great potential in risk estimation of internal radiobiology studies, as organ-on-a-chip can provide human-specific, tissue-level and organ-level data in vitro. Despite this potential, only a few studies have focused on this application domain, as far as we know.6–9 The exposure to low-dose internal irradiation is an important issue, since it is unavoidable in nature and in the accidents of nuclear plants, as well as in radiation therapy. In this paper, we will briefly review the state-of-the-art in organ-on-a-chip studies, analyze the key points in low-dose internal radiobiology studies, and discuss the possible breakthroughs when applying organs-on-chips in radiobiology in the near future.Organ-on-a-chip
The concept of “organ-on-a-chip” as microfluidic in vitro cell culture systems that emulate the physiology of the key functional units of an organ, was established first by Huh et al., in their publication of a “lung-on-a-chip” in 2010.10 This field was then developing rapidly and organ-on-a-chip is becoming a popular technology for biological research, due to the high expectations of simulating key aspects of the human physiology environment and the potential for replacement of animal models. Applying organs-on-chips in drug development might save years for therapeutic compounds to enter clinical trials, because it would relieve the predominant reliance on animal tests to assess the kinetics, efficacy and safety of drug candidates.11 Besides, culturing human cells on chips enables studies of pharmacokinetics in specific individuals.5 Here, we will briefly summarize the state-of-the-art and possibilities for organ-chip technology in different applications areas (Fig. 1).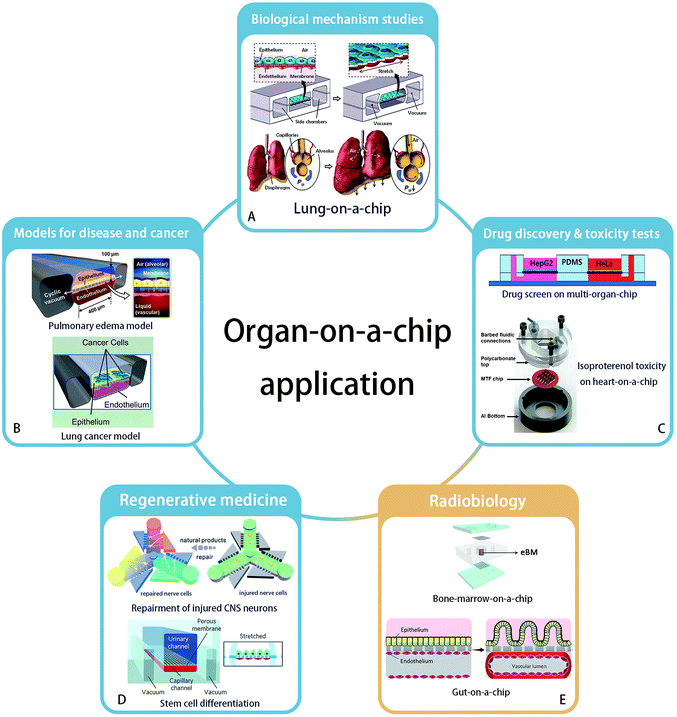 | ||
| Fig. 1 Application areas of organ-on-a-chip technology. (A) Biological mechanism – lung-on-a-chip. Reprinted with permission from ref. 10 (Copyright© 2010, American Association for the Advancement of Science). (B) Disease model – pulmonary edema and lung cancer model – could be established by organ-on-a-chip. Reprinted with permission from ref. 16 (Copyright© 2012, American Association for the Advancement of Science) and ref. 28 (Copyright© 2017, Cell Press) (C) organ-on-a-chip has ability to accomplish new drug discovery and toxicity tests. Reprinted with permission from ref. 37 (Copyright© 2017, John Wiley & Sons) and ref. 31 (Copyright© 2013, Royal Society of Chemistry) (D) regenerative medicine such as neuron recovery and stem cell differentiation. Reprinted with permission from ref. 37 (Copyright© 2018, Royal Society of Chemistry) and ref. 46 (Copyright© 2017, Springer Nature). And (E) the great potential in radiobiology studies is developing. Bone marrow-on-a-chip and gut-on-a-chip has been proved their strength. Drawn refering to ref. 9 and reprinted with permission from ref. 6 (Copyright ©2018, Springer Nature). | ||
Biological mechanism studies
Organ-on-a-chip technology enables studies of the structure and function of a specific organ, as well as the interactions between organs.12 For example, to confirm the regulating ability of Transforming Growth Factor (TGF)-β1 to a four-tissue/organ-system including liver, lung, kidney and adipose tissue, an elementary “human-on-a-chip” was reported by Zhang, et al.,13 providing a possibility to mimic the real human microenvironment in vitro. Huh et al. built the first “organ-on-a-chip”, reconstructing the lung function with cyclic mechanical strains of the alveolar capillary interface, which led to an increased uptake of external substances into epithelial and endothelial cells and transportation of these substances to the microvascular compartment, similar to how uptake takes place in a real lung alveolus.10 A key example of the application of this system was shown by the increased absorption of nanoparticles when the lung alveolus interface was exposed to mechanical strain. Besides the reaction of organs and tissues to the external environment, the interactions between organs also can be studied. Sances et al. proved the auxiliary action of brain microvascular endothelial cells in the maturation of human neurons by co-culturing these two cell types on a chip.14 In addition, a multi-organ-chip system established by Skardal and his team members considered the inter-organ responses, to obtain a more accurate therapeutic reaction.15Models for disease and cancer
Organ-on-a-chip technology offers an excellent platform for disease modeling, like pulmonary edema,16 protein-induced lung inflammation,17 central nervous system disease18 and type 2 diabetes.19 For example, Benam et al. studied chronic obstructive pulmonary disease on a chip and developed the potentials to identify new biomarkers of the disease progression.20 Another example is the application of blood–brain barrier (BBB)-on-chip systems to study the effect of inflammation on barrier function, and how to interfere with this process.21–24As the second largest risk of death with a high global fatality rate,25 tumor and cancer biology are a major area of interest to study in organ-on-a-chip systems. Based on the bioprinting technology, three-dimensional tumor model with spheroids' shape has been established in vitro.26,27 Hassell, et al. recreated a lung-on-a-chip with non-small-cell lung cancer and observed the growth of tumor cells in a controlled microenvironment. They observed clinically relevant processes like tumor dormancy and the response to therapeutical tyrosine kinase inhibitors. Moreover, benefiting from the mechanical movement on the chip, the role of mechanical strain in affecting the sensitivity of cancer to therapeutics was revealed. It was found that signal transduction of epidermal growth factor receptor and MET protein kinase may affect the sensitivity.28 In addition, organ-on-a-chip systems have also been applied in anti-cancer drug screens in a breast ductal carcinoma in situ model,29 as well as in studies on the mechanism of metastasis of breast cancer cells by an in vitro 3D bone-on-a-chip model.30
Drug discovery and toxicity tests
As organ-on-a-chip technology develops, models are increasingly being used for drug screening. A heart-on-a-chip model built by Agarwal et al. recapitulated a curative effect of isoproterenol.31 Besides, an intestine-chip,32 liver-chip,33 kidney-chip34 and many other types of organ-chips were used to test substance toxicity and to screen drugs. Furthermore, a multi-organ-chip platform integrating intestine, liver, skin and kidney was established by Maschmeyer, et al. to test repeated dose systemic toxicity.35 Recently, multi-organ-chips were built to identify anti-angiogenic and anti-tumor drugs,36 and to demonstrate anti-cancer activity of a flavonoid luteolin with integrated liver and cancer tissues.37Regenerative medicine
The reconstruction of organs can be of great use in regenerative medicine, which has a great potential to cure or replace directly damaged tissues and organs without the existing complications of using organ allografts.38 Organ-on-a-chip technology provides a broad platform to understand key aspects of how various fabrication strategies affect cell viability and tissue function in the context of regenerative medicine. For example, a neuron recovery study was carried out on a microfluidic chip before the emergence of the organs-on-chips concept.39 Neurons were also cultured on a 3D chip to assess the response to medicine and the regeneration of injured central nervous system neurons.40 With the help of 3D bioprinting technology,41 a heart-chip model was printed and constructed with endothelialized human myocardium for regenerative medicine to substitute an unhealthy heart.42Organ-on-a-chip systems allow stem cell culture and differentiation, and the targeted differentiation of stem cells has created high expectations on the regenerations of organs.43,44 A microfluidic chip as a primary organ chip was designed by Park et al. to explore the different osteogenic capacity of human bone marrow- and adipose tissue-derived mesenchymal stem cells.45 In another work, Musah et al. realized the differentiation of induced pluripotent stem cells to functional human podocytes, resulting in the regeneration of kidney glomerular-capillary-wall function on-chip.46
Radiobiology
The application of organ-on-chip technology in radiobiology is still a newly developed area. In 2011, a microfluidic chip with liver tissue was used for the first time in this research field by Snyder et al. to confirm the liver tissue injury from space-like radiation and to evaluate treatment effects by the pro-drug amifostine.7 Then Torisawa et al. validated the effects of γ-radiation on the hematopoietic function of bone marrow in a bone marrow-on-a-chip system. This system was used to confirm the beneficial effects of Granulocyte-Colony Stimulating Factor (G-CSF) and bactericidal/permeability-increasing protein (BPI) to recovery from radiation separately in 2014 and 2016.8,9 Very recently, Jalili-Firoozinezhad et al. applied the “gut on chip” for radiobiology studies. Their results indicated that γ-ray radiation exacerbates the generation of reactive oxygen species, cytotoxicity, apoptosis and DNA fragmentation, and damages the tight junctions and integrity of the intestinal barrier, as well as blunts the villus-like morphology. In addition, this gut model proved that dimethyloxaloylglycine (DMOG) may help to prevent the aforementioned damage caused by radiation.6Overall, although the organ-on-a-chip technology has not been widely used in clinical studies, it showed success potentials in studies of disease mechanisms, drug screening and even experimental regeneration studies. Similar to its success in pharmacological studies, we think organs-on-chips will also become of great use in the field of radiobiology, particularly by enabling the development of protective therapies after radiation exposure and by stimulating the optimization of personal radiotherapy.
Review of radiobiology studies
In the 1900s, it took a decade for people to realize the danger of radiation effects on any type of living being including humans, as exemplified by the application of radium in objects for everyday use.47 Nowadays, understanding the bio-effects induced by radiation is still a critical question, although the exposure to radiation in people's lives is limited to specific occasions, such as biomedical diagnosis, treatment and even including the leaking of nuclear power plants.In this paper, we focus on the internal low-dose radiation and we discuss the opportunities of using organs-on-chips in radiobiology studies. Internal low-dose radiation is defined as the radiation from inside of the body when nuclides have been inhaled or ingested, as a consequence of which nuclides can exist in the body for long periods, producing persistent radiation damage to organs. Radiotherapy is another typical way in which radiation nuclides are introduced in the body. On the one side, the release of energetic photons, ions, electrons and the resulting radiolysis of water are helpful to kill cancer cells. On the other side, the side-effects of radiation therapy are clearly being seen, such as inducing salivary gland swelling and pain when the radioactive iodine targets thyroid gland for treating differentiated papillary and follicular cancers.48,49 Still, the radiation doses are moderate or low in these scenarios.
The epidemiological studies demonstrated the deterministic effects of damage and diseases correlated to the dose absorbed. For example, the minimum detectable dose of radiation inducing leukemia and solid cancers are 100 and 200 mSv.50,51 While, under the low dose domain (<100 mSv), the dose–response and disease relationship for doses below 100 mSv are not completely clear yet.52 However, it unavoidable for us to be exposed to natural and artificial sources, like radionuclide in our food supply, medical diagnostics, clinical treatment and industrial radiography. Therefore, it is especially important to study the dose–response relationship of correlated diseases and their relevant biological mechanisms. Here we summarize some unsolved key issues in low-dose radiobiology studies.
Linear non-threshold model of risk estimation
Risk estimation under low-dose radiation is extrapolated from a linear non-threshold (LNT) model, obtained from epidemiological data at doses higher than 100 mSv.52 However, the LNT model encounters challenges at low-dose radiation due to the unclear dose–response relationships, as both adverse effects (“bystander” effects and genomic instability) and beneficial effects (adaptive effects and low dose-hypersensitive) were found to be induced by doses lower than 100 mSv.53–57 Low-dose radiation induces bystander effects, in which the response of cells to radiation is greater than the volume that is actually irradiated; this effect may contribute to increase cancer risk of low dose radiation.57–59 In contrast, radiation hormesis, represented as repair stimulation at low doses and repair inhibition at high doses, is considered to be a beneficial effect.60,61 It includes adaptive protection response in mammalian cells after single or protracted exposures to low doses of X- or γ-radiation. Molecular studies demonstrated the relevant mechanism might be associated with the detection of DNA damage, subsequent transmission of DNA damage signals and activation of DNA repair pathways.62,63 Adaptive effects can keep cells alive through preventing and repairing DNA damage or by removing damaged cells via apoptosis to reduce genomic instability and eliminate mutated cells.64,65The current LNT model used in radiation protection assumes that the cellular response of low-dose radiation is linear with dose and the same as at the high dose. The mechanism of health risks related to low-dose radiation is not completely clear; as we can see several different models have been proposed as candidates for replacing the LNT model as shown in Fig. 2.
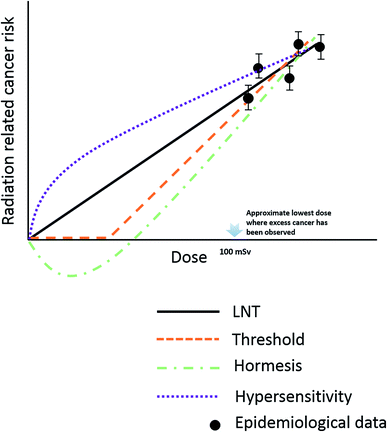 | ||
| Fig. 2 Schematic representation of different model used to estimate radiation risks down to very low dose. | ||
Individual susceptibility
Various individual responses were continuously reported from the first decade of the 20th century,66 which show significant variation in the response to radiotherapy. Individual differences in the radiosensitivity of human skin were found in breast cancer patients receiving radiotherapy because the tumor originated from different defections in genes.67 A genotype dependent genomic instability was triggered by bystander effects after radiation of human lymphocytes.68 Therefore, a biomarker used to distinguish individual radiosensitivity is helpful for optimizing the normal tissue tolerance dose in radiotherapy or an acceptable dose in radiation protection. Although, G2 chromosomal radio-sensitivity assay,69,70 micronucleus (MN) yields71 and DNA misrepair assays72 are commonly used to analyze cancer proneness after radiation exposure, these indicators are also changed when the organism is under other toxicant stress. The integration of ‘-omics’ approaches such as genomics, proteomics, metabolomics may facilitate the goal of understanding the variation between individuals in radio-susceptibility to cancer.73Linear energy transfer difference
Different sources of radiation generate different kinds of particles with different radiation effects. The energy absorption of radiation with low linear energy transfer (LET) such as X-rays or gamma-rays decreases exponentially with the penetration depth. In contrast, most of the high LET radiation deposits energies at the end of penetration depth forming a Bragg peak.74 High LET radiation is likely to produce high ionization along its trajectories and induces complex and irreparable clustered DNA damage.75,76 The foci levels of the phosphorylated histone, gamma-H2AX, a marker of DNA double-stranded breaks (DSB), remain significantly higher in α-irradiated cells than in gamma-irradiated cells,77 since the repair of DSB is inhibited as a function of LET (up to 200 keV μm−1). The cellular response to genetic damage shows differences between high-LET and low-LET radiation, for instance the Activating Transcription Factor 3 (ATF3) was upregulated and remained at a high level only in a group exposed to high-LET irradiation.78 Theoretical studies also point to the same conclusion of the differences between high and low radiation damage to molecules.79 However, the prediction of radiation effects on large scale (such as cells, tissues, organs) and the associated temporal evolution is a major challenge for theoretical studies.Internal exposure
Most of the exposure to low-dose radiation is internal, which may cause serious health problems. As reported, excess of childhood leukaemia was found in the Cumbrian coastal village nearby the Sellafield nuclear installations.80 An increased risk of leukaemia and liver cancer were manifested in patients who were injected with medium Thorotrast containing 232Th.81 Besides the ionizing effects due to the particle emitted from radionuclide, the chemical properties of the specific radionuclide also contribute to the damage effects of organisms. Lanthanum, which has a similar chemical characteristic of the radionuclide actinide, but no radioactivity, can impair memory and learning ability via altering DNA concentration, protein/DNA ratios and inhibition to activities of antioxidant enzymes in the brain.82,83 To acquire the health risk of radionuclide, it is essential to identify the metabolism, translocation of radionuclide between tissues and the underlying mechanism related to its dysfunction after it is absorbed by human body. However, non-human organisms are commonly and directly used as models to study the metabolism of radionuclides. For example, in risk estimation of inhalation of uranium, animal models do present with numbers of cancers per radiation dose that is comparable to the rates in human beings, but they have different preferential cancer occurrence for local sites.84 The difference might be attributed to the different distribution of doses in the lung airways because of the structural differences in the lungs of human versus, for example, rat.85–87 Recently, to overcome the challenge of species differences when using animal models, organs-on-chips have emerged as a new platform to assay radio-biological effects, as we discussed above. Moreover, as the effective doses in tissues are difficult to be measured directly in live organs, it is not easy to build an precise model to estimate the dose–response relationships for internal exposure. Thus, it is critical, but also is a challenge to clearly understand the risk of internal exposure in low-dose radiation-induced bio-effects.Challenges in radiobiology studies
Although the safety of low-dose radiation has been studied for decades, many important questions still remain unanswered. The major challenges in the risk estimation are summarized below, to establish a deep insight into the response of cell, tissue, organ and individuals to ionizing irradiation, based on the current methods of in vitro cellular experiments and in vivo non-human models.Discussions and conclusions
Benefits from organ-on-chip
The emerging of organ-on-a-chip technologies provides new platforms for radiobiology studies, which can be very useful for new drug discovery and might also be a possible solution in studies of the effects of internal exposure with low dose. This latter application domain is possibly even more helpful than the new drug discoveries, since radiation experiments cannot be performed directly in humans, in contrast to the clinical testing of drug candidates. Below we summarize several advantages of using organ-on-chip technology in studying radiobiology. A comparison of features between traditional methods and organ-on-a-chip in radiobiology studies is summarized in Table 1.| Features | Traditional method | Organ-on-a-chip | ||
|---|---|---|---|---|
| 2D culture in vitro | Animal model | |||
| Advantages | Mimicking the micro-environment of tissues and organs | No | Yes | Yes |
| High level of integration and in vivo characterization | ||||
| Internal kinetics and distribution of the radionuclide in tissue and organs | Limited | Limited | Yes | |
| Require less radionuclide materials and generates less contaminated experimental wastes | ||||
| Personal radiosensitivity and radiotherapy | Limited | No | Yes | |
| Dependent on the advance of biological materials used for building tissue in vitro | ||||
| Data availability for human health risk estimation by radiations | Limited. Cellular property varies after long culture in vitro | Limited. Species differences barrier its direct application | Yes | |
| Limitations | Study on chronic diseases induced by internal radionuclide | No | Yes | Limited |
| Investigation on cognitive impairment | No | Yes | No | |
Potential and limitations
As organs-on-chips can provide a unique platform for the study of radiobiology, we now list some key points of the potential impact these systems may have in the research on low-dose radiobiology.The migration and metabolism of radioactive elements in tissues and organs is an important issue that determines where the radionuclide accumulates and acts after ingested by the body. The characterization of the dynamic migration is a challenge, however it can possibly be done by multi-organ-chip and body-on-chip technology. By using biomarkers or other visualizing techniques, it may be possible to perform characterization of the migration of radionuclides based on their temporal distribution in various artificial tissues/organs. A clear model of the metabolism/migration of radionuclides is also crucial to calculate the effective dose in tissues and organs and predict the morbidity risk. The accuracy of mimicking the internal microenvironment will be important, which might directly determine the metabolism of radioactive elements and the resulting bio-effects.
Organs-on-chips enable the culture of primary cells originated from persons or specific group of people, to perform studies of personal damage response. This is not only valuable for understanding the mechanisms of how low-dose radiation induces bio-effects, but could also be used to improve the personal chemotherapies. Judging the level of radiation damage on the chip, and the correlation between cellular responses in the chip to the real organs are technical key points. Biomarkers,10 genomic analysis108 and physiological responses109 can be helpful for the analysis of radiation damage, which can be integrated in the chip.
Organ-on-a-chip technology has found successful application in multiple research fields. We strongly believe it can also provide unique and innovative tools for radiobiology studies, based on its mimicry of human organ functions, its potential for microfluidic integration and the compatibility with multiple endpoint assays, thereby paving the way for the next generation of precision radiobiology studies.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the support from Analytics and Testing Centre in NPU and financial support from NSFC (Grant No. U1730133, U1732143), and the Fundamental Research Funds for the Central Universities (Grant No. 3102017jc01001), and the Presidential Funds of China Academy of Engineering Physics (Grant No. 201501001).References
- Y. Ma, M. Lin, G. Huang, Y. Li, S. Wang, G. Bai, T. J. Lu and F. Xu, Adv. Mater., 2018, 30, 1705911 CrossRef.
- D. E. Ingber, Cell, 2016, 164, 1105–1109 CrossRef CAS.
- C. Luni, E. Serena and N. Elvassore, Curr. Opin. Biotechnol., 2014, 25, 45–50 CrossRef CAS.
- A. Polini, L. Prodanov, N. S. Bhise, V. Manoharan, M. R. Dokmeci and A. Khademhosseini, Expert Opin. Drug Discovery, 2014, 9, 335–352 CrossRef CAS.
- T. Takebe, B. Zhang and M. Radisic, Cell Stem Cell, 2017, 21, 297–300 CrossRef CAS.
- S. Jalili-Firoozinezhad, R. Prantil-Baun, A. Jiang, R. Potla, T. Mammoto, J. C. Weaver, T. C. Ferrante, H. J. Kim, J. M. S. Cabral, O. Levy and D. E. Ingber, Cell Death Dis., 2018, 9, 223 CrossRef.
- J. E. Snyder, Q. Hamid, C. Wang, R. Chang, K. Emami, H. Wu and W. Sun, Biofabrication, 2011, 3, 034112 CrossRef CAS.
- Y. S. Torisawa, C. S. Spina, T. Mammoto, A. Mammoto, J. C. Weaver, T. Tat, J. J. Collins and D. E. Ingber, Nat. Methods, 2014, 11, 663 CrossRef CAS.
- Y. S. Torisawa, T. Mammoto, E. Jiang, A. Jiang, A. Mammoto, A. L. Watters, A. Bahinski and D. E. Ingber, Tissue Eng., Part C, 2016, 22, 509–515 CrossRef.
- D. Huh, B. D. Matthews, A. Mammoto, M. Montoya-Zavala, H. Y. Hsin and D. E. Ingber, Science, 2010, 328, 1662–1668 CrossRef CAS.
- H. Nau, Environ. Health Perspect., 1986, 113–129 CrossRef CAS.
- M. Nikolic, T. Sustersic and N. Filipovic, Front. Bioeng. Biotechnol., 2018, 6, 120 CrossRef.
- C. Zhang, Z. Zhao, N. A. Abdul Rahim, D. van Noort and H. Yu, Lab Chip, 2009, 9, 3185–3192 RSC.
- S. Sances, R. Ho, G. Vatine, D. West, A. Laperle, A. Meyer, M. Godoy, P. S. Kay, B. Mandefro, S. Hatata, C. Hinojosa, N. Wen, D. Sareen, G. A. Hamilton and C. N. Svendsen, Stem Cell Rep., 2018, 10, 1222–1236 CrossRef CAS.
- A. Skardal, S. V. Murphy, M. Devarasetty, I. Mead, H.-W. Kang, Y.-J. Seol, Y. Shrike Zhang, S.-R. Shin, L. Zhao, J. Aleman, A. R. Hall, T. D. Shupe, A. Kleensang, M. R. Dokmeci, S. Jin Lee, J. D. Jackson, J. J. Yoo, T. Hartung, A. Khademhosseini, S. Soker, C. E. Bishop and A. Atala, Sci. Rep., 2017, 7, 8837 CrossRef.
- D. Huh, D. C. Leslie, B. D. Matthews, J. P. Fraser, S. Jurek, G. A. Hamilton, K. S. Thorneloe, M. A. McAlexander and D. E. Ingber, Sci. Transl. Med., 2012, 4, 159ra147 CrossRef.
- T. H. Punde, W. H. Wu, P. C. Lien, Y. L. Chang, P. H. Kuo, M. D. T. Chang, K. Y. Lee, C. D. Huang, H. P. Kuo, Y. F. Chan, P. C. Shih and C. H. Liu, Integr. Biol., 2015, 7, 162–169 CrossRef CAS.
- Y. Yi, J. Park, J. Lim, C. J. Lee and S. H. Lee, Trends Biotechnol., 2015, 33, 762–776 CrossRef CAS.
- S. Bauer, C. Wennberg Huldt, K. P. Kanebratt, I. Durieux, D. Gunne, S. Andersson, L. Ewart, W. G. Haynes, I. Maschmeyer, A. Winter, C. Ämmälä, U. Marx and T. B. Andersson, Sci. Rep., 2017, 7, 14620 CrossRef.
- K. H. Benam, R. Villenave, C. Lucchesi, A. Varone, C. Hubeau, H. H. Lee, S. E. Alves, M. Salmon, T. C. Ferrante, J. C. Weaver, A. Bahinski, G. A. Hamilton and D. E. Ingber, Nat. Methods, 2015, 13, 151 CrossRef.
- Y. B. Arık, M. W. v. d. Helm, M. Odijk, L. I. Segerink, R. Passier, A. v. d. Berg and A. D. v. d. Meer, Biomicrofluidics, 2018, 12, 042218 CrossRef.
- J. A. Brown, S. G. Codreanu, M. Shi, S. D. Sherrod, D. A. Markov, M. D. Neely, C. M. Britt, O. S. Hoilett, R. S. Reiserer, P. C. Samson, L. J. McCawley, D. J. Webb, A. B. Bowman, J. A. McLean and J. P. Wikswo, J. Neuroinflammation, 2016, 13, 306 CrossRef.
- L. M. Griep, F. Wolbers, B. de Wagenaar, P. M. ter Braak, B. B. Weksler, I. A. Romero, P. O. Couraud, I. Vermes, A. D. van der Meer and A. van den Berg, Biomed. Microdevices, 2013, 15, 145–150 CrossRef CAS.
- A. Herland, A. D. van der Meer, E. A. FitzGerald, T.-E. Park, J. J. F. Sleeboom and D. E. Ingber, PLoS One, 2016, 11, e0150360 CrossRef.
- C. Fitzmaurice, C. Allen, R. M. Barber, L. Barregard, Z. A. Bhutta and H. Brenner, JAMA Oncol., 2017, 3, 524–548 CrossRef.
- Y. Fan, N. G. Avci, D. T. Nguyen, A. Dragomir, Y. M. Akay, F. Xu and M. Akay, IEEE J. Transl. Eng. Health Med., 2015, 3, 1–8 Search PubMed.
- K. Ling, G. Huang, J. Liu, X. Zhang, Y. Ma, T. Lu and F. Xu, Engineering, 2015, 1, 269–274 CrossRef CAS.
- B. A. Hassell, G. Goyal, E. Lee, A. Sontheimer-Phelps, O. Levy, C. S. Chen and D. E. Ingber, Cell Rep., 2017, 21, 508–516 CrossRef CAS.
- Y. Choi, E. Hyun, J. Seo, C. Blundell, H. C. Kim, E. Lee, S. H. Lee, A. Moon, W. K. Moon and D. Huh, Lab Chip, 2015, 15, 3350–3357 RSC.
- S. Hao, L. Ha, G. Cheng, Y. Wan, Y. Xia, D. M. Sosnoski, A. M. Mastro and S. Y. Zheng, Small, 2018, 14, 1702787 CrossRef.
- A. Agarwal, J. A. Goss, A. Cho, M. L. McCain and K. K. Parker, Lab Chip, 2013, 13, 3599–3608 RSC.
- A. Bein, W. Shin, S. Jalili-Firoozinezhad, M. H. Park, A. Sontheimer-Phelps, A. Tovaglieri, A. Chalkiadaki, H. J. Kim and D. E. Ingber, Cell. Mol. Gastroenterol. Hepatol., 2018, 5, 659–668 CrossRef.
- E. M. Materne, A. G. Tonevitsky and U. Marx, Lab Chip, 2013, 13, 3481–3495 RSC.
- K. J. Jang, A. P. Mehr, G. A. Hamilton, L. A. McPartlin, S. Chung, K. Y. Suh and D. E. Ingber, Integr. Biol., 2013, 5, 1119–1129 CrossRef CAS.
- I. Maschmeyer, A. K. Lorenz, K. Schimek, T. Hasenberg, A. P. Ramme, J. Hübner, M. Lindner, C. Drewell, S. Bauer, A. Thomas, N. S. Sambo, F. Sonntag, R. Lauster and U. Marx, Lab Chip, 2015, 15, 2688–2699 RSC.
- D. T. T. Phan, X. Wang, B. M. Craver, A. Sobrino, D. Zhao, J. C. Chen, L. Y. N. Lee, S. C. George, A. P. Lee and C. C. W. Hughes, Lab Chip, 2017, 17, 511–520 RSC.
- H. Lee, D. S. Kim, S. K. Ha, I. Choi, J. M. Lee and J. H. Sung, Biotechnol. Bioeng., 2017, 114, 432–443 CrossRef CAS.
- A. S. Mao and D. J. Mooney, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 14452–14459 CrossRef CAS.
- S. X. Guo, F. Bourgeois, T. Chokshi, N. J. Durr, M. A. Hilliard, N. Chronis and A. Ben-Yakar, Nat. Methods, 2008, 5, 531 CrossRef CAS.
- Y. Tang, Q. F. Qiu, F. L. Zhang, M. Xie and W. H. Huang, Lab Chip, 2018, 18, 971–978 RSC.
- G. Zhao, X. Zhang, T. J. Lu and F. Xu, Adv. Funct. Mater., 2015, 25, 5726–5738 CrossRef CAS.
- Y. S. Zhang, A. Arneri, S. Bersini, S.-R. Shin, K. Zhu, Z. Goli-Malekabadi, J. Aleman, C. Colosi, F. Busignani, V. Dell'Erba, C. Bishop, T. Shupe, D. Demarchi, M. Moretti, M. Rasponi, M. R. Dokmeci, A. Atala and A. Khademhosseini, Biomaterials, 2016, 110, 45–59 CrossRef CAS.
- Y. L. Han, S. Wang, X. Zhang, Y. Li, G. Huang, H. Qi, B. Pingguan-Murphy, Y. Li, T. J. Lu and F. Xu, Drug Discovery Today, 2014, 19, 763–773 CrossRef.
- S. Naskar, V. Kumaran and B. Basu, Regener. Eng. Transl. Med., 2019, 5, 99–127 CrossRef CAS.
- S. H. Park, W. Y. Sim, B. H. Min, S. S. Yang, A. Khademhosseini and D. L. Kaplan, PLoS One, 2012, 7, e46689 CrossRef CAS.
- S. Musah, A. Mammoto, T. C. Ferrante, S. S. F. Jeanty, M. Hirano-Kobayashi, T. Mammoto, K. Roberts, S. Chung, R. Novak, M. Ingram, T. Fatanat-Didar, S. Koshy, J. C. Weaver, G. M. Church and D. E. Ingber, Nat. Biomed. Eng., 2017, 1, 0069 CrossRef CAS.
- J. Prisco, Radium Girls: The dark times of luminous watches, https://edition.cnn.com/style/article/radium-girls-radioactive-paint/index.html Search PubMed.
- G. Sunavala-Dossabhoy, Oral Dis., 2018, 24, 198–201 CrossRef CAS.
- C. Alexander, J. B. Bader, A. Schaefer, C. Finke and C. Kirsch, J. Nucl. Med., 1998, 39, 1551–1554 CAS.
- J. D. Mathews, A. V. Forsythe, Z. Brady, M. W. Butler, S. K. Goergen, G. B. Byrnes, G. G. Giles, A. B. Wallace, P. R. Anderson, T. A. Guiver, P. McGale, T. M. Cain, J. G. Dowty, A. C. Bickerstaffe and S. C. Darby, BMJ [Br. Med. J.], 2013, 346, f2360 Search PubMed.
- M. S. Pearce, J. A. Salotti, M. P. Little, K. McHugh, C. Lee, K. P. Kim, N. L. Howe, C. M. Ronckers, P. Rajaraman, A. W. Craft, L. Parker and A. Berrington de González, Lancet, 2012, 380, 499–505 CrossRef.
- J. D. Boice, Int. J. Radiat. Biol., 2017, 93, 1079–1092 CrossRef CAS.
- L. Cai and S. Z. Liu, Int. J. Radiat. Biol., 1990, 58, 187–194 CrossRef CAS.
- M. C. Joiner, B. Marples, P. Lambin, S. C. Short and I. Turesson, Int. J. Radiat. Oncol., Biol., Phys., 2001, 49, 379–389 CrossRef CAS.
- W. F. Morgan, Radiat. Res., 2003, 159, 567–580 CrossRef CAS.
- W. F. Morgan, Radiat. Res., 2003, 159, 581–596 CrossRef CAS.
- C. B. Seymour and C. Mothersill, Nat. Rev. Cancer, 2004, 4, 158 CrossRef.
- S. Bright and M. Kadhim, Int. J. Radiat. Biol., 2018, 94, 727–736 CrossRef CAS.
- W. F. Morgan and M. B. Sowa, Mutat. Res., 2007, 616, 159–164 CrossRef CAS.
- B. R. Scott, J. Cell Commun. Signal., 2014, 8, 341–352 CrossRef.
- E. J. Calabrese and L. A. Baldwin, Annu. Rev. Pharmacol. Toxicol., 2003, 43, 175–197 CrossRef CAS.
- F. R. Tang and W. K. Loke, Int. J. Radiat. Biol., 2015, 91, 13–27 CrossRef CAS.
- C. J. Bakkenist and M. B. Kastan, Nature, 2003, 421, 499 CrossRef CAS.
- G. A. R. Laura Torres Royo, M. Á. Pianetta and M. Arenas Prat, Rep. Pract. Oncol. Radiother., 2020, 25, 250–254 CrossRef.
- L. E. Feinendegen, Br. J. Radiol., 2005, 78, 3–7 CrossRef CAS.
- N. Foray, C. Colin and M. Bourguignon, Radiology, 2012, 264, 627–631 CrossRef.
- S. L. Tucker, I. Turesson and H. D. Thames, Eur. J. Cancer, 1992, 28, 1783–1791 CrossRef.
- R. L. Munira, A. Kadhim, S. R. Moore, D. A. Macdonald, K. L. Chapman, G. Patel and K. M. Prise, Mutat. Res., 2010, 688, 91–94 CrossRef.
- K. Baria, C. Warren, S. A. Roberts, C. M. West and D. Scott, Br. J. Cancer, 2001, 84, 892 CrossRef CAS.
- A. C. Riches, P. E. Bryant, C. M. Steel, A. Gleig, A. J. Robertson, P. E. Preece and A. M. Thompson, Br. J. Cancer, 2001, 85, 1157 CrossRef CAS.
- D. Scott, J. B. P. Barber, E. L. Levine, W. Burrill and S. A. Roberts, Br. J. Cancer, 1998, 77, 614 CrossRef CAS.
- P. A. Jeggo and M. Löbrich, Oncogene, 2007, 26, 7717 CrossRef CAS.
- C. Herskind, C. J. Talbot, S. L. Kerns, M. R. Veldwijk, B. S. Rosenstein and C. M. L. West, Cancer Lett., 2016, 382, 95–109 CrossRef CAS.
- D. Schulz-Ertner, O. Jäkel and W. Schlegel, Semin. Radiat. Oncol., 2006, 16, 249–259 CrossRef.
- S. Timm, Y. Lorat, B. Jakob, G. Taucher-Scholz and C. E. Rübe, Radiother. Oncol., 2018, 129, 600–610 CrossRef CAS.
- M. Hada and A. G. Georgakilas, J. Radiat. Res., 2008, 49, 203–210 CrossRef CAS.
- E. L. Leatherbarrow, J. V. Harper, F. A. Cucinotta and P. O'Neill, Int. J. Radiat. Biol., 2006, 82, 111–118 CrossRef CAS.
- N. Hamada, T. Imaoka, S. i. Masunaga, T. Ogata, R. Okayasu, A. Takahashi, T. A. Kato, Y. Kobayashi, T. Ohnishi, K. Ono, Y. Shimada and T. Teshima, J. Radiat. Res., 2010, 51, 365–383 CrossRef CAS.
- W. Friedland, M. Dingfelder, P. Kundrát and P. Jacob, Mutat. Res., 2011, 711, 28–40 CrossRef CAS.
- W. Richard, J. Labelled Compd. Radiopharm., 2007, 50, 388–391 CrossRef.
- Y. Ishikawa, I. Wada and M. Fukumoto, Int. Congr. Ser., 2002, 1236, 191–194 CrossRef.
- L. Feng, H. Xiao, X. He, Z. Li, F. Li, N. Liu, Z. Chai, Y. Zhao and Z. Zhang, Neurotoxicol. Teratol., 2006, 28, 119–124 CrossRef CAS.
- X. He, Z. Zhang, H. Zhang, Y. Zhao and Z. Chai, Toxicol. Sci., 2008, 103, 354–361 CrossRef CAS.
- R. Winkler-Heil, M. Hussain and W. Hofmann, Radiat. Environ. Biophys., 2015, 54, 225 CrossRef CAS.
- L. Koblinger, Phys. Med. Biol., 1985, 30, 541 CrossRef CAS.
- L. Koblinger and W. Hofmann, J. Aerosol Med., 1995, 8, 21–32 CrossRef.
- L. Koblinger, W. Hofmann, R. C. Graham and R. R. Mercer, J. Aerosol Med., 1995, 8, 7–19 CrossRef.
- L. Zhang, M. P. Schwartz, Z. Hou, Y. Bai, H. Ardalani, S. Swanson, J. Steill, V. Ruotti, A. Elwell, B. K. Nguyen, J. Bolin, R. Stewart, J. A. Thomson and W. L. Murphy, Stem Cell Rep., 2017, 8, 907–918 CrossRef.
- K. L. Schmeichel and M. J. Bissell, J. Cell Sci., 2003, 116, 2377–2388 CrossRef CAS.
- T. Takahashi, T. Ogasawara, Y. Asawa, Y. Mori, E. Uchinuma, T. Takato and K. Hoshi, Tissue Eng., 2007, 13, 1583–1592 CrossRef CAS.
- J. Glowacki, S. Mizuno and J. S. Greenberger, Cell Transplant., 1998, 7, 319–326 CrossRef CAS.
- M. B. Sowa, W. B. Chrisler, K. D. Zens, E. J. Ashjian and L. K. Opresko, Mutat. Res., 2010, 687, 78–83 CrossRef CAS.
- A. Asaithamby, B. Hu, O. Delgado, L.-H. Ding, M. D. Story, J. D. Minna, J. W. Shay and D. J. Chen, Nucleic Acids Res., 2011, 39, 5474–5488 CrossRef CAS.
- E. Behravesh, K. Emami, H. Wu and S. Gonda, Adv. Space Res., 2005, 35, 260–267 CrossRef CAS.
- D. J. Brenner, R. Doll, D. T. Goodhead, E. J. Hall, C. E. Land, J. B. Little, J. H. Lubin, D. L. Preston, R. J. Preston, J. S. Puskin, E. Ron, R. K. Sachs, J. M. Samet, R. B. Setlow and M. Zaider, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 13761–13766 CrossRef CAS.
- A. C. Economos, Physiologist, 1979, 22, S71–S72 CAS.
- J. H. Lin, Drug Metab. Dispos., 1995, 23, 1008–1021 CAS.
- M. Sutton and S. R. Burastero, Chem. Res. Toxicol., 2004, 17, 1468–1480 Search PubMed.
- M. Fouillit, G. Grillon, P. Fritsch, G. Rateau, D. Pavé, J. Delforge and B. Le Gall, Int. J. Radiat. Biol., 2004, 80, 683–689 CrossRef CAS.
- M. Nedergaard, Science, 2013, 340, 1529–1530 CrossRef CAS.
- M. W. Van der Helm, A. D. Van der Meer, J. C. T. Eijkel, A. Van den Berg and L. I. Segerink, Tissue Barriers, 2016, 4, e1142493 CrossRef.
- E. Ron, J. H. Lubin, R. E. Shore, K. Mabuchi, B. Modan, L. M. Pottern, A. B. Schneider, M. A. Tucker and J. J. D. Boice, Radiat. Res., 1995, 141, 259–277 CrossRef CAS.
- M. C. Jansen-van der Weide, M. J. W. Greuter, L. Jansen, J. C. Oosterwijk, R. M. Pijnappel and G. H. de Bock, Eur. Radiol., 2010, 20, 2547–2556 CrossRef.
- T. Mammoto, A. Mammoto, Y. S. Torisawa, T. Tat, A. Gibbs, R. Derda, R. Mannix, M. de Bruijn, C. Yung, D. Huh and D. E. Ingber, Dev. Cell, 2011, 21, 758–769 CrossRef CAS.
- S. L. Irons, V. Serra, D. Bowler, K. Chapman, S. Militi, F. Lyng and M. Kadhim, Int. J. Radiat. Biol., 2012, 88, 735–742 CrossRef CAS.
- M. H. P. Rajaraman, S. Bouffler and A. Wojcik, Ann. ICRP, 2018, 47, 126–141 CrossRef.
- E. Ron, B. Modan, D. Preston, E. Alfandary, M. Stovall and J. J. D. Boice, Radiat. Res., 1989, 120, 516–531 CrossRef CAS.
- G. H. W. Sanders and A. Manz, TrAC, Trends Anal. Chem., 2000, 19, 364–378 CrossRef CAS.
- I. Dalle-Donne, R. Rossi, R. Colombo, D. Giustarini and A. Milzani, Clin. Chem., 2006, 52, 601–623 CrossRef CAS.
Footnote |
| † These authors equally contributed to this work. |
| This journal is © The Royal Society of Chemistry 2020 |
