Open Access Article
Open Access ArticlePotential role of medicinal plants and their constituents in the mitigation of SARS-CoV-2: identifying related therapeutic targets using network pharmacology and molecular docking analyses†
Eman Shawky *,
Ahmed A. Nada and
Reham S. Ibrahim
*,
Ahmed A. Nada and
Reham S. Ibrahim
Department of Pharmacognosy, Faculty of Pharmacy, Alexandria University, Alexandria 21521, Egypt. E-mail: shawkyeman@yahoo.com; eman.m.shawky@alexu.edu.eg; Tel: +20 1005294669
First published on 27th July 2020
Abstract
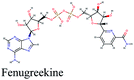
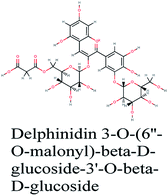
Since the outbreak of Coronavirus disease (COVID-19) caused by SARS-CoV-2 in December 2019, there has been no vaccine or specific antiviral medication for treatment of the infection where supportive care and prevention of complications is the current management strategy. In this work, the potential use of medicinal plants and more than 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 of their constituents was investigated within two suggested therapeutic strategies in the fight against SARS-CoV-2 including prevention of SARS-CoV-2 RNA synthesis and replication, through targeting vital proteins and enzymes as well as modulation of the host's immunity through production of virulence factors. Molecular docking studies on the viral enzymes 3Clpro, PLpro and RdRp suggested rocymosin B, verbascoside, rutin, caftaric acid, luteolin 7-rutinoside, fenugreekine and cyanidin 3-(6′′-malonylglucoside) as promising molecules for further drug development. Meanwhile, the medicinal plants Glycyrrhiza glabra, Hibiscus sabdariffa, Cichorium intybus, Chrysanthemum coronarium, Nigella sativa, Anastatica hierochuntica, Euphorbia species, Psidium guajava and Epilobium hirsutum were enriched in compounds with the multi-targets PTGS2, IL2, IL1b, VCAM1 and TNF such as quercetin, ursolic acid, kaempferol, isorhamnetin, luteolin, glycerrhizin and apigenin. Enriched pathways of the molecular targets included cytokine–cytokine receptor interaction, TNF signaling pathway, NOD-like receptor signaling pathway, Toll-like receptor signaling pathway, NF-kappa B signaling pathway and JAK-STAT3 signaling pathway which are all closely related to inflammatory, innate and adaptive immune responses. The present study identified natural compounds targeting SARS-CoV-2 for further in vitro and in vivo studies and emphasizes the potential role of medicinal plants in the mitigation of SARS-CoV-2.
500 of their constituents was investigated within two suggested therapeutic strategies in the fight against SARS-CoV-2 including prevention of SARS-CoV-2 RNA synthesis and replication, through targeting vital proteins and enzymes as well as modulation of the host's immunity through production of virulence factors. Molecular docking studies on the viral enzymes 3Clpro, PLpro and RdRp suggested rocymosin B, verbascoside, rutin, caftaric acid, luteolin 7-rutinoside, fenugreekine and cyanidin 3-(6′′-malonylglucoside) as promising molecules for further drug development. Meanwhile, the medicinal plants Glycyrrhiza glabra, Hibiscus sabdariffa, Cichorium intybus, Chrysanthemum coronarium, Nigella sativa, Anastatica hierochuntica, Euphorbia species, Psidium guajava and Epilobium hirsutum were enriched in compounds with the multi-targets PTGS2, IL2, IL1b, VCAM1 and TNF such as quercetin, ursolic acid, kaempferol, isorhamnetin, luteolin, glycerrhizin and apigenin. Enriched pathways of the molecular targets included cytokine–cytokine receptor interaction, TNF signaling pathway, NOD-like receptor signaling pathway, Toll-like receptor signaling pathway, NF-kappa B signaling pathway and JAK-STAT3 signaling pathway which are all closely related to inflammatory, innate and adaptive immune responses. The present study identified natural compounds targeting SARS-CoV-2 for further in vitro and in vivo studies and emphasizes the potential role of medicinal plants in the mitigation of SARS-CoV-2.
Introduction
Coronaviruses (CoV) are a family of enveloped viruses containing a large positive-sense single-stranded RNA. They are distributed in a wide range of animals and can infect humans, primarily afflicting the pulmonary system and gastrointestinal tract. Four genera of CoV exist: alpha-, beta, gamma- and delta coronavirus. Alpha- and beta-coronavirus stem from mammals, particularly bats, while gamma- and delta-coronaviruses are of avian origin.1In recent years, the world experienced two pandemics due to CoV. These outbreaks include the severe acute respiratory syndrome coronavirus (SARS-CoV) and the Middle East respiratory syndrome coronavirus (MERS-CoV). During late December of 2019, a cluster of hospital admitted patients were diagnosed with severe pneumonia of unknown etiology. This cluster was epidemiologically linked to a local seafood market in Wuhan, Hubei Province, People's Republic of China.2,3 These patients were found to be infected with the novel coronavirus strain, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2 – previously named 2019-nCoV) which belongs to the beta-coronavirus genera.4
Infection with SARS-CoV-2 can be asymptomatic or gives rise to a symptomatic disease termed Coronavirus Disease 2019 (COVID-19) which varies in severity and may lead to fatality.5 The median incubation period is reported to be 5.1 days and almost all infected symptomatic patients show symptoms in no more than 12 days of infection.6 In cases of mortality, the number of days from symptom onset to death was reported to range from 6 to 41 days with an average of 14 days.7 The most commonly occurring symptoms of COVID-19 include fever, dry cough, and fatigue.8,9 Initially, some patients may present with gastrointestinal upset in the form of diarrhea and nausea prior to onset of fever.9 Other symptoms include dyspnoea, sputum production, headache, haemptyosis10 and lymphopenia.8 The most common complication during hospitalization is pneumonia, preceding acute respiratory distress syndrome (ARDS).8
Another side of SARS-CoV-2 infection is the cytokine storm syndrome which is brought upon the body as a result of the immune system's excessive response. The culprits, cytokines and chemokines are expressed and secreted upon pathogen encounter and although their importance in the body's immunity is well-documented, dysregulation and abundance in their production does more harm than good.11 COVID-19 patients were found to have an elevated level of interleukin 1B (IL-1B), IL-6, IL-10, interferon gamma (IFN-γ), tumor necrosis factor α (TNF-α), interferon gamma-induced protein (IP10), granulocyte colony-stimulating factor (GCSF) and MCP1 (monocyte chemoattractant protein-1). In addition, ICU admitted patients showed higher levels of plasma cytokines compared to their non-ICU admitted counterparts; indicating that the severity of COVID-19 illness is amplified by cytokine levels.10,12 Moreover, serum levels of IL-6, IL-10 and TNF-α in non-ICU admitted patients showed negative correlation with T-cell count. This infers that higher levels of these cytokines elicit a negative regulatory response in the production of T-cell or their survival.12 Hence, inhibition of specific pro-inflammatory cytokines may lessen the hyper-inflammation involved in COVID-19 pathogenesis.13
In patients suffering from COVID-19, SARS-CoV-2 binds to epithelial cells present in the alveoli, thus invoking and activating the body's innate and adaptive immune system. This leads to the production and release of cytokines in order to further stimulate the immune system. As a result, an increase in blood vessel permeability occurs and subsequently, blood cells and fluids seep into the alveoli and this results in dyspnea.14,15 If uncontrolled, a ramification of continued hyper-inflammation and diffuse damage of the alveoli is ARDS.16
Currently, there are no vaccines or specific antiviral medication to treat SARS-CoV-2 infections.10 Therefore, supportive care and prevention of complications is an important management strategy to minimize harm. Suggested therapeutic strategies in the fight against SARS-CoV-2 can be split into 4 different categories: (1) prevention of SARS-CoV-2 RNA synthesis and therefore replication, through action on vital proteins and enzymes, (2) impeding virus-cell receptor binding or suppression of virus self-assembly, (3) stimulation of the host's immunity through production of virulence factors and (4) blocking the entry of the virus into host cells through action on the host's enzymes or cellular entry receptor.17
Nonstructural proteins (Nsp) are functional proteins of high importance to CoV. They take part in the process of virus replication and host infection through their role in RNA transcription and translation and the subsequent processing and modification of synthesized proteins.17 Nsp are produced through proteolytic cleavage of replicase polyprotein 1a (pp1a) and replicase polyprotein 1ab (pp1ab) by the action of viral papain-like protease (PLpro) on the N-termini resulting in 3 products18 and 3-chymotrypsin-likeprotease (3CLpro, also named main protease) at central region and c-termini which gives rise to a minimum of 12 products, 3CLpro included.19 From the aforementioned cleavage, a total of 16 Nsp are produced which are named Nsp1–Nsp16.20 Nsp12 is an RNA-dependent RNA polymerase (RdRp) enzyme that is vital in the lifecycle of CoV as it is responsible for the replication–transcription complex.21,22 However, despite the significant role that RdRp plays in RNA viruses, there is little data on RdRp inhibitors in any CoV. Therefore, it is clear that these key viral enzymes are of great importance to CoV. And owing to their essential active sites, it is hypothesized that small inhibitory molecules are of high therapeutic potential. Other targets include viral spike surface glycoprotein (S) which has a critical role in virus-cell receptor binding. However, this is a target for larger molecules such as lipopeptide EK1C4 as reported by Xia, S. et al.23
For centuries, natural products have been a rich source of medicinally active constituents that have had a pivotal role in treating and preventing an infinite number of diseases. Natural products are a cornerstone of Traditional Chinese Medicine (TCM), which encompasses a tremendous wealth of ancient knowledge. The importance of TCM in emerging pandemics was made evident when it was effectively partnered with conventional medicine in the management of the SARS outbreak in 2003.24 Therefore, due to their notable contributions in the SARS-CoV pandemic, the National Health Commission of the People's Republic of China employed TCM in the COVID-19 treatment guidelines.25 At a press conference held on February 17, an official reported that 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 107 COVID-19 cases (85.2% of total cases at the time) were treated with TCM.26
107 COVID-19 cases (85.2% of total cases at the time) were treated with TCM.26
Additionally, in an investigation covering 23 Chinese provinces, it was reported that the most frequently prescribed Chinese herbs used in the prevention of COVID-19 were (in descending order): Radix astragali, Glycyrrhizae Radix Et Rhizome, Radix Saposhnikoviae, Rhizoma Atractylodis macrocephalae, Lonicerae japonicae Flos, Fructus forsythia, Atractylodis Rhizoma, Radix Platycodonis, Pogostemonis Herba, Cyrtomium fortunei J. Sm.27
Within this context, the potential of the use of medicinal plants in the mitigation of the novel SARS-CoV-2 infection has been investigated. A database comprising more than 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compounds was compiled and docked against the crucial viral proteins; 3-chymotypsin protease (3CLpro), papain-like protease (PLpro) and RNA-dependent RNA polymerase (RdRp) to find potential inhibitors for these enzymes. Furthermore, network pharmacology analysis of all the compounds in the database was attempted to speculate those that can target the inflammatory and immunity-related pathways at the molecular level and revealing those plants with potential multi-components and multi-targets that may help regulate the body functions and possibly will play a therapeutic role in the mitigation of the disease.
000 compounds was compiled and docked against the crucial viral proteins; 3-chymotypsin protease (3CLpro), papain-like protease (PLpro) and RNA-dependent RNA polymerase (RdRp) to find potential inhibitors for these enzymes. Furthermore, network pharmacology analysis of all the compounds in the database was attempted to speculate those that can target the inflammatory and immunity-related pathways at the molecular level and revealing those plants with potential multi-components and multi-targets that may help regulate the body functions and possibly will play a therapeutic role in the mitigation of the disease.
Materials and methods
Molecular docking studies
The Protein Data Bank (PDB) was utilized to retrieve the crystal structure of the three SARS-CoV-2 viral proteins; main proteinase or chymotrypsin-like protease (3CLpro, PDB ID: 5R7Y), papain-like protease (PLpro, PDB ID: 6W9C) and RNA-Dependent RNA Polymerase (RdRp, PDB ID: 6M71). The proteins structures were prepared by means of protein preparation module of Schrodinger's Maestro Molecular modeling suit (Schrödinger Release 2018-1). The proteins were first preprocessed by assigning bond orders and hydrogens in addition to creation of zero order bonds to metals and disulphide bonds, all the water molecules were deleted beyond 5 Å from the active site, hydrogen bonds were assigned using PROPKA at PH = 7 followed by energy minimization using OPLS 3 force field until the relative mean standard deviation (RMSD) of the minimized structure compared to the crystal structure exceeded 0.30 Å.28 Location of the binding site for the docking experiments was determined using the receptor grid generation module and the boxes enclosing the centroids of co-crystallized ligands were set as the grids.The 3D structures of the compounds were imported as SDF file into the LigPrep module of the Maestro molecular modeling package to deliver low energy structures of compounds. Ionization states were set to generate all possible states at pH 7. Molecular docking simulations were performed using Glide docking program of Maestro molecular modeling package implementing SP- and XP-Glide module. Compound–target interactions including hydrogen bond, ion pair interactions, hydrophobic interactions and the binding modes were analyzed in Maestro interface. The docking scores of the top 50 interacting compounds were utilized for construction of another compound–target network using Cytoscape 3.5.1 (http://www.cytoscape.org/) software.
Study design of network pharmacology analysis
The 2D structures of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 compounds from medicinal plants were imported from different resources including NANPDB (http://african-compounds.org/nanpdb) public database, Dictionary of Natural Products (DNP) and two in-house database.28,29 The chemical structures of the compounds were confirmed through ChEMBL (https://www.ebi.ac.uk/chembl/) and PubChem (https://pubchem.ncbi.nlm.nih.gov/). Schrodinger software (2018-1) was used to convert the 2D chemical structures of the compounds to SMILES format. Qikprop software (Schrodinger suite 2018-1) was utilized for drug-likeness study to filter out the compounds by calculation of absorption, distribution, metabolism, and excretion (ADME) criteria and Lipinski's rule of five.
500 compounds from medicinal plants were imported from different resources including NANPDB (http://african-compounds.org/nanpdb) public database, Dictionary of Natural Products (DNP) and two in-house database.28,29 The chemical structures of the compounds were confirmed through ChEMBL (https://www.ebi.ac.uk/chembl/) and PubChem (https://pubchem.ncbi.nlm.nih.gov/). Schrodinger software (2018-1) was used to convert the 2D chemical structures of the compounds to SMILES format. Qikprop software (Schrodinger suite 2018-1) was utilized for drug-likeness study to filter out the compounds by calculation of absorption, distribution, metabolism, and excretion (ADME) criteria and Lipinski's rule of five.
Identifying potential targets/genes of the filtered chemical compounds was achieved through the public database STITCH DB (http://stitch.embl.de/, ver. 5.0) and Swiss Target Predication (http://www.swisstargetprediction.ch) with the ‘Homo sapiens’, while STRING database (https://string-db.org/) was used to draw a protein–protein interaction network (PPI network). The identified targets were further filtered by retaining only those correlated to inflammation or immunity through searches in the UNIPROT and Therapeutic Target Database (TTD).
Information about gene ontology including the highly-associated pathways, biological processes and functional annotation of the target protein was retrieved from Database for Annotation, Visualization and Integrated Discovery (DAVID) ver. 6.8 (https://david.ncifcrf.gov/) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (http://www.genome.jp/kegg/pathway.html). Pathways with P-values < 0.05 were only retained.
The obtained results were employed for construction of Compound–Target, Compound–Target–Pathway and Compound–Target–Pathway-Plant visual networks using Cytoscape software. The network parameters were calculated using the network analyzer plug-in in Cytoscape. The importance of nodes in each network was judged through their combined score of interactions.
Results and discussion
Identifying potential medicinal plants constituents against viral targets
A medicinal plant/natural products library comprised of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 823 phytoconstituents was screened in silico via molecular docking against three proven protein targets in SARS-CoV-2 striving for potent anti-COVID-19 natural compounds. 3-Chymotrypsin-like protease (3CLpro), papain-like protease (PLpro) and RNA-dependent RNA polymerase (RdRp) are elemental for viral replication and therefore, impending targets for anti-coronaviruses screening. Lopinavir, nelfinavir and remdesivir are commonly used antiviral drugs acting on viral proteins that are presently available in the market and undergoing clinical trials for repurposing in treatment of SARS-CoV-2 infection. These drugs were also analyzed against the three selected key protein targets. Meanwhile, other drugs like hydroxychloroquine and dexamathasone don't act on the viral proteins but rather on the immune system reaction caused by the viral infection so they were not involved in the docking study.
823 phytoconstituents was screened in silico via molecular docking against three proven protein targets in SARS-CoV-2 striving for potent anti-COVID-19 natural compounds. 3-Chymotrypsin-like protease (3CLpro), papain-like protease (PLpro) and RNA-dependent RNA polymerase (RdRp) are elemental for viral replication and therefore, impending targets for anti-coronaviruses screening. Lopinavir, nelfinavir and remdesivir are commonly used antiviral drugs acting on viral proteins that are presently available in the market and undergoing clinical trials for repurposing in treatment of SARS-CoV-2 infection. These drugs were also analyzed against the three selected key protein targets. Meanwhile, other drugs like hydroxychloroquine and dexamathasone don't act on the viral proteins but rather on the immune system reaction caused by the viral infection so they were not involved in the docking study.
Molecular docking against 3-chymotrypsin-like protease (3CLpro)
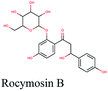
The main protease (Mp) or 3-chymotrypsin-like protease (3CLpro) or Nsp5 has a critical role in cleavage of the polyprotein at eleven distinct sites to generate various Nsps that are essential for viral replication.30 The maturation of Nsps, which is crucial in the virus life cycle is directly mediated via 3CLpro. The detailed inspection on catalytic mechanism of 3CLpro makes it an attractive target for anti-COVID-19 drug development.31Virtual screening of our database using Glide Xp protocol against 3CLpro crystalline structure (5R7Y) deposited in March in complex with co-crystallized ligand disclosed that rutin from Anastatica hierochuntica, Glycyrrhiza glabra and Allium myrianthum followed by rocymosin b from Glycyrrhiza glabra and subsequently verbascoside from Cichorium intybus, Olea europaea and Marrubium vulgare were the top three scoring compounds (Table 1) which endorsed us to investigate their binding modes. The compounds bound to the Cys–His catalytic dyad (Cys-145 and His-41)32 along with the other residues, with notable docking scores (Table 1). It was discerned that the active site of the main protease in which the three top scoring compounds bind enclosed hydrophobic residues such as Met49, Leu141, Cys145, Met165, Leu167 and Pro168 in addition to amino acids polar contributions such as Thr25, Thr26, His41, Asn142, Ser144, His163 and Hie164. Hydrogen bonding with Leu141 and Glu166 also stabilized their conformations (Fig. 1).
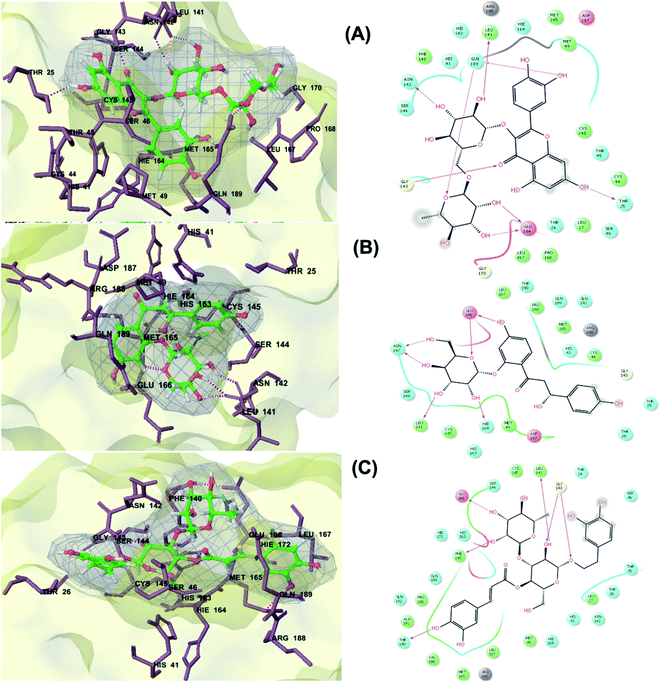 | ||
| Fig. 1 3D (to the left) and 2D (to the right) interaction diagrams of (A) rutin, (B) rocymosin b and (C) verbascoside with the crystal structure of 3CLpro (PDB ID: 5R7Y). | ||
Molecular docking against papain-like protease (PLpro)
PLpro is the proteolytic enzyme that cleaves the N-terminus of viral replicase poly-protein at three sites to release mature Nsp1, 2 and 3. This process is fundamental for correcting virus replication.18 Moreover, PLpro also has a critical role in antagonizing the host's innate immunity.33 As a result of its imperative function in the process of coronavirus replication and host infection, PLpro has emerged as a prevalent target for coronavirus inhibitors. However, no PLpro inhibitor has reached the FDA marketing approval till this moment.After conducting the extra precision Glide docking protocol on our database and the three chosen antiviral repurposed drugs, the interactions between the three top scoring constituents and PLpro crystalline structure (PDB ID: 6W9C) released on first of April 2020, were analyzed (Fig. 2 and S2†). It was observed that the specific pocket in which the top scoring compounds bind contained non-polar amino acids like Leu162, Val202, Met206, Met208 and Ile222 in addition to amino acids polar contributions such as Ser170 and Asn156.
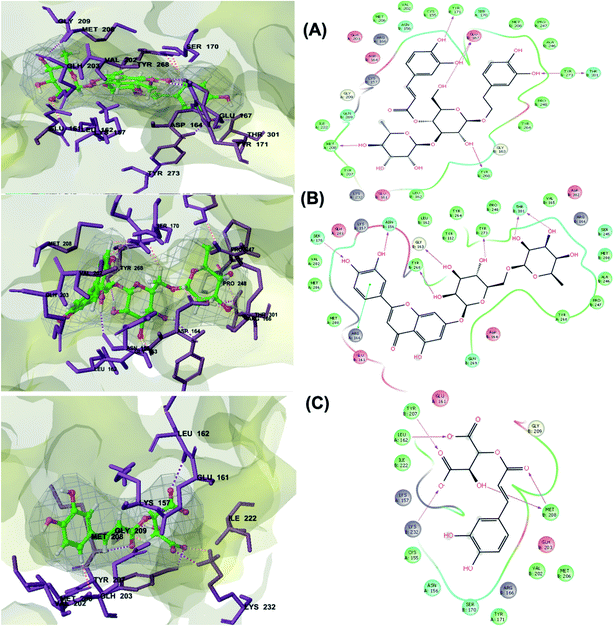 | ||
| Fig. 2 3D (to the left) and 2D (to the right) interaction diagrams of (A) verbascoside, (B) luteolin-7-rutinoside and (C) caftaric acid with the crystal structure of PLpro (PDB ID: 6W9C). | ||
Verbascoside from Cichorium intybus, Olea europaea and Marrubium vulgare which achieved the highest XP docking score (Table 1) was furthermore bound to PLpro active site through hydrogen bonding with Glu167, Tyr171, Met208, Tyr268, Tyr273 and Thr301 and additionally through charged negative interactions with Glh203 and Asp164 (Fig. 2A).
It was also discerned that verbascoside had bound to the two selected proteases PLpro and 3CLpro with high XP docking scores. These dual targets inhibition advocates verbascoside as a potential druggable anti-COVID-19.
The second active top scoring compound against PLpro; luteolin-7-rutinoside from Cynara scolymus (Table 1) interacted with numerous hydrogen bonds with the amino acids Asn156, Gly163, Ser170, Tyr268, Tyr273 and Thr301 as well as π–π stacking interaction with Arg166 and negatively charged interactions with Glh161, Asp164, Glh203 and Asp302 (Fig. 2B).
Analysis of binding modes of caftaric acid; the third top scoring compound, with PLpro revealed that besides the polar and non-polar amino acids contributions in the active site, caftaric acid exhibited four H-bonding with Leu162, Tyr207, Met208 and Lys232 (Fig. 2C).
Molecular docking against RNA-dependent RNA polymerase (RdRp)
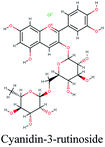
RNA-dependent RNA polymerase (RdRp) or nonstructural protein 12 (Nsp12) is the crucial enzyme for coronavirus replication/transcription complex. It catalyzes the replication of RNA from an RNA template upon binding to cofactors. The RdRp domain of polymerase is located at the C-terminus and possesses a conserved Ser–Asp–Asp motif.33 The crystal structure of SARS-Cov-2 RdRp in complex with cofactors (PDB ID: 6M71) has been recently released in RCSB PDB on the first of April 2020 which provided an excellent ground for structure-based drug discovery.34 Targeted inhibition of RdRp could not cause host cells significant side effects or toxicity.35 Fast drug repurposing attempts have recognized the antiviral remdesivir, a RdRp inhibitor, as a promising anti-COVID-19.36 Clinical trials are currently continuing to investigate the full efficiency spectrum of remdesivir in COVID-19 patients.37Molecular docking studies of the compiled database against RdRp crystal structure (PDB ID: 6M71) revealed that the glycoside “cyanidin 3-(6''-malonylglucoside)” present in Cichorium intybus showed the lowest binding energy with XPG score of −11.541 kcal mol−1. As shown in Fig. 3A, cyanidin 3-(6′′-malonylglucoside) fitted well into RdRp active site. Eight hydrogen bonds were predicted between Lys545, Arg553, Arg555, Tyr619, Asp623, Asp760 and the carbonyl and hydroxyl groups of the compound. In addition, charged negative interactions were also observed with Asp 618, 623, 760 and 761.
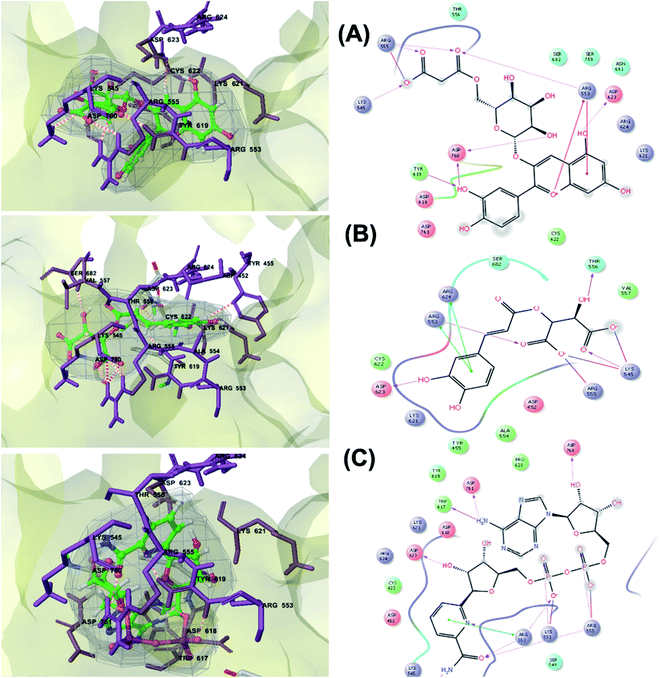 | ||
| Fig. 3 3D (to the left) and 2D (to the right) interaction diagrams of (A) cyanidin 3-(6′′-malonylglucoside), (B) caftaric acid and (C) fenugreekine with the crystal structure of RdRp (PDB ID: 6M71). | ||
The second top scoring compound “caftaric acid”, a constituent of Cichorium intybus, Olea europaea and Marrubium vulgare, showed a dual SARS-CoV-2 target inhibition; PLpro as previously described with RdRp. It possessed a docking score of −10.664 kcal mol−1. The binding modes of its interaction with RdRp are depicted in Fig. 3B, in which many of the hydrophobic amino acid residues such as Tyr455, Ala554, Val557 and Cys622 compose a relatively hydrophobic environment to accommodate the compound. Moreover, π–π stacking interactions with Arg553, Arg624, hydrogen bonding with Lys545, Arg553, Thr556, Asp623 and polar interaction with Ser682 stabilize its conformation within the active pocket.
Fenugreekine (from Trigonella foenum-graecum) was the third scoring compound with binding energy of −9.894 kcal mol−1. Analysis of binding modes (Fig. 3C) revealed numerous hydrogen bonds between fenugreekine and RdRp active site residues such as Trp617, Asp623, Asp760, Asp761, Lys551, Arg553, Arg555 and Thr556 in addition to Van der Waals forces with Val557, Cys622 and Trp619 residues. Polar interaction with Ser549, Thr556 and Ser682 as well as charged negative interactions with Asp542, Asp618, Asp623 and Asp761 were also perceived.
It is noteworthy that these natural top-scored phytoconstituents surpassed in their binding free energies (Table 1) the three selected antiviral drugs adopted from FDA drug repurposing criteria; remdesivir; the proven RdRp inhibitor which is currently in stage III clinical trial, and the anti-HIV drugs; lopinavir and nelfinavir. The 3D and 2D interaction diagrams of the three repurposed anti-COVID-19 drug candidates with RdRp, PLpro and 3CLpro crystalline structures are displayed in the ESI Fig. S1–S3.† The difference in stereochemistry observed in the docked poses is due to the ligand preparation module of Schrodinger suite which generates the chemical structure of the compound at physiological pH which may cause change in ionization or stereochemistry of the compound.
Our analyses revealed that theses top scoring compounds of natural compounds might serve as potential druggable anti- SARS-CoV-2 agents for further in vitro and in vivo studies and for drug repositioning, optimization and development to combat COVID-19.
Multi-target constituents and medicinal plants identification through docking network
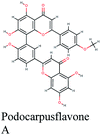
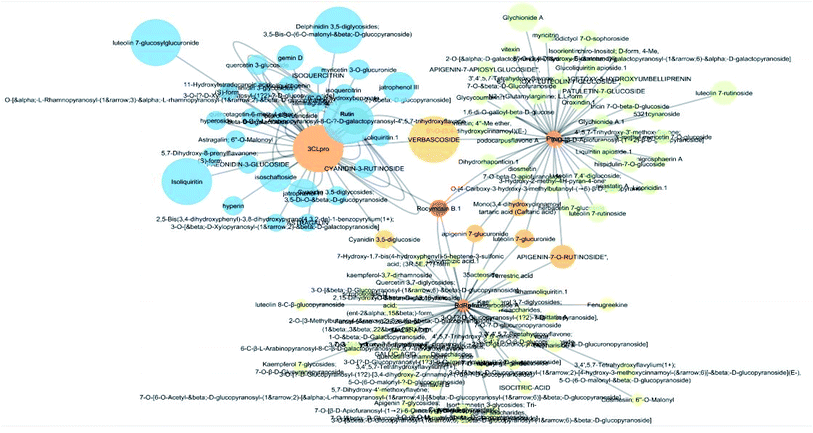
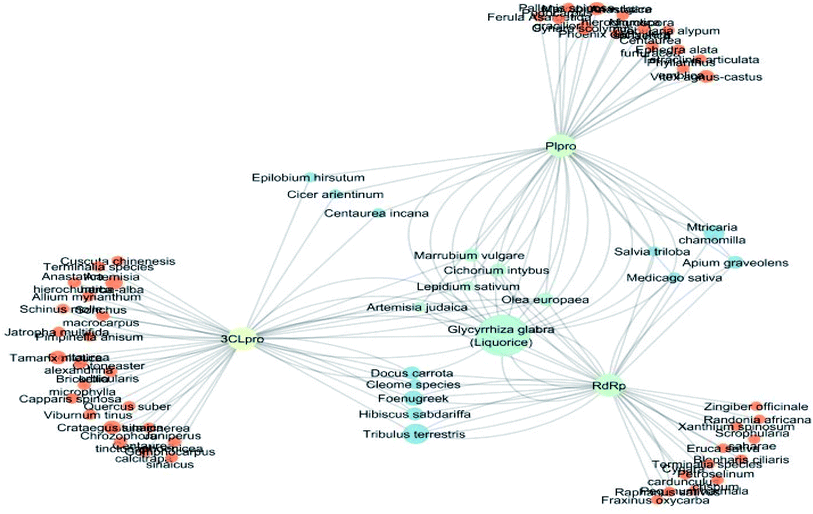
The docking sores of the top 50 compounds against each individual antiviral target protein were exploited for construction of a visual network (Fig. 4). The nodes were sized according to their relative docking scores. As indicated from the size of its node shown in figure, 3CLpro enzyme was the protein target with the lowest docking scores and favorable binding energies followed by PLpro and finally RdRp. It can be observed that rocymosin B (a dihydrochalcone glycoside) was the only compound commonly observed in the top 20 scoring compounds of all three mutual antiviral targets; 3CLpro, PLpro and RdRp with docking scores of −11.844, −10.593 and −9.361, respectively. Meanwhile, verbascoside (acetoside, a caffeoyl phenylethanoid glycoside) exhibited remarkably low binding energy towards both 3-chymotrypsin-like and papain protease enzymes (Fig. 4) with docking scores of −11.721 and −14.041, respectively.Investigating the plant sources of the top 50 scoring compounds in the docking study against the three antiviral protein targets revealed that the plants Glycyrrhiza glabra (liquorice), Cichorium intybus (chicory), Olea europaea (olive leaves), Marrubium vulgare (white horehound), Lepidium sativum (garden cress) and Artemisia judaica (Judean wormwood or Shih Balady) enclosed compounds that are mutually active against the three anti-viral targets (Fig. 5). The node size of Glycyrrhiza glabra (liquorice) indicated that it was enriched in the highest number of compounds possessing favorable binding poses in all three viral proteins, followed by Cichorium intybus, Olea europaea and Marrubium vulgare. Meanwhile Epilobium hirsutum (hairy willow-herb), Cicer arietinum (chickpea) and Centaurea incana (Kantarioun) possessed compounds with mutual targets against 3CLpro and PLpro. On the other hand, Hibiscus sabdariffa (Roselle), Tribulus terrestris, Trigonella foenum-graecum, Cleome species and Daucus carota possessed compounds that are mutually active against the two viral proteins 3CLpro and RdRp. Moreover, a dual PLpro, RdRp target binding was achieved by compounds from Matricaria chamomilla (German chamomile), Apium graveolens (celery), Medicago sativa (alfalfa) and Salvia triloba.
Identifying potential targets of medicinal plants in inflammation and immune regulation pathways
Huge inflammatory response is observed in COVID-19 and in several clinical trials, it has been shown that combination of anti-inflammatory and antiviral drugs may be more effective than each one individually.38 One of the main mechanisms involved in the SARS-CoV-2 induced-ARDS is the cytokine storm, the deadly uncontrolled systemic inflammatory response resulting from the release of large amounts of pro-inflammatory cytokines (IFN-α, IFN-γ, IL-1β, IL-6, IL-12, IL-18, IL-33, TNF-α, TGFβ, etc.). Therefore, arbitration of the immune evasion of SARS-CoV-2 is imperative in its treatment and specific drug development.39–41 Targeting this approach by medicinal plants and their constituents and identifying their potential inflammation and immunity-correlated target proteins and pathways was the focus of this part of the study.About 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 compounds from more than 1300 plant sources were compiled from different database including the North African plants database, Dictionary of Natural products and some in-house available databases. Duplicate compounds were removed and the resulting compounds were filtered using ADME screening based on their oral bioavailability (OB) and Lipinski's rule “rule of five”. Compounds having OB of less than 30% and disobeying two or more of the Lipinski's rule “rule of five” criteria were excluded from further analysis resulting in a database of 10
500 compounds from more than 1300 plant sources were compiled from different database including the North African plants database, Dictionary of Natural products and some in-house available databases. Duplicate compounds were removed and the resulting compounds were filtered using ADME screening based on their oral bioavailability (OB) and Lipinski's rule “rule of five”. Compounds having OB of less than 30% and disobeying two or more of the Lipinski's rule “rule of five” criteria were excluded from further analysis resulting in a database of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 823 compounds.
823 compounds.
Screening the compounds for possible target was performed through the STITCH 5.0 public databases and Swiss Target Prediction database, while the correlation of the identified targets to inflammation and immunity were retrieved from UniProt and TTD.
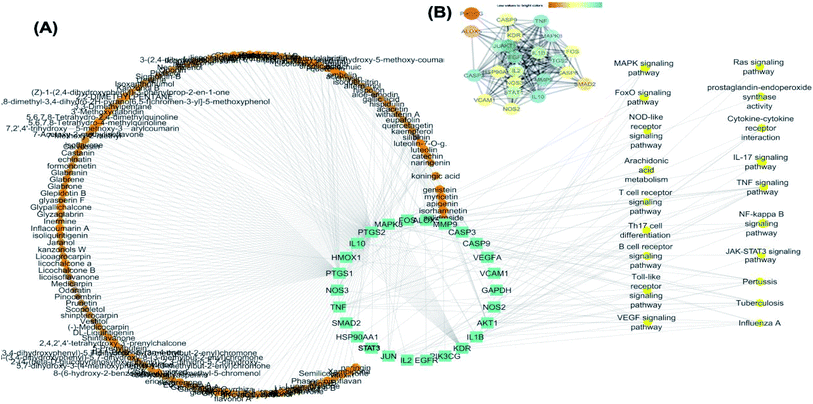
25 common proteins related to inflammatory and immunity pathways were identified as potential targets for 121 plants constituents (Table 2) through 1073 interactions in the constructed compound–target network with an average of 7.2 targets for each constituent (Fig. 6A). Protein correlation results indicated that the identified potential target proteins in the anti-inflammatory effects were correlated to and regulated each other (Fig. 6B).
| Protein | Interacting compound (s) | Involved pathways |
|---|---|---|
| AKT1 | Isorhamnetin, salidroside, apigenin, myricetin, genistein | VEGF signaling pathway, Toll-like receptor signaling pathway, B cell receptor signaling pathway, TNF signaling pathway, tuberculosis, influenza A |
| NOS2 | Isorhamnetin | Pertussis, tuberculosis |
| MAPK8 | Luteolin, quercetagetin, isorhamnetin | Toll-like receptor signaling pathway, NOD-like receptor signaling pathway, RIG-I-like receptor signaling pathway, TNF signaling pathway, pertussis, tuberculosis |
| PTGS2 | Catechin, eupafolin, withaferin A, apigenin, medicocarpin, DL-liquiritigenin, licofuranocoumarin, astragalin, calycosin, formononetin, 6-prenylated eriodictyol | NF-kappa B signaling pathway, VEGF signaling pathway, TNF signaling pathway, prostaglandin-endoperoxide synthase activity |
| ALOX5 | Protocatechuic acid, kaempferol, quercetin | Arachidonic acid metabolism |
| CASP3 | Alternariol, apigenin, kaempferol, salidroside, gallic acid, isoquercitrin, myricetin, chrysin, hispolon | TNF signaling pathway, pertussis, tuberculosis |
| CASP8 | Aloe-emodin | NOD-like receptor signaling pathway |
| CASP9 | Aloe-emodin, alternariol | VEGF signaling pathway, tuberculosis, influenza A |
| FOS | Luteolin | Toll-like receptor signaling pathway, T cell receptor signaling pathway, B cell receptor signaling pathway, TNF signaling pathway, pertussis |
| HSP90AA1 | Aloe-emodin | NOD-like receptor signaling pathway |
| IL1B | Quercetin, ursolic acid, isorhmnetin, | Cytokine–cytokine receptor interaction, NF-kappa B signaling pathway, TNF signaling pathway, MAPK signaling pathway, IL-17 signaling pathway, Th17 cell differentiation, pertussis, influenza A, tuberculosis |
| IL2 | Quercetin, ursolic acid | T cell receptor signaling pathway, Th17 cell differentiation, cytokine–cytokine receptor interaction |
| IL10 | Salidroside | FoxO signaling pathway |
| STAT1 | Acacetin, quercetin | Cytokine–cytokine receptor interaction |
| JUN | Acacetin, gallic acid, kaempferol, luteolin | Toll-like receptor signaling pathway, T cell receptor signaling pathway, B cell receptor signaling pathway, TNF signaling pathway |
| KDR | 1-Methoxyphaseollidin, astragalin, gancaonin B, glycyrin, licochalcone G | Cytokine–cytokine receptor interaction, VEGF signaling pathway, MAPK signaling pathway, ras signaling pathway |
| PTGS2 | Astragalin, calycosin, glyzaglabrin, isoliquiritigenin, isorhamnetin, kaempferol, naringenin, quercetin, ursolic acid | Arachidonic acid metabolism |
| TNF | Glycyrrhizin, kaempferol, naringin, quercetin, ursolic acid | Cytokine–cytokine receptor interaction, NF-kappa B signaling pathway, TNF signaling pathway, MAPK signaling pathway, IL-17 signaling pathway, HTLV-I infection, pertussis, human papillomavirus infection, T cell receptor signaling pathway, influenza A, tuberculosis |
| SMAD2 | Luteolin | FoxO signaling pathway |
| VCAM1 | Isoliquiritigenin, kaempferol, quercetin | NF-kappa B signaling pathway, TNF signaling pathway |
| VEGFA | Acacetin, hispidulin, quercetin, ursolic acid | NF-kappa B signaling pathway, VEGF signaling pathway, MAPK signaling pathway, human papillomavirus infection, PI3K-Akt signaling pathway |
| PIK3CG | Myricetin | TNF signaling pathway |
| NOS3 | Genistein kaempferol, luteolin, luteolin-7-O-g, silibinin | VEGF signaling pathway, HIF-1 signaling pathway |
| GADPH | Koningic acid | HIF-1 signaling pathway |
| MMP9 | Luteolin, gallic acid, hispolon | TNF signaling pathway |
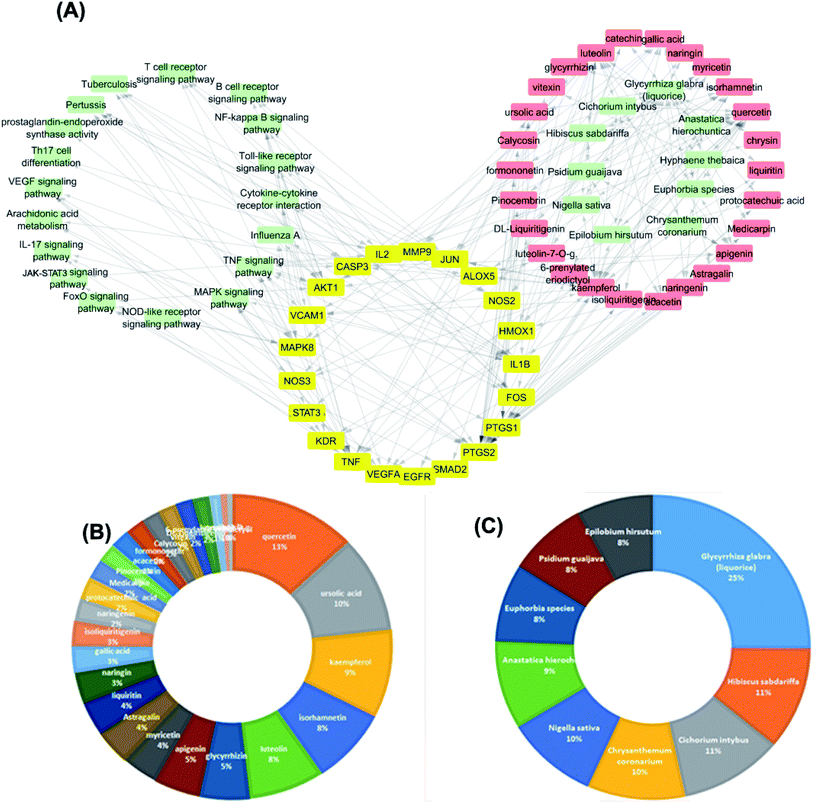
The targets which possessed the highest combined score were presumed to be the most enriched in the network and were revealed to be PTGS2, IL2, IL1b, VCAM1 and TNF. Meanwhile, the constituents with the highest combined score were quercetin, ursolic acid, kaempferol, isorhamnetin, luteolin, glycerrhizin and apigenin (Fig. 7).
PTGS2 (prostaglandin-endoperoxide synthase 2) is a key enzyme in prostaglandin biosynthesis and is responsible for production of inflammatory prostaglandins. It has been recently hypothesized that the lipid mediator prostaglandin E2 may play a crucial role in SARS-CoV-2 infection progress and can be considered a pliable treatment intervention.42
IL2 (Interleukin-2 receptor subunit beta) and ILb (Interleukin-1 beta) are potent pro-inflammatory cytokines which are considered as the major endogenous pyrogen. They induce the synthesis of prostaglandins, neutrophil influx, T-cell and B-cell activation and cytokine production. Meanwhile, TNF (tumor necrosis factor) cytokine is mainly secreted by macrophages and is a potent pyrogen causing fever by direct action or by stimulation of interleukin-1 secretion and impairs regulatory T-cells function. VCAM1 (vascular cell adhesion protein 1) has an important role in leukocyte-endothelial cell adhesion and may play a pathophysiologic role both in immune responses and in leukocyte migration to sites of inflammation. The identified molecular targets are strongly related to the pathophysiology of SARS-CoV-2 infection where it has been shown that most patients with severe SARS-CoV-2 exhibit-noticeably high serum levels of the pro-inflammatory cytokines IL-6 and IL-1β, as well as IL-2, IL-8, IL-17, and TNF, which leads to what is known as the ‘cytokine storm’. It is postulated that small-molecule inhibitors of their downstream signaling components may offer a potential for blocking cytokine storm-related immunopathology.40,43,44
Furthermore, literature review on the most enriched compounds in the compound–target–pathway network indicated their strong correlation to the identified molecular targets in in vitro and in vivo studies. For instance, quercetin was shown to play a modulating, biphasic and regulatory action on inflammation and immunity as it could inhibit lipopolysaccharide (LPS)-induced mRNA levels of TNF-α and interleukin (IL)-1α. Quercetin effectively mitigated rhinovirus-induced progression of lung disease in a mouse model of COPD through prevention of accumulation of neutrophils, macrophages and T cells in the lungs.45 Quercetin inhibited serum necrosis factor α, interleukin 1β, and interleukin 6, and nitric oxide (NO) in LPS-induced acute lung injury,46 and suppressed LPS-induced lung inflammation through a heme oxygenase-1-dependent pathway.47 Similar suggested mechanisms were shown for the flavonoids kaempferol, isorhamnetin, luteolin and apigenin.
Ursolic acid showed inhibitory effect on pulmonary tissues damage in mice through suppression of inflammatory cytokine and oxidative enzymes.48 It reduced inflammatory factors TNF-α, IL-6 and IL-1β secretion in macrophages in response to LPS stimulation.49 β glycyrrhetinic acid, isoliquiritigenin, and ursolic acid inhibited the gene expressions of TNF-α, and iNOS, partly through inhibiting NF-κB expression and attenuating NF-κB nuclear translocation.50
In addition, glycyrrhizin was revealed to have anti-inflammatory and protective effects on LPS-induced ALI in mice through inhibition of pro-inflammatory cytokines playing a key role in the initial phase of inflammatory response of pulmonary inflammation, which suggests that inhibition of the TLR-4/NF-κB signal pathway would be a possible mechanism underlying its action.51
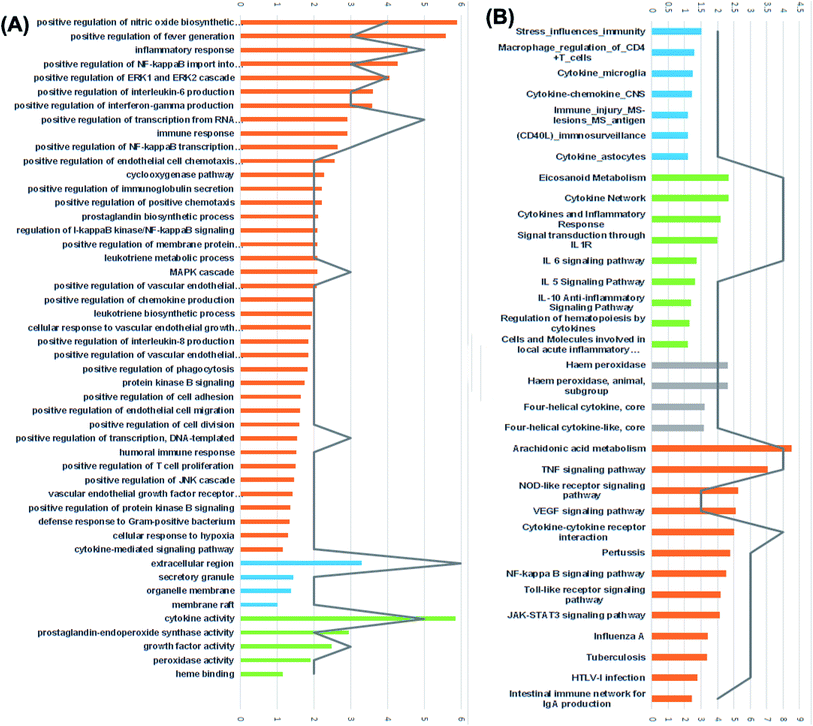
For the identification of the related pathways and GO functional analyses, the identified 26 targets were forwarded to DAVID online database. The obtained results were utilized for the construction of compound–target–pathway network (Table 3, Fig. 8). It could be observed that enriched pathways of the molecular targets were cytokine–cytokine receptor interaction, TNF signaling pathway, NOD-like receptor signaling pathway, Toll-like receptor signaling pathway, NF-kappa B (Nuclear Factor kappa-light-chain-enhancer of activated B cells) signaling pathway, pertussis, JAK-STAT3 signaling pathway, influenza A, tuberculosis, and HTLV-I infection.
| # term ID | Term description | Observed gene count | False discovery rate | Matching proteins in network |
|---|---|---|---|---|
| hsa04668 | TNF signaling pathway | 11 | 1.30 × 10−17 | AKT1, CASP3, CASP8, FOS, IL1B, JUN, MAPK8, MMP9, PTGS2, TNF, VCAM1 |
| hsa04657 | IL-17 signaling pathway | 10 | 2.27 × 10−16 | CASP3, CASP8, FOS, HSP90AA1, IL1B, JUN, MAPK8, MMP9, PTGS2, TNF |
| hsa05161 | Hepatitis B | 10 | 1.08 × 10−14 | AKT1, CASP3, CASP8, CASP9, FOS, JUN, MAPK8, MMP9, STAT1, TNF |
| hsa05152 | Tuberculosis | 10 | 6.08 × 10−14 | AKT1, CASP3, CASP8, CASP9, IL10, IL1B, MAPK8, NOS2, STAT1, TNF |
| hsa04010 | MAPK signaling pathway | 9 | 2.33 × 10−10 | AKT1, CASP3, FOS, IL1B, JUN, KDR, MAPK8, TNF, VEGFA |
| hsa05133 | Pertussis | 8 | 3.76 × 10−13 | CASP3, FOS, IL10, IL1B, JUN, MAPK8, NOS2, TNF |
| hsa04620 | Toll-like receptor signaling pathway | 8 | 3.94 × 10−12 | AKT1, CASP8, FOS, IL1B, JUN, MAPK8, STAT1, TNF |
| hsa04659 | Th17 cell differentiation | 8 | 3.94 × 10−12 | FOS, HSP90AA1, IL1B, IL2, JUN, MAPK8, SMAD2, STAT1 |
| hsa04210 | Apoptosis | 8 | 2.75 × 10−11 | AKT1, CASP3, CASP8, CASP9, FOS, JUN, MAPK8, TNF |
| hsa05168 | Herpes simplex infection | 8 | 2.33 × 10−10 | CASP3, CASP8, FOS, IL1B, JUN, MAPK8, STAT1, TNF |
| hsa05166 | HTLV-I infection | 8 | 2.31 × 10−9 | AKT1, FOS, IL2, JUN, MAPK8, SMAD2, TNF, VCAM1 |
| hsa04151 | PI3K-Akt signaling pathway | 8 | 2.24 × 10−8 | AKT1, CASP9, HSP90AA1, IL2, KDR, NOS3, PIK3CG, VEGFA |
| hsa05321 | Inflammatory bowel disease (IBD) | 7 | 1.14 × 10−11 | IL10, IL1B, IL2, JUN, SMAD2, STAT1, TNF |
| hsa04621 | NOD-like receptor signaling pathway | 7 | 4.91 × 10−9 | CASP8, HSP90AA1, IL1B, JUN, MAPK8, STAT1, TNF |
| hsa05164 | Influenza A | 7 | 5.12 × 10−9 | AKT1, CASP9, IL1B, JUN, MAPK8, STAT1, TNF |
| hsa05165 | Human papillomavirus infection | 7 | 2.72 × 10−7 | AKT1, CASP3, CASP8, PTGS2, STAT1, TNF, VEGFA |
| hsa04370 | VEGF signaling pathway | 6 | 7.66 × 10−10 | AKT1, CASP9, KDR, NOS3, PTGS2, VEGFA |
| hsa04660 | T cell receptor signaling pathway | 6 | 1.09 × 10−8 | AKT1, FOS, IL10, IL2, JUN, TNF |
| hsa04060 | Cytokine–cytokine receptor interaction | 6 | 1.85 × 10−6 | IL10, IL1B, IL2, KDR, TNF, VEGFA |
| hsa04658 | Th1 and Th2 cell differentiation | 5 | 2.91 × 10−7 | FOS, IL2, JUN, MAPK8, STAT1 |
| hsa05144 | Jak-STAT signaling pathway | 4 | 1.28 × 10−6 | AKT1, IL10, IL2, STAT1 |
| hsa04064 | NF-kappa B signaling pathway | 4 | 1.41 × 10−5 | IL1B, PTGS2, TNF, VCAM1 |
| hsa04066 | HIF-1 signaling pathway | 4 | 1.66 × 10−5 | AKT1, NOS2, NOS3, VEGFA |
| hsa04068 | FoxO signaling pathway | 4 | 4.41 × 10−5 | AKT1, IL10, MAPK8, SMAD2 |
| hsa05160 | Hepatitis C | 4 | 4.41 × 10−5 | AKT1, MAPK8, STAT1, TNF |
| hsa04630 | Malaria | 4 | 8.87 × 10−5 | IL10, IL1B, TNF, VCAM1 |
| hsa05169 | Epstein–Barr virus infection | 4 | 0.00018 | AKT1, IL10, JUN, MAPK8 |
| hsa04014 | Ras signaling pathway | 4 | 0.00029 | AKT1, KDR, MAPK8, VEGFA |
| hsa04662 | B cell receptor signaling pathway | 3 | 0.00018 | AKT1, FOS, JUN |
| hsa04920 | Adipocytokine signaling pathway | 3 | 0.00018 | AKT1, MAPK8, TNF |
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | 3 | 0.00024 | AKT1, KDR, VEGFA |
| hsa04611 | Platelet activation | 3 | 0.00078 | AKT1, NOS3, PIK3CG |
| hsa05310 | Asthma | 2 | 0.001 | IL10, TNF |
| hsa04672 | Intestinal immune network for IgA production | 2 | 0.0022 | IL10, IL2 |
| hsa00590 | Arachidonic acid metabolism | 2 | 0.0038 | ALOX5, PTGS2 |
| hsa04750 | Inflammatory mediator regulation of TRP channels | 2 | 0.0078 | IL1B, MAPK8 |
| hsa05322 | Systemic lupus erythematosus | 2 | 0.0081 | IL10, TNF |
| hsa04670 | Leukocyte transendothelial migration | 2 | 0.0111 | MMP9, VCAM1 |
| hsa04062 | Chemokine signaling pathway | 2 | 0.0236 | AKT1, STAT1 |
The identified pathways are all closely related to inflammatory, innate and adaptive immune responses. TNF signaling pathway plays an important role in the regulation of immune cells. It is capable of inhibiting viral replication via IL1 and IL6 producing cells. Meanwhile, Toll-like receptors (TLRs) and Nod-like receptors (NLRs) are important mediators of immune recognition. They initiate innate immune responses as well as activation of adaptive immune responses through identification of pathogen-associated molecular patterns. NOD (nucleotide-binding oligomerization domain)-like receptor signaling pathway is responsible for detecting specific pathogen-associated molecules or host-derived damage signals in the cytosol and initiating the innate immune response. NF-kB signaling pathway is involved in stress responses and is activated by pro-inflammatory cytokines like IL-1b and TNF-a. JAK-STAT3 signaling pathway (Janus kinase (JAK)-signal transducer and activator of transcription) plays critical roles in organizing the immune system, especially cytokine receptors and is involved in the modulation of the polarization of T helper cells.
Most researches have shown that SARS-CoV-2 infection can lead to uncontrolled inflammatory innate responses and impaired adaptive immune responses resulting in harmful tissue damage. Drugs directed against key inflammatory cytokines or other aspects of the innate immune response are considered a potential class of adjunctive therapies for SARS-CoV-2 infection.52,53 Corona virus infection of monocytes and macrophages results in their activation and secretion of IL-6 and other inflammatory cytokines which downstream signal transduction is mediated by JAKs and STAT3.41,54–56
Furthermore, results of GO enrichment analysis (Fig. 8) revealed that these targets are involved in a variety of biological processes, including inflammatory response, positive regulation of fever generation, positive regulation of NF-kappaB import into nucleus, positive regulation of interleukin-6 production, positive regulation of interferon-gamma production, prostaglandin biosynthetic process, leukotriene metabolic process, positive regulation of T cell and B cell proliferation in the cellular components extracellular region, secretory granule, organelle membrane and membrane raft.
The 5 enriched molecular functions of the target proteins were mainly associated with cytokine activity, growth factor activity, prostaglandin-endoperoxide synthase activity and heme binding.
The dissemination of the 1073 compound–target interactions on the plants was then inspected (Table S1†) and the plants were ordered in terms of their combined score of compound–target interactions and a merged plant-constituent-target–pathway network was constructed for the top interacting plants (Fig. 7A). The plants with the highest percentage of C–T interactions were Glycyrrhiza glabra (liquorice), Hibiscus sabdariffa, Cichorium intybus, Chrysanthemum coronarium, Nigella sativa, Anastatica hierochuntica, Euphorbia species, Psidium guajava and Epilobium hirsutum.
The plant Glycyrrhiza glabra (liquorice) has a long history of use for the treatment of several inflammatory disease and its effect has been verified through different in vitro and in vivo models. In one study, Glycyrrhiza glabra suppressed the ability of LPS to induce mRNA and protein expressions of tumor necrosis factor-α (TNF-α), interleukin 1β (IL-1β), and IL-6 productions.57 In another study, Glycyrrhiza glabra displayed preventive and therapeutic effects on pulmonary fibrosis.58 The flavones of liquorice markedly decreased pro-inflammatory cytokines expression levels through modulation of NF-kB/MAPK pathway.59
Meanwhile, Hibiscus sabdariffa was revealed to possess potent anti-inflammatory activity as it maintained the ratio of IL-1β/IL-1ra in the plasma and hippocampus of Wistar rats model.60 In another study, Hibiscus sabdariffa and its isolate delphinidin in a cell model reduced the levels of inflammatory mediators including iNOS, IL-6, and TNF-α induced by LPS and downregulated NF-κB pathway. In animal model, they reduced the production of IL-6, and TNF-α and attenuated mouse paw edema induced by LPS.61
Cichorium intybus L. (chicory) is commonly used for the treatment of inflammatory conditions. Chicory roots demonstrated significant dose-dependent decrease in paw edema in carrageenan-induced paw edema method. Chicory roots diminished the serum TNF-α, IL-6, and IL-1 levels62 and its anti-inflammatory action was shown to be due to a direct modulation of cytokine expression.63 The plant Chrysanthemum coronarium was shown to decrease TNF-α synthesis in the cells of monocyte origin activated with LPS, and decreased cytokine levels in vitro.64
The plant Nigella sativa has a long history of use as an immunomodulatory and anti-inflammatory herb. In an in vitro study, it differently modulated the pro-inflammatory cytokines including Interleukin (IL)-1 alpha, IL-1 beta, IL-6.65 Nigella sativa also is known to possess the potential to modulate adaptive immunity and may play a critical role in modulating the balance of Th1/Th2 lymphocytes, leading to altered Th1/Th2 cytokine profiles.66
Conclusion
Many of the medicinal plants and their constituents have a potential for use in the mitigation of the new SARS-CoV-2 infection. Herein, a database comprised of more than 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 compounds was screened against the three viral targets 3CLpro, PLpro and RdRp, and several constituents identified may inhibit SARS-CoV-2 activity through inhibition of virus replication. A network pharmacology analysis was performed for all the plants constituents and revealed that several compounds possessed multi-targets and enriched pathways of the molecular targets including cytokine–cytokine receptor interaction, TNF signaling pathway, Toll-like receptor signaling pathway, NF-kappa B signaling pathway, and JAK-STAT3 signaling pathway. These results suggest a potential role of medicinal plants in the management of the current SARS-CoV-2 infection. A special emphasis is paid to the plant Glycyrrhiza glabra (liquorice) which has a long history of use in treatment of several viral infections. In fact, randomized controlled trials confirmed that the Glycyrrhiza glabra demonstrated a reduction of mortality and viral activity in SARS-CoV-2 related coronavirus.67 Meanwhile, the plants Hibiscus sabdariffa and Cichorium intybus are rich sources of caffeic acid derivatives which have been widely investigated concerning their antiviral potential.68–70
500 compounds was screened against the three viral targets 3CLpro, PLpro and RdRp, and several constituents identified may inhibit SARS-CoV-2 activity through inhibition of virus replication. A network pharmacology analysis was performed for all the plants constituents and revealed that several compounds possessed multi-targets and enriched pathways of the molecular targets including cytokine–cytokine receptor interaction, TNF signaling pathway, Toll-like receptor signaling pathway, NF-kappa B signaling pathway, and JAK-STAT3 signaling pathway. These results suggest a potential role of medicinal plants in the management of the current SARS-CoV-2 infection. A special emphasis is paid to the plant Glycyrrhiza glabra (liquorice) which has a long history of use in treatment of several viral infections. In fact, randomized controlled trials confirmed that the Glycyrrhiza glabra demonstrated a reduction of mortality and viral activity in SARS-CoV-2 related coronavirus.67 Meanwhile, the plants Hibiscus sabdariffa and Cichorium intybus are rich sources of caffeic acid derivatives which have been widely investigated concerning their antiviral potential.68–70
The performed network pharmacology analysis revealed the synergistic nature of the compounds within each medicinal plant as shown in Fig. 6 and 7. Combination of the most enriched plants in the created network i.e. Glycyrrhiza glabra (liquorice), Hibiscus sabdariffa, Cichorium intybus, could be chosen for mitigation of SARS-CoV-2.
Further molecular dynamic simulation studies, in vitro and in vivo analyses are required to confirm the results of our study, however, the insights provided by the present study may substantiate valuable exploration and development of anti-SARS-CoV-2 therapeutic agents from natural origin.
Conflicts of interest
None.References
- P. C. Y. Woo, S. K. P. Lau, C. S. F. Lam, C. C. Y. Lau, A. K. L. Tsang, J. H. N. Lau, R. Bai, J. L. L. Teng, C. C. C. Tsang, M. Wang, B.-J. Zheng, K.-H. Chan and K.-Y. Yuen, Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavi, J. Virol., 2012, 86, 3995–4008 CrossRef CAS.
- WHO, Novel Coronavirus – China, https://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/ Search PubMed.
- WHO, Pneumonia of unknown cause – China, https://www.who.int/csr/don/05-january-2020-pneumonia-of-unkown-cause-china/en/ Search PubMed.
- N. Zhu, D. Zhang, W. Wang, X. Li, B. Yang, J. Song, X. Zhao, B. Huang, W. Shi, R. Lu, P. Niu, F. Zhan, X. Ma, D. Wang, W. Xu, G. Wu, G. F. Gao and W. Tan, A Novel Coronavirus from Patients with Pneumonia in China, 2019, N. Engl. J. Med., 2020, 382, 727–733 CrossRef CAS.
- J. Yang, Y. Zheng, X. Gou, K. Pu, Z. Chen, Q. Guo, R. Ji, H. Wang, Y. Wang and Y. Zhou, Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis, Int. J. Infect. Dis., 2020, 94, 91–95 CrossRef CAS.
- S. A. Lauer, K. H. Grantz, Q. Bi, F. K. Jones, Q. Zheng, H. R. Meredith, A. S. Azman, N. G. Reich and J. Lessler, The incubation period of coronavirus disease 2019 (CoVID-19) from publicly reported confirmed cases: Estimation and application, Ann. Intern. Med., 2020, 172, 577–582 CrossRef.
- W. Wang, J. Tang and F. Wei, Updated understanding of the outbreak of 2019 novel coronavirus (2019-nCoV) in Wuhan, China, J. Med. Virol., 2020, 92, 441–447 CrossRef CAS.
- W. Guan, Z. Ni, Y. Hu, W. Liang, C. Ou, J. He, L. Liu, H. Shan, C. Lei, D. S. C. Hui, B. Du, L. Li, G. Zeng, K.-Y. Yuen, R. Chen, C. Tang, T. Wang, P. Chen, J. Xiang, S. Li, J. Wang, Z. Liang, Y. Peng, L. Wei, Y. Liu, Y. Hu, P. Peng, J. Wang, J. Liu, Z. Chen, G. Li, Z. Zheng, S. Qiu, J. Luo, C. Ye, S. Zhu and N. Zhong, Clinical characteristics of 2019 novel coronavirus infection in China, medRxiv, 2020, 382, 1708–1720 CAS.
- D. Wang, B. Hu, C. Hu, F. Zhu, X. Liu, J. Zhang, B. Wang, H. Xiang, Z. Cheng, Y. Xiong, Y. Zhao, Y. Li, X. Wang and Z. Peng, Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China, J. Am. Med. Assoc., 2020, 323, 1061–1069 CrossRef CAS.
- C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao, Y. Hu, L. Zhang, G. Fan, J. Xu, X. Gu, Z. Cheng, T. Yu, J. Xia, Y. Wei, W. Wu, X. Xie, W. Yin, H. Li, M. Liu, Y. Xiao, H. Gao, L. Guo, J. Xie, G. Wang, R. Jiang, Z. Gao, Q. Jin, J. Wang and B. Cao, Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China, Lancet, 2020, 395, 497–506 CrossRef CAS.
- P. Mehta, D. F. McAuley, M. Brown, E. Sanchez, R. S. Tattersall and J. J. Manson, Lancet, 2020, 395, 1033–1034 CrossRef CAS.
- B. Diao, C. Wang, Y. Tan, X. Chen, Y. Liu, L. Ning, L. Chen, M. Li, Y. Liu, G. Wang, Z. Yuan, Z. Feng, Y. Wu and Y. Chen, Reduction and Functional Exhaustion of T Cells in Patients with Coronavirus Disease 2019 (COVID-19), medRxiv, 2020, 11, 827 Search PubMed.
- G. Schett, M. Sticherling and M. F. Neurath, COVID-19: risk for cytokine targeting in chronic inflammatory diseases?, Nat. Rev. Immunol., 2020, 20, 271–272 CrossRef CAS.
- K. Brune, J. Frank, A. Schwingshackl, J. Finigan and V. K. Sidhaye, Pulmonary epithelial barrier function: some new players and mechanisms, Am. J. Physiol.: Lung Cell. Mol. Physiol., 2015, 308, L731–L745 CrossRef CAS PubMed.
- M. M. Leiva-Juárez, J. K. Kolls and S. E. Evans, Lung epithelial cells: therapeutically inducible effectors of antimicrobial defense, Mucosal Immunol., 2018, 11, 21–34 CrossRef.
- Z. Xu, L. Shi, Y. Wang, J. Zhang, L. Huang, C. Zhang, S. Liu, P. Zhao, H. Liu, L. Zhu, Y. Tai, C. Bai, T. Gao, J. Song, P. Xia, J. Dong, J. Zhao and F.-S. Wang, Pathological findings of COVID-19 associated with acute respiratory distress syndrome, Lancet Respir. Med., 2020, 8, 420–422 CrossRef CAS PubMed.
- C. Wu, Y. Liu, Y. Yang, P. Zhang, W. Zhong, Y. Wang, Q. Wang, Y. Xu, M. Li, X. Li, M. Zheng, L. Chen and H. Li, Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods, Acta Pharm. Sin. B, 2020, 10, 766–788 CrossRef.
- B. H. Harcourt, D. Jukneliene, A. Kanjanahaluethai, J. Bechill, K. M. Severson, C. M. Smith, P. A. Rota and S. C. Baker, Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity, J. Virol., 2004, 78, 13600–13612 CrossRef CAS.
- J. Ziebuhr, E. J. Snijder and A. E. Gorbalenya, Virus-encoded proteinases and proteolytic processing in the Nidovirales, J. Gen. Virol., 2000, 81, 853–879 CrossRef CAS.
- E. J. Snijder, P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. M. Poon, Y. Guan, M. Rozanov, W. J. M. Spaan and A. E. Gorbalenya, Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage, J. Mol. Biol., 2003, 331, 991–1004 CrossRef CAS.
- M. A. Marra, S. J. M. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. N. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton and R. L. Roper, The Genome Sequence of the SARS-Associated Coronavirus, Science, 2003, 300, 1399–1404 CrossRef CAS PubMed.
- A. O. Adedeji, B. Marchand, A. J. W. Te Velthuis, E. J. Snijder, S. Weiss, R. L. Eoff, K. Singh and S. G. Sarafianos, Mechanism of nucleic acid unwinding by SARS-CoV helicase, PLoS One, 2012, 7, e36521 CrossRef CAS.
- S. Xia, M. Liu, C. Wang, W. Xu, Q. Lan, S. Feng, F. Qi, L. Bao, L. Du, S. Liu, C. Qin, F. Sun, Z. Shi, Y. Zhu, S. Jiang and L. Lu, Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion, Cell Res., 2020, 30, 343–355 CrossRef CAS.
- Z. Chen and T. Nakamura, Statistical evidence for the usefulness of Chinese medicine in the treatment of SARS, Phytother. Res., 2004, 18, 592–594 CrossRef PubMed.
- K. K. Sahu, A. K. Mishra and A. Lal, Comprehensive update on current outbreak of novel coronavirus infection (2019-nCoV), Ann. Transl. Med., 2020, 8, 393 CrossRef.
- D. Zhang, X. Zhang, B. Peng, S. Deng, Y. Wang, L. Yang, K. Zhang, C. Lin and K. Wu, Molecular basis for treating COVID-19 with official Chinese herbal formula LCTE, 2020, DOI:10.21203/rs.3.rs-20828/v1.
- H. Luo, Q.-L. Tang, Y.-X. Shang, S.-B. Liang, M. Yang, N. Robinson and J.-P. Liu, Can Chinese Medicine Be Used for Prevention of Corona Virus Disease 2019 (COVID-19)? A Review of Historical Classics, Research Evidence and Current Prevention Programs, Chin. J. Integr. Med., 2020, 26, 243–250 CrossRef CAS.
- H. M. Dawood, R. S. Ibrahim, E. Shawky, H. M. Hammoda and A. M. Metwally, Integrated in silico–in vitro strategy for screening of some traditional Egyptian plants for human aromatase inhibitors, J. Ethnopharmacol., 2018, 224, 359–372 CrossRef CAS.
- R. S. Ibrahim, R. S. R. Mahrous, H. M. Fathy, A. A. Omar and R. M. Abu EL-Khair, Anticoagulant Activity Screening of an In-House Database of Natural Compounds for Discovering Novel Selective Factor Xa Inhibitors; A Combined In Silico and In Vitro Approach, Med. Chem. Res., 2020, 29, 707–726 CrossRef CAS.
- K. Anand, J. Ziebuhr, P. Wadhwani, J. R. Mesters and R. Hilgenfeld, Coronavirus main proteinase (3CLpro) Structure: Basis for design of anti-SARS drugs, Science, 2003, 300, 1763–1767 CrossRef CAS PubMed.
- T. Pillaiyar, M. Manickam, V. Namasivayam, Y. Hayashi and S. H. Jung, J. Med. Chem., 2016, 59, 6595–6628 CrossRef CAS.
- M. T. ul Qamar, S. M. Alqahtani, M. A. Alamri and L.-L. Chen, Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants, J. Pharm. Anal. DOI:10.1016/j.jpha.2020.03.009.
- L. Yuan, Z. Chen, S. Song, S. Wang, C. Tian, G. Xing, X. Chen, Z. X. Xiao, F. He and L. Zhang, P53 degradation by a coronavirus papain-like protease suppresses type I interferon signaling, J. Biol. Chem., 2015, 290, 3172–3182 CrossRef CAS.
- Y. Gao, L. Yan, Y. Huang, F. Liu, Y. Zhao, L. Cao, T. Wang, Q. Sun, Z. Ming, L. Zhang, J. Ge, L. Zheng, Y. Zhang, H. Wang, Y. Zhu, C. Zhu, T. Hu, T. Hua, B. Zhang, X. Yang, J. Li, H. Yang, Z. Liu, W. Xu, L. W. Guddat, Q. Wang, Z. Lou and Z. Rao, Structure of the RNA-dependent RNA polymerase from COVID-19 virus, Science, 2020, 368, 779–782 CrossRef CAS.
- C. K. Chu, S. Gadthula, X. Chen, H. Choo, S. Olgen, D. L. Barnard and R. W. Sidwell, Antiviral activity of nucleoside analogues against SARS-coronavirus (SARS-CoV), Antiviral Chem. Chemother., 2006, 17, 285–289 CrossRef CAS.
- M. Wang, R. Cao, L. Zhang, X. Yang, J. Liu, M. Xu, Z. Shi, Z. Hu, W. Zhong and G. Xiao, Cell Res., 2020, 30, 269–271 CrossRef CAS.
- Y. Wang, D. Zhang, G. Du, R. Du, J. Zhao, Y. Jin, S. Fu, L. Gao, Z. Cheng, Q. Lu, Y. Hu, G. Luo, K. Wang, Y. Lu, H. Li, S. Wang, S. Ruan, C. Yang, C. Mei, Y. Wang, D. Ding, F. Wu, X. Tang, X. Ye, Y. Ye, B. Liu, J. Yang, W. Yin, A. Wang, G. Fan, F. Zhou, Z. Liu, X. Gu, J. Xu, L. Shang, Y. Zhang, L. Cao, T. Guo, Y. Wan, H. Qin, Y. Jiang, T. Jaki, F. G. Hayden, P. W. Horby, B. Cao and C. Wang, Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial, Lancet, 2020, 395, 1569–1578 CrossRef CAS.
- B. Vellingiri, K. Jayaramayya, M. Iyer, A. Narayanasamy, V. Govindasamy, B. Giridharan, S. Ganesan, A. Venugopal, D. Venkatesan, H. Ganesan, K. Rajagopalan, P. K. S. M. Rahman, S.-G. Cho, N. S. Kumar and M. D. Subramaniam, COVID-19: A promising cure for the global panic, Sci. Total Environ., 2020, 138277 CrossRef CAS.
- N. Vaninov, In the eye of the COVID-19 cytokine storm, Nat. Rev. Immunol., 2020, 20, 277 CrossRef CAS.
- W. Wang, J. He, P. Lie, L. Huang, S. Wu, Y. Lin and X. Liu, The definition and risks of Cytokine Release Syndrome-Like in 11 COVID-19-Infected Pneumonia critically ill patients: Disease Characteristics and Retrospective Analysis, medRxiv, 2020 DOI:10.1101/2020.02.26.20026989.
- P. Conti, G. Ronconi, A. Caraffa, C. Gallenga, R. Ross, I. Frydas and S. Kritas, Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by COVID-19: anti-inflammatory strategies, J. Biol. Regul. Homeostatic Agents, 2020, 34, 11–15 Search PubMed.
- J. Smeitink, X. Jiang, S. Pecheritsyna, H. Renkema, R. Van Maanen and J. Beyrath, Hypothesis: mPGES-1-derived Prostaglandin E2, a so far missing link in COVID-19 pathophysiology?, 2020, DOI:10.20944/preprints202004.0180.v1.
- S. F. Pedersen and Y.-C. Ho, SARS-CoV-2: A Storm is Raging, J. Clin. Invest., 2020, 130, 2202–2205 CrossRef CAS PubMed.
- C. Zhang, Z. Wu, J.-W. Li, H. Zhao and G.-Q. Wang, The cytokine release syndrome (CRS) of severe COVID-19 and Interleukin-6 receptor (IL-6R) antagonist Tocilizumab may be the key to reduce the mortality, Int. J. Antimicrob. Agents, 2020, 105954 CrossRef CAS.
- M. Farazuddin, R. Mishra, Y. Jing, V. Srivastava, A. T. Comstock and U. S. Sajjan, Quercetin prevents rhinovirus-induced progression of lung disease in mice with COPD phenotype, PLoS One, 2018, 13, e0199612 CrossRef.
- L. Wang, J. Chen, B. Wang, D. Wu, H. Li, H. Lu, H. Wu and Y. Chai, Protective effect of quercetin on lipopolysaccharide-induced acute lung injury in mice by inhibiting inflammatory cell influx, Exp. Biol. Med., 2014, 239, 1653–1662 CrossRef.
- K. Takashima, M. Matsushima, K. Hashimoto, H. Nose, M. Sato, N. Hashimoto, Y. Hasegawa and T. Kawabe, Protective effects of intratracheally administered quercetin on lipopolysaccharide-induced acute lung injury, Respir. Res., 2014, 15, 150 CrossRef.
- C. Li, J. Chen, W. Yuan, W. Zhang, H. Chen and H. Tan, Preventive effect of ursolic acid derivative on particulate matter 2.5-induced chronic obstructive pulmonary disease involves suppression of lung inflammation, IUBMB Life, 2020, 72, 632–640 CrossRef CAS.
- J. Zhao, H. Zheng, Z. Sui, F. Jing, X. Quan, W. Zhao and G. Liu, Ursolic acid exhibits anti-inflammatory effects through blocking TLR4-MyD88 pathway mediated by autophagy, Cytokine, 2019, 123, 154726 CrossRef CAS PubMed.
- J.-X. Zhou and M. Wink, Evidence for Anti-Inflammatory Activity of Isoliquiritigenin, 18β Glycyrrhetinic Acid, Ursolic Acid, and the Traditional Chinese Medicine Plants Glycyrrhiza glabra and Eriobotrya japonica, at the Molecular Level, Medicines, 2019, 6, 55 CrossRef CAS PubMed.
- S. A. Lee, S. H. Lee, J. Y. Kim and W. S. Lee, Effects of glycyrrhizin on lipopolysaccharide-induced acute lung injury in a mouse model, J. Thorac. Dis., 2019, 11, 1287–1302 CrossRef PubMed.
- Y. Shi, Y. Wang, C. Shao, J. Huang, J. Gan, X. Huang, E. Bucci, M. Piacentini, G. Ippolito and G. Melino, COVID-19 infection: the perspectives on immune responses, Cell Death Differ., 2020, 27, 1451–1454 CrossRef CAS.
- D. Wu and X. O. Yang, TH17 responses in cytokine storm of COVID-19: An emerging target of JAK2 inhibitor Fedratinib, J. Microbiol., Immunol. Infect., 2020, 53(3), 368–370 CrossRef CAS PubMed.
- D. McGonagle, K. Sharif, A. O'Regan and C. Bridgewood, The Role of Cytokines including Interleukin-6 in COVID-19 induced Pneumonia and Macrophage Activation Syndrome-Like Disease, Autoimmun. Rev., 2020, 102537 CrossRef CAS PubMed.
- B. Hanley, S. B. Lucas, E. Youd, B. Swift and M. Osborn, Autopsy in suspected COVID-19 cases, J. Clin. Pathol., 2020, 73(5), 239–242 CrossRef.
- D. McGonagle, K. Sharif, A. O'Regan and C. Bridgewood, Interleukin-6 use in COVID-19 pneumonia related macrophage activation syndrome, Autoimmun. Rev., 2020, 102537 CrossRef CAS.
- C. Li, T. Eom and Y. Jeong, Glycyrrhiza glabra L. extract inhibits LPS-induced inflammation in RAW macrophages, J. Nutr. Sci. Vitaminol., 2015, 61, 375–381 CrossRef CAS.
- M. Samareh Fekri, H. R. Poursalehi, F. Sharififar, A. Mandegary, F. Rostamzadeh and R. Mahmoodi, The effects of methanolic extract of Glycyrrhiza glabra on the prevention and treatment of bleomycin-induced pulmonary fibrosis in rat: experimental study, Drug Chem. Toxicol. DOI:10.1080/01480545.2019.1606232.
- L. Frattaruolo, G. Carullo, M. Brindisi, S. Mazzotta, L. Bellissimo, V. Rago, R. Curcio, V. Dolce, F. Aiello and A. R. Cappello, Antioxidant and anti-inflammatory activities of flavanones from Glycyrrhiza glabra L. (licorice) leaf phytocomplexes: Identification of licoflavanone as a modulator of NF-kB/MAPK pathway, Antioxidants, 2019, 8, 186 CrossRef CAS.
- M. I. Garbi, S. F. Mohammed, A. A. Magzoub, R. M. Hassabelrasoul, M. S. Saleh, A. M. Badri, I. T. Ibrahim, A. A. Elshikh, A. S. Kabbashi, C. Mohamed and I. Garbi, In vitro anti-inflammatory properties of methanolic extract of Hibiscus sabdariffa flowers, Int. J. Home Sci., 2017, 3, 234–237 Search PubMed.
- T. Sogo, N. Terahara, A. Hisanaga, T. Kumamoto, T. Yamashiro, S. Wu, K. Sakao and D. X. Hou, Anti-inflammatory activity and molecular mechanism of delphinidin 3-sambubioside, a Hibiscus anthocyanin, BioFactors, 2015, 41, 58–65 CrossRef CAS.
- W. Rizvi, M. Fayazuddin, S. Shariq, O. Singh, S. Moin, K. Akhtar and A. Kumar, Anti-inflammatory activity of roots of Cichorium intybus due to its inhibitory effect on various cytokines and antioxidant activity, Anc. Sci. Life, 2014, 34, 44 CrossRef.
- L. Rezagholizadeh, Y. Pourfarjam, A. Nowrouzi, M. Nakhjavani, A. Meysamie, N. Ziamajidi and P. S. Nowrouzi, Effect of Cichorium intybus L. on the expression of hepatic NF-κB and IKKβ and serum TNF-aα in STZ- and STZ+ niacinamide-induced diabetes in rats, Diabetol. Metab. Syndr., 2016, 8, 11 CrossRef PubMed.
- M. Strzelecka, M. Bzowska, J. Kozieł, B. Szuba, O. Dubiel, D. Rivera Nunez, M. Heinrich and J. Bereta, J. Physiol. Pharmacol., 2005, 56, 139–156 Search PubMed.
- L. Bordoni, D. Fedeli, C. Nasuti, F. Maggi, F. Papa, M. Wabitsch, R. De Caterina and R. Gabbianelli, Antioxidant and anti-inflammatory properties of nigella sativa oil in human pre-adipocytes, Antioxidants, 2019, 8, 51 CrossRef CAS.
- A. F. Majdalawieh and M. W. Fayyad, Int. Immunopharmacol., 2015, 28, 295–304 CrossRef CAS.
- C. Fiore, M. Eisenhut, R. Krausse, E. Ragazzi, D. Pellati, D. Armanini and J. Bielenberg, Phyther Res., 2008, 22, 141–148 CrossRef CAS PubMed.
- H. Shen, A. Yamashita, M. Nakakoshi, H. Yokoe, M. Sudo, H. Kasai, T. Tanaka, Y. Fujimoto, M. Ikeda, N. Kato, N. Sakamoto, H. Shindo, S. Maekawa, N. Enomoto, M. Tsubuki and K. Moriishi, Inhibitory effects of caffeic acid phenethyl ester derivatives on replication of hepatitis C virus, PLoS One, 2013, 8, e82299 CrossRef.
- Y. Xie, B. Huang, K. Yu and W. Xu, Further discovery of caffeic acid derivatives as novel influenza neuraminidase inhibitors, Bioorg. Med. Chem., 2013, 21, 7715–7723 CrossRef CAS PubMed.
- Y.-H. Wu, B.-Y. Zhang, L.-P. Qiu, R.-F. Guan, Z.-H. Ye and X.-P. Yu, Structure Properties and Mechanisms of Action of Naturally Originated Phenolic Acids and Their Derivatives against Human Viral Infections, Curr. Med. Chem., 2017, 24, 3995–4008 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra05126h |
| This journal is © The Royal Society of Chemistry 2020 |