Open Access Article
Open Access ArticleBiofilm formation potential and chlorine resistance of typical bacteria isolated from drinking water distribution systems†
Zebing Zhuab,
Lili Shan *a,
Fengping Hu*a,
Zehua Lia,
Dan Zhongb,
Yixing Yuanb and
Jie Zhangb
*a,
Fengping Hu*a,
Zehua Lia,
Dan Zhongb,
Yixing Yuanb and
Jie Zhangb
aSchool of Civil Engineering and Architecture, East China Jiao Tong University, Nanchang, 330013, China. E-mail: hitshanlili@163.com; hufengping1968@126.com
bState Key Laboratory of Urban Water Resource and Environment, School of Environment, Harbin Institute of Technology, Harbin, 150090, China
First published on 24th August 2020
Abstract
Biofilms are the main carrier of microbial communities throughout drinking water distribution systems (DWDSs), and strongly affect the safety of drinking water. Understanding biofilm formation potential and chlorine resistance is necessary for exploring future disinfection strategies and preventing water-borne diseases. This study investigated biofilm formation of five bacterial strains isolated from a simulated DWDS at different incubation times (24 h, 48 h, and 72 h), then evaluated chlorine resistance of 72 h incubated biofilms under chlorine concentrations of 0.3, 0.6, 1, 2, 4, and 10 mg L−1. All five bacterial strains had biofilm formation potential when incubated for 72 h. The biofilm formation potential of Acinetobacter sp. was stronger than that of Bacillus cereus, Microbacterium sp. and Sphingomonas sp. were moderate, and that of Acidovorax sp. was weak. In contrast, the order of chlorine resistance was Bacillus sp. > Sphingomonas sp. > Microbacterium sp. > Acidovorax sp. > Acinetobacter sp. Thus, the chlorine resistance of a single-species biofilm has little relation with the biofilm formation potential. The biofilm biomass is not a major factor affecting chlorine resistance. Moreover, the chlorine resistance of a single-species biofilm is highly related to the physiological state of bacterial cells, such as their ability to form spores or secrete extracellular polymeric substances, which could reduce the sensitivity of the single-species biofilm to a disinfectant or otherwise protect the biofilm.
Introduction
Characterizing microbial communities and ensuring the microbiological safety of drinking water is a major public health challenge. In drinking water distribution systems (DWDSs), microorganisms can present in the bulk water (planktonic bacteria) and along pipe walls (loose deposits and biofilm bacteria).1,2 Most of the bacterial biomass is present in biofilms, in which active bacteria account for about 95% of all bacteria.3,4 Biofilms formed in DWDSs can consume disinfectant and increase the bacterial resistance to disinfection, resulting in bacterial regrowth leading to color, turbidity, odor, and corrosion problems, higher pathogen concentrations and outbreaks of water-borne diseases.4,5 Generally, the disinfection strategies for drinking water are based on disinfecting bulk water samples; however, this is an insufficient basis to inhibit the microbial contamination caused by biofilms in a DWDS.6 Given that microorganisms in a biofilm will resuspend into the bulk water,7 an effective strategy for monitoring and controlling biofilm development in DWDSs is highly desirable.In recent years, complex microbial communities have been increasingly found in DWDSs by high-throughput genetic sequencing. The phyla associated with the Proteobacteria (α-, β- and γ-Proteobacteria), Actinobacteria, Firmicutes, and Bacteroidetes are ubiquitous in DWDSs, and the genera Sphingomonas, Pseudomonas, Acidovorax, Brevundimonas, Acinetobacter, Methylobacterium, Microbacterium, and Bacillus also are ubiquitous in DWDSs.8–12 Such microbial communities can survive in extreme conditions in DWDSs.13 In a DWDS, growth of bacteria in a biofilm helps reduce the sensitivity of bacterial cells and helps them survive in oligotrophic conditions and in the presence of high chlorine concentration.14,15
In an actual DWDS, the biofilms are usually consisted of complex communities.16 Biofilm studies in situ are not common due to the complex systems in DWDSs and difficult accessibility.16 Typical approaches for establishing multispecies biofilm communities to utilize strains isolated from similar environments and assumed to coexist in the same environment are applied in vitro studies. Generally, these large-scale studies focus on the overall diversity but often lack of the single-species microhabitat and spatial information, which are driving factors for microbial interaction and diversity maintaining.17 Simplified single-species biofilms are important tools to understand the contributions of individual species to community functions in a variety of environments.18 Sun et al.19 and Zhang et al.20 found that Sphingomonas sp. survived in an extremely high concentration of chlorine, and that some were sterilized at a residual chlorine concentration of 4 mg L−1 and contact time of 240 min with only approximately 5% of viability reduction.19 Szabo et al.21 found that the log reduction of Bacillus sp. was just 1.4-log at a 25 mg L−1 free chlorine (pH 7) for 44 h. There are many similar in-depth studies of biofilms in DWDSs, but the results vary from study to study. It is difficult to compare the formation potential and chlorine resistance of different biofilms cultured in vitro under different conditions because they are frequently influenced by their environments, such as nutrient conditions, incubation time, hydraulic conditions, biofilm structure, and spatial location.22–24 Therefore, more accurate results can be provided by simultaneously compare multiple bacteria isolated from similar DWDSs.
Biofilm studies in situ are not common due to the complex systems in DWDSs and difficult accessibility.
Hence, the objective of this study was to characterize the contributions of individual species to biofilm formation in DWDS using five simplified single-species isolated from a previously simulated DWDS.25 The five species (Sphingomonas sp., Acidovorax defluvii, Acinetobacter sp., Bacillus cereus, and Microbacterium laevaniformans) are common and abundant in drinking water systems worldwide8–12 and could be considered to represent simplified model bacteria to study the contributions of individual species to biofilm formation in the drinking water environment.18 Biofilm studies in situ are not common16 while the in vitro biofilm formation assays performed in 96-well microtiter plates are common in biofilm studies.16,26 A single-species-biofilm-based model was introduced to assess formation potential and chlorine resistance of different biofilms cultured in vitro under same conditions in this study. The single-species biofilm formation was investigated at different incubation times (24 h, 48 h, and 72 h). Then the 72 h incubated biofilms were treated with different chlorine concentrations (0.3, 0.6, 1, 2, 4, and 10 mg L−1), the inactivation efficiency of the different bacteria was analysed,27,28 and the biological mechanism of bacteria with strong chlorine resistance was further studied. As there is still in a phase of reinforcing previous findings in the study of single-species biofilms of bacteria found in DWDSs, the results of the present study could provide valuable insights on the ability in initial biofilm formation and chlorine resistance.
Materials and methods
Bacterial strains
Five bacterial strains were isolated from a previous simulated DWDS with cast iron and 304 stainless steel materials, and the biofilms incubated for 3 months at chlorine concentrations of 1 mg L−1, 3 mg L−1 and 4.2 mg L−1.25 Table S1† summarizes the basic information about the five tested bacterial strains: Sphingomonas sp., Acidovorax defluvii, Acinetobacter sp., Bacillus cereus, and Microbacterium laevaniformans. The bacteria affiliated with Proteobacteria were Gram-negative (G−), while the bacteria affiliated with Firmicutes and Actinobacteridae were Gram-positive (G+). All sequences of tested bacterial strains were deposited in Genbank of the National Center for Biotechnology Information (NCBI) under accession numbers MT279967 to MT279971. Scanning electron microscope images of the bacterial strains are shown in Fig. S1.† All bacteria were rod shaped and differed only in morphology (short, long, straight and irregular).Biofilm formation
Biofilm formation was conducted using the microtiter plate method and modified.29 R2A broth (Teknova, CA, USA) was used as growth medium. R2A broth, which is a low nutrient growth medium with a high content of nutrients, was widely used to incubate bacteria presented in drinking water.2,18,27,30 Briefly, a volume of 200 μL of a standardized bacterial suspension (1.0 × 106 cells per mL in R2A broth) in mid-log phase was revived in biofilm-inducing media and dispensed in a sterile 96-well microtiter plate (Corning Incorporated, USA). R2A broth without bacterium was used as the control. The plates were incubated for 24, 48 and 72 h in a shaking incubator at 150 rpm at 25 °C with capping. Fresh R2A broth was gently exchanged at a clean bench every 24 h during the incubation period. For biofilm sampling, R2A broth was carefully removed from plates with a pipette, and then rinsed three times with 250 μL sterilized phosphate buffered saline (PBS, pH 7.0) to remove non-adherent and loosely attached planktonic bacteria in each well. The plates were air-dried at a clean bench for 30 minutes for further experiment and analyses.Biofilm chlorination experiment
Generally, biofilm formation could reach a relatively stable state after 72 h inoculation in microtiter plates.16,26,31 To understand the chlorine resistance of the five different bacteria, the 72 h incubated biofilms were used in chlorination experiments. The chlorination experiments were performed in 250 μL PBS (pH 7.0) free from chlorine. Briefly, NaClO (Sinopharm Chemical Reagent Co., Ltd, Shanghai, China) was diluted with PBS to create a working solution with chlorine concentrations of 0.3 mg L−1, 0.6 mg L−1, 1 mg L−1, 2 mg L−1, 4 mg L−1 and 10 mg L−1, which were calibrated using a residual chlorine meter (PC II, HACH, USA). The negative control comprised of only 250 μL of PBS, and the positive control was each species of biofilm cultured in 250 μL of PBS without chlorine. Biofilm chlorination experiments were conducted in a shaking incubator at 150 rpm at 25 °C to ensure sufficient contact between the working solution and biofilm. Fresh working solution was gently changed every 20 min during the chlorination period of 1 h. Wells were rinsed two times with 250 μL 0.5% (w/v) NaS2O3 to stop the chlorination reaction, and then rinsed one time with sterilized PBS to remove non-adherent and loosely attached planktonic bacteria in each well. The plates were then air-dried at a clean bench for 30 minutes for further analyses.Analytical methods
The crystal violet (CV) assays have been commonly suggested as a method for the estimation of biomass quantity.26,29,31 Biofilm biomass was determined by the method described by Stepanovic29 and modified. Briefly, each biofilm was fixed with 250 μL 98% methanol (v/v) for 15 minutes, then removed and air-dried. The fixed biofilm was stained for 5 min with 250 μL of CV. The dyed biofilm was then rinsed with sterilized distilled water to remove excess CV and air-dried. Following washing, 250 μL 33% glacial acetic acid (v/v) was used to release and dissolve epibiotic CV, then 100 μL aliquots were transferred to a fresh microtiter plate for absorbance measurement at 570 nm (OD570) using a microtiter plate reader (Model-680, Bio-Rad, Hercules, CA, USA). Each biofilm was analyzed in triplicate.Biofilm activity was determined using the method described by Simoes32 and modified. Briefly, 200 μL XTT (Sigma, USA) and menadione (Sigma, USA) was added to each biofilm, confirming a final concentration of 50 μg mL−1 of XTT–menadione. The plates were incubated on a shaker at 150 rpm at 25 °C for 3 h in dark conditions. Then, 100 μL of supernatant was absorbed into a new microporous plate, and the OD value was determined at 490 nm (OD490). Each biofilm was analyzed in triplicate. For the low value of OD490, the activity of a biofilm was described as the specific respiratory activity of the biofilm using OD490/570 (OD490/OD570, OD490 is biofilm activity while OD570 is biofilm biomass).
To better compare chlorine resistance of the different bacteria, the inactivation rate of single-species biofilm for 60 min was investigated at different chlorine concentrations in the present study. The 72 h incubated biofilms were used for bacterial counts, which were measured by counting heterotrophic bacteria on plates of R2A solid medium (HPC-R2A). The determination process involved adding 200 μL sterilized distilled water to each well, and then resuspending the attached biofilm using a sterilized toothpick. To detach bacteria from the attached surface (suspended solids, loose deposits, and pipe specimens), the biofilm samples were placed in an ultrasonic ice water bath for 5 min (oscillated for 1 minute and letting stand for 1 minute, repeated three times).3,4,16,33 Then the suspension was oscillated in a whirlpool for 30 seconds to assure that the bacteria from the biofilm were evenly distributed in the suspension. This diluted suspension with R2A solid medium was incubated and counted. Each biofilm was analyzed in triplicate.
Biofilm formation potential
The biofilm formation potential is mainly determined based on the OD570 value of the biofilm biomass. The determination method described by Stepanovic et al.29 was used as follows: no biofilm formation potential (0): OD570 ≤ ODc; weak biofilm formation potential (+): ODc < OD570 ≤ 2 ODc; medium biofilm formation potential (++): 2 ODc < OD570 ≤ 4 ODc; and strong biofilm formation potential (+++): OD570 > 4 ODc. This classification was based upon the cut-off of the optical density (ODc) value defined as three standard deviation values above the mean OD of the negative control. The negative control is culture R2A broth without bacteria. As long as it is measured on the microtiter plate reader, there will be a certain value, which may be very low.Efficiency and mechanism of chlorination
The inactivation efficiency of the five different bacteria was calculated with the following three parameters: biofilm biomass reduction, biofilm activity reduction and logarithmic reduction of biofilm.27,28
 | (1) |
 | (2) |
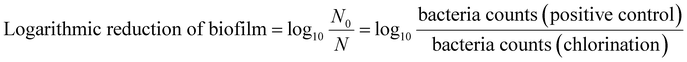 | (3) |
The biofilm biomass reduction and the biofilm activity reduction of the biofilms were defined as 100% when the absorbance was less than that of the negative control. The logarithmic reduction of a biofilm was expressed as the base-10 logarithm of bacteria count of the positive control (log10(N0)), when its bacteria count was lower than the detection limit of the R2A plate count (5 CFU mL−1). The lower biofilm biomass reduction or the lower logarithmic reduction shows the stronger resistance to chlorine.
The spore counts of each biofilm were measured using a modified Szabo34 method. Briefly, the fully mixed suspension was incubated for 10 minutes at 80 °C to kill the vegetative cells of the biofilm. The resulting suspension was then subjected to HPC-R2A plate counting, which defined the spore count. The spore proportion of each biofilm was counted before and after treatment at 80 °C.
Statistical analysis
The statistical analysis was applied by the method described by Vaz-Moreira35 and modified. Data of biofilm biomass, biofilm activity, and HPC-R2A at different incubation time were compared using one-way or two-way analysis of variance (ANOVA) and a post hoc Tukey test. The relationship between biofilm biomass and HPC-R2A in 72 h incubated biofilms was assessed based on a Pearson correlation analysis. Statistical calculations were based on a confidence level ≥ 95%, assuming a significance level for the separation set at p < 0.05. These analysis were supported by SPSS software 16.0 for Windows.Results
Biofilm biomass and biofilm formation potential of single-species
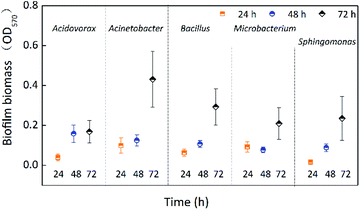
The single-species biofilm formation was investigated free from chlorine at different incubation times (24 h, 48 h, and 72 h). Fig. 1 shows the biofilm biomass of five species incubated for 24 h, 48 h, and 72 h. The biofilm biomass of all five species was significantly (p < 0.05) affected by incubation time (Table 1), and all biofilms obtained the largest biomass at 72 h. Except for Microbacterium laevaniformans, the biofilm biomass gradually increased as a function of incubation time. The biofilm biomass of Acinetobacter sp., Bacillus cereus, and Microbacterium laevaniformans at 72 h was significantly (p < 0.05) larger than at 24 h and 48 h. The biofilm biomass of Acidovorax defluvii at 72 h was significantly (p < 0.05) larger than at 24 h, while the biofilm biomass at 48 h was not significantly (p > 0.05) different from that at 24 h. Acinetobacter sp. had a larger biofilm biomass at 24 h and 72 h than the other four species of biofilms, and it was significantly (p < 0.05) higher than that of Sphingomonas sp. at 24 h, but not significantly (p > 0.05) different from the other three species of biofilm at 24 h. Acidovorax defluvii had the largest biofilm biomass at 48 h, and it was significantly (p < 0.05) larger than others at 48 h. Interestingly, the biofilm biomass of Acidovorax defluvii was the smallest of the five biofilms at 72 h. This might be due to the biofilm maturity of Acidovorax defluvii at 48 h. The order of biofilm biomass at 24 h was Acinetobacter sp. > Microbacterium laevaniformans > Bacillus cereus > Acidovorax defluvii > Sphingomonas sp. The order of biofilm biomass at 48 h was Acidovorax defluvii > Acinetobacter sp. > Bacillus cereus > Sphingomonas sp. > Microbacterium laevaniformans. The order of biofilm biomass at 72 h was Acinetobacter sp. > Bacillus cereus > Sphingomonas sp. > Microbacterium laevaniformans > Acidovorax defluvii.| Parameters | Effect | DFa | F valueb | P-Value |
|---|---|---|---|---|
| a Degree of freedom.b Test for comparing model with residual (error) variance.c sSignificant at P < 0.05.d nsNot significant at P > 0.05. | ||||
| Biofilm biomass | A – Bacteria | 4 | 4.409 | 0.006sc |
| B – Incubation time | 2 | 46.442 | <0.001sc | |
| AB | 8 | 2.885 | 0.017sc | |
| Specific respiratory activity | A | 4 | 10.937 | <0.001sc |
| B | 2 | 4.772 | 0.057nsd | |
| AB | 8 | 2.064 | 0.072nsd | |
| Bacterial counts (HPC) | A | 4 | 10.850 | 0.001sc |
| Biofilm biomass reduction | A | 4 | 26.342 | <0.001sc |
| C – Chlorine | 5 | 47.035 | <0.001sc | |
| AC | 20 | 1.735 | 0.052nsd | |
| Biofilm activity reduction | A | 4 | 18.884 | <0.001sc |
| C | 5 | 112.121 | <0.001sc | |
| AC | 20 | 5.580 | <0.001sc | |
| Inactivation rate | A | 4 | 70.582 | <0.001sc |
| C | 5 | 141.615 | <0.001sc | |
| AC | 20 | 8.363 | <0.001sc | |
| Spore | C | 6 | 11.147 | <0.001sc |
| Vegetative cell + spore | C | 6 | 11.147 | <0.001sc |
Table 2 indicates the biofilm formation potential of the five species as a function of incubation time. The biofilm formation potential can be divided into four types: no potential, weak potential, medium potential, and strong potential.29 As presented in Table 2, the biofilm formation potential gradually increased as a function of incubation time. Acinetobacter sp. and Microbacterium laevaniformans showed weak biofilm formation potential when incubated for 24 h, while the other species exhibited no biofilm formation potential. Except for Microbacterium laevaniformans, the biofilm formation potential of the other four species became apparent only when incubated for 48 h. All five tested species had biofilm formation potential when incubated for 72 h; Acinetobacter sp. especially showed strong biofilm formation potential.
| Bacteria | Incubation time (h) | ||
|---|---|---|---|
| 24 | 48 | 72 | |
| a “0” shows non-biofilm forming abilities; “+” shows the biofilm forming abilities: weak biofilm formation potential (+); medium biofilm formation potential (++); strong biofilm formation potential (+++). | |||
| Acidovorax defluvii | 0 | + | + |
| Acinetobacter sp. | + | + | +++ |
| Bacillus cereus | 0 | + | ++ |
| Microbacterium laevaniformans | + | 0 | ++ |
| Sphingomonas sp. | 0 | + | ++ |
Specific respiratory activity of single-species biofilm
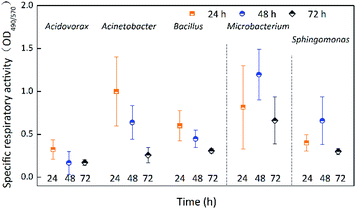
For the low values of OD490 (most values < 0.09) (Fig. S2†), the activity of a biofilm was described as the specific respiratory activity of biofilm using OD490/570. Fig. 2 shows the specific respiratory activity of the five species incubated for 24 h, 48 h, and 72 h. Except for Acinetobacter sp., the specific respiratory activity of the biofilms was not significantly (p > 0.05) affected by incubation time (Table 1). The specific respiratory activity of Acinetobacter sp. at 72 h was significantly (p < 0.05) lower than at 24 h and 48 h. The specific respiratory activity of Acidovorax defluvii was the lowest at 48 h, while the other four species had the lowest specific respiratory activity at 72 h. The specific respiratory activity of Microbacterium laevaniformans and Sphingomonas sp. increased first and then decreased as incubation time increased, while that of Acinetobacter sp. and Bacillus cereus steadily decreased. In the initial stage of incubation (24 h), the specific respiratory activity of all species of biofilm was not significantly (p > 0.05) different, but were significantly (p < 0.05) different at incubation times of 48 h and 72 h. The specific respiratory activity of Microbacterium laevaniformans at 48 h and 72 h was significantly (p < 0.05) higher than that of the other four species, for which the specific respiratory activity was not significantly (p > 0.05) different. The order of specific respiratory activity of the biofilms at 24 h was Acinetobacter sp. > Microbacterium laevaniformans > Bacillus cereus > Sphingomonas sp. > Acidovorax defluvii. The order of specific respiratory activity of biofilms at 48 h was Microbacterium laevaniformans > Sphingomonas sp. > Acinetobacter sp. > Bacillus cereus > Acidovorax defluvii. The order of specific respiratory activity of biofilms at 72 h was Microbacterium laevaniformans > Bacillus cereus > Sphingomonas sp. > Acinetobacter sp. > Acidovorax defluvii.Bacterial counts of single-species biofilm
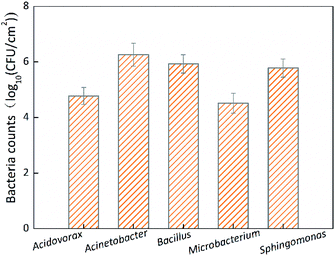
Only the bacterial counts of biofilms incubated for 72 h were determined due to the huge workload required in plate counting. Fig. 3 shows the HPC of the single-species biofilms incubated for 72 h. The HPC of Acinetobacter sp. was 1.81 × 106 CFU cm−2, which was significantly (p < 0.05) higher than that of the other four species of biofilms, while the HPCs of the other four species of biofilms were <9 × 105 CFU cm−2 and were not significantly (p > 0.05) different. The order of HPCs at 72 h was Acinetobacter sp. > Bacillus cereus > Sphingomonas sp. > Acidovorax defluvii > Microbacterium laevaniformans. Among the HPCs, that of Acinetobacter sp. was the largest and was about 55 times higher than the smallest HPC (Microbacterium laevaniformans).Biofilm biomass reduction of single-species
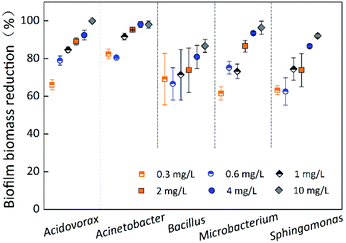
The contact time for chlorination experiments was 60 min at different chlorine concentrations (0.3 mg L−1, 0.6 mg L−1, 1 mg L−1, 2 mg L−1, 4 mg L−1, and 10 mg L−1). Fig. 4 indicates the removal of single-species biofilm biomass at different chlorine concentrations. The biofilm biomass reduction for all five species was significantly (p < 0.05) affected by the chlorine concentration (Table 1). Microbacterium laevaniformans had the strongest chlorine resistance at chlorine concentration of 0.3 mg L−1, followed by Sphingomonas sp. The chlorine resistance of Sphingomonas sp. and Bacillus cereus gradually stronger than the other three bacteria with increasing chlorine concentration; at 2 mg L−1 of chlorine and higher the chlorine resistance of these two species of biofilms was significantly (p < 0.05) higher than that of the other three species of biofilms. Bacillus cereus had relatively strong chlorine resistance because the biofilm biomass reduction was not significantly (p > 0.05) affected by chlorine concentration, while the resistance of the other four species of biofilms was significantly (p < 0.05) different. Among the four species of biofilms (excluding Bacillus cereus), the removal of Acidovorax defluvii and Acinetobacter sp. biofilm biomass was not significantly (p > 0.05) different at chlorine concentrations of 2 mg L−1 and higher, but were significantly (p < 0.05) different at chlorine concentration of 1 mg L−1 and lower. The removal of Sphingomonas sp. and Microbacterium laevaniformans biofilm biomass was significantly (p < 0.05) different at chlorine concentrations of 2 mg L−1 and 4 mg L−1.At chlorine concentration of 10 mg L−1, the removal of Bacillus cereus biofilm biomass was less than 87%, while removal of biomass for all other species exceeded 92%. Because the removal of Acinetobacter sp. biofilm biomass was the largest of the five species under the different chlorine concentrations evaluated, it was likely that Acinetobacter sp. was highly sensitive to chlorine. Acidovorax defluvii was also sensitive to chlorine at concentrations higher than 0.6 mg L−1. In general, the order of chlorine resistance was Bacillus cereus > Sphingomonas sp. > Microbacterium laevaniformans > Acidovorax defluvii > Acinetobacter sp. As described in Fig. 4 and S2† and Table 3, the biofilm biomass was not significantly (p > 0.05) affected by chlorine resistance and bacteria species (biofilm biomass reduction) (Table 1). For example, Acinetobacter sp. had the largest biofilm biomass and bacterial cell counts (Table 3), but was very highly sensitive to chlorine.
| Bacteria | Biofilm biomass (OD570 ± SD) | Biofilm activity (OD490 ± SD) | Bacterial counts (log10 CFU cm−2 ± SD) |
|---|---|---|---|
| Acidovorax defluvii | 0.169 ± 0.055 | 0.029 ± 0.012 | 4.72 ± 0.21 |
| Acinetobacter sp. | 0.431 ± 0.141 | 0.103 ± 0.015 | 6.23 ± 0.39 |
| Bacillus cereus | 0.293 ± 0.090 | 0.089 ± 0.019 | 5.90 ± 0.27 |
| Microbacterium laevaniformans | 0.209 ± 0.080 | 0.124 ± 0.023 | 4.49 ± 0.28 |
| Sphingomonas sp. | 0.236 ± 0.110 | 0.068 ± 0.024 | 5.75 ± 0.20 |
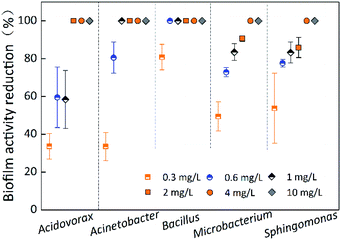
Biofilm activity reduction of single-species
Fig. 5 indicates the biofilm activity reduction of single-species biofilms under different chlorine concentrations at a contact time of 60 min. The biofilm activity reduction for all five species was 100% at chlorine concentrations of 4 mg L−1 and 10 mg L−1. The OD490 of most of the single-species biofilms was less than that of the control, and the OD490 of all five species of biofilms was less than 0.125 before the chlorination experiments began (Table 3). Unfortunately, biofilm activity determined by the XTT–menadione method was limited due to low values, low discrimination ability and large errors. Therefore, use of the XTT–menadione method to determine the biofilm activity reduction during chlorination experiments cannot be recommended.Inactivation rate of single-species biofilm
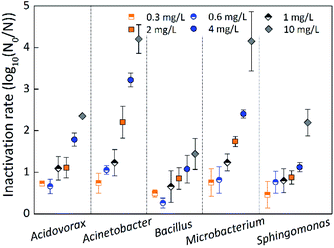
The inactivation rate was measured by the logarithmic reduction log10(N0/N), where N0 and N represent the number of bacteria before and after inactivation.33,36 Fig. 6 indicates the inactivation rate of single-species biofilms under different chlorine concentrations at a contact time of 60 min. The inactivation rate of Bacillus cereus was slightly greater than that of Sphingomonas sp. at the low chlorine concentration of 0.3 mg L−1, while Bacillus cereus was the least inactivated at all other chlorine concentrations, suggesting that Bacillus cereus had a strong resistance to chlorine. The inactivation rate of Microbacterium laevaniformans was greatest at chlorine concentrations of 0.3 mg L−1 and 1 mg L−1, but inactivation rate of Acinetobacter sp. was greatest at the other chlorine concentrations, suggesting that these two species of biofilms were sensitive to chlorine.The inactivation rate of the single-species biofilms was not significantly (p > 0.05) different at low chlorine concentrations of 0.3 mg L−1 and 1 mg L−1, and inactivation rate of Bacillus cereus was significantly (p < 0.05) less than that of the other four species of biofilms at chlorine concentration of 0.6 mg L−1. When the chlorine concentration increased to 2 mg L−1 or higher, the inactivation rate of the five species of biofilms was significantly (p < 0.05) different. Bacillus cereus and Sphingomonas sp. showed the least inactivation rate, and were significantly (p < 0.05) different from the other three species of biofilms. In contrast, at chlorine concentrations of 2 mg L−1 or higher, Microbacterium laevaniformans and Acinetobacter sp. were the most inactivated, and the extents of inactivation rate were significantly (p < 0.05) greater than that of the other three species of biofilms.
The inactivation rate of single-species biofilms was not significantly (p > 0.05) different at chlorine concentrations of 1 mg L−1 or lower. When the chlorine concentration increased from 1 mg L−1 to 10 mg L−1, the inactivation rate of Acinetobacter sp. increased significantly (p < 0.05) from 1.23-log to 4.20-log. When the chlorine concentration increased from 2 mg L−1 to 10 mg L−1, the inactivation rate of Acidovorax defluvii and Microbacterium laevaniformans increased significantly (p < 0.05) from 1.11-log to 2.35-log and from 1.74-log to 4.15-log, respectively. The inactivation rate of Sphingomonas sp. and Bacillus cereus was significantly (p < 0.05) different only at chlorine concentrations from 4 mg L−1 to 10 mg L−1, increasing from 1.12-log to 2.20-log and from 1.08-log to 1.44-log, respectively.
Similar to results determined using the crystal violet (CV) method, the order of biofilm resistance to chlorine was Bacillus cereus > Sphingomonas sp. > Acidovorax defluvii > Microbacterium laevaniformans > Acinetobacter sp. These results showed that Acinetobacter sp. and Microbacterium laevaniformans were sensitive to chlorine, while Acidovorax defluvii, Sphingomonas sp., and Bacillus cereus had a relatively strong resistance to chlorine. In addition, the relatively positive correlation between the inactivation rate and the HPC bacterial counts of single-species biofilms was observed at a chlorine concentration of 2 mg L−1 (r = 0.52; P = 0.044), while the inactivation rate was not significantly (p > 0.05) affected by the bacterial cell counts at other chlorine concentrations.
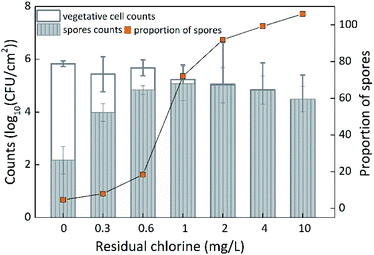
Spore counts and proportions of Bacillus cereus
To better understand the mechanism of chlorine resistance, the vegetative cell counts and spore counts of Bacillus cereus were determined because the Bacillus cereus exhibited the strongest chlorine resistance of all single-species biofilms examined. Based on the method described by Szabo et al.,34 the vegetative cell counts and spore counts of Bacillus cereus before and after disinfection were determined, and the proportion of spores was also analyzed (Fig. 7). The counts and the proportion of spores did not significantly increase (p > 0.05) at a low chlorine concentration of 0.3 mg L−1, but the spore counts did significantly increase (p < 0.05) at a chlorine concentration of 0.6 mg L−1, and both the counts and the proportion of spores significantly increased at a chlorine concentration of 1 mg L−1 (p < 0.05). When the chlorine concentration increased to 2 mg L−1, the spore counts gradually decreased, while the proportion of spores gradually increased. In fact, the spore counts gradually decreased to 3.1 × 104 CFU cm−2, which was significantly (p < 0.05) higher than that before disinfection and at a chlorine concentration of 0.3 mg L−1. The proportion of spores was 105% (due to standard error) so the proportion of spores was considered to be 100%. Bacillus cereus was sensitive to a chlorine concentration of 0.6 mg L−1. This is likely because the vegetative cells (which were sensitive to chlorine) were mainly incubated in R2A broth before disinfection, and changed to spores as the chlorine concentration increased. The spores had strong chlorine resistance.Discussion
All five bacterial strains examined in this study had biofilm formation potential when incubated for 72 h (Fig. 1). The specific respiratory activity of the five species of biofilms was negatively correlated with the biofilm biomass to some extent (Fig. 1 and 2). The bacterial cells adhered to the material surface, which led to the increased production of complex extracellular polymeric substances (EPS), and the proportion of biofilm biomass associated with non-metabolically active biomass also increased; as a result, the specific respiratory activity of the biofilms decreased.37,38 At 72 h of incubation, Acinetobacter sp. showed strong biofilm formation potential; Bacillus cereus, Microbacterium laevaniformans, and Sphingomonas sp. exhibited medium biofilm formation potential; and Acidovorax defluvii showed weak biofilm formation potential. Acinetobacter sp. has been found to co-aggregate with bacteria commonly found in drinking water, and has been shown to be responsible for promoting biofilm formation.7,39 These results corresponded to the bacterial counts in the biofilm (Fig. 1 and 3). Microbacterium laevaniformans showed weak biofilm formation potential when incubated for 24 h, but it showed no biofilm formation potential in the following 24 h. Previously, researchers showed that the initially strong adsorption and biofilm formation potential of bacteria on pipes does not mean that they will have strong biofilm formation potential later; in other words, a bacterium's initial performance cannot ultimately determine the characteristics of the biofilm it forms.37,40The chlorine resistance of the single-species biofilms as determined using the CV method was similar to that determined using the HPC method. Bacillus cereus showed the strongest chlorine resistance (except at a chlorine concentration of 0.3 mg L−1). Previous researchers reported that the chlorine resistance of G− bacteria is stronger than that of G+ bacteria, even though the latter has a thick cell wall. However, the cell wall of G+ bacteria has a simple composition (mainly containing peptidoglycan and a certain amount of teichoic acid) without an extracellular membrane, and contains multiple sites that are sensitive to disinfectants. While the G− bacteria have a thinner cell wall than that of G+ bacteria, the wall is relatively complex in its composition, having an extracellular membrane as a special feature containing phospholipid, lipopolysaccharide and protein, which enables the bacteria to tolerate disinfectants.27,41 However, opposing results are available that show some G+ bacteria have stronger chlorine resistance than G− bacteria, and this resistance is also associated with cell wall and membrane construction. To achieve sterilization, the effective chlorine concentration for G+ bacteria has been shown to be 0.3–0.5 mg L−1, while that for G− bacteria is much lower (≈0.05 mg L−1).42 However, a similar tendency was not observed in this study. Bacillus cereus (affiliated with G+ bacteria) had the strongest chlorine resistance, and the Sphingomonas sp. (affiliated with G− bacteria) also had strong chlorine resistance. In view of the lack of consensus in previous research, as well as the fact that both the G+ bacteria and G− bacteria had strong chlorine resistance in this study, it must be accepted that the chlorine resistance of some G− bacteria is stronger than that of G+ bacteria. However, the main reason for bacterial resistance to chlorine is complex. The chlorine resistance of a bacterial biofilm could be due mainly to some special structures, such as glycocalyx included in the capsular and mucous film, spores and other features.
Sphingomonas sp. can secrete extracellular material (mucopolysaccharide/glycocalyx),38 which can mix with proteins, nucleic acids and extracellular adsorption materials to form EPS.43 Microbial cells that have EPS consisting of glycocalyx as the main component can be expected to be more easily adsorbed on the surface of materials, relatively secluded from the interference of an unfavorable environment.7,43–45 Sun et al.19 and Zhang et al.20 found that Sphingomonas sp. can survive in an extremely high concentration of chlorine, and some can be sterilized only at a chlorine concentration of 4 mg L−1 and contact time of 240 min with low inactivation rate (0.03-log).19 Bacteria associated with Bacillus will form spores in the later period of their growth or in an unfavorable environment.46 Spores are the most resistant structure in the living world, and are extremely prominent in imparting heat resistance, chemical disinfectant resistance and radiation resistance.21,47,48 Thus, Bacillus has strong chlorine resistance due to its large proportion of spores.46,48 Spores can withstand the effects of disinfectants owing principally to the cumulative effect of spores in respect of their structure, chemistry and biochemistry.47,48
The spores of Bacillus were examined before and after disinfection in this study. The proportion of spores was only 0.02% before disinfection and the vegetative cell accounted for the vast majority of Bacillus. Both the spore counts and proportion of spores gradually increased with increasing chlorine concentration until most of the vegetative cells were inactivated by chlorine and many were transformed into spores. As one vegetative cell could be transformed into only one spore, the spores could not reproduce. Therefore, the inactivation rate of vegetative cells of Bacillus greatly increased with increasing chlorine concentration while at the same time the number of vegetative cells transformed into spores decreased; therefore, the spore counts initially increased and then decreased as the chlorine concentration increased.
Based on the initial spore counts, the optimum chlorine concentration for spore germination was between 0.6 mg L−1 and 1 mg L−1, which is the range of chlorine concentration used in most water treatment plants. Thus, the Bacillus was not inactivated by chlorine; rather, the spore counts and proportion of spores were increased by the chlorine. This finding highlights a serious challenge for the control of Bacillus sp. in DWDSs. Interestingly, although the Bacillus showed the strongest chlorine resistance of the five single-species biofilms examined, it had only medium biofilm formation potential. On the other hand, Acinetobacter sp. exhibited strong biofilm formation potential, but was very highly sensitive to chlorine. Thus, the results from this study (Fig. 1 and 4 and Table 3) showed that there was no positive correlation between biofilm formation potential and chlorine resistance of the five biofilms examined. These results indicated that the chlorine resistance of a single-species biofilm was mainly associated with the intrinsic mechanism of chlorine resistance by individual bacterial cells and showed little relation to the overall biofilm biomass.
From the results of this study, the chlorine resistance of single-species biofilms can be summarized as one of three types:
(a) The single-species biofilm associated with spore-formation exhibited the strong chlorine resistance. Spores can protect the single-species biofilm against a disinfectant and subsequently transform into vegetative cells under suitable environmental conditions.
(b) The single-species biofilm associated with secretions of EPS also showed strong chlorine resistance. EPS can shield a single-species biofilm from the interference of an unfavorable environment.
(c) The single-species biofilm that did not have special structures (e.g., neither spores nor EPS) was often sensitive to disinfectant.
In summary, the special structures of single-species biofilms had a significant influence on the chlorine resistance of the biofilms; and single-species biofilm that formed spores or secreted EPS exhibited a stronger chlorine resistance. The chlorine resistance of single-species biofilm had little relation with the biofilm formation potential; biofilm biomass was not a major factor affecting chlorine resistance.
Conclusions
The five tested strains isolated from the simulated DWDS had different biofilm formation potentials. Acinetobacter sp. exhibited strong biofilm formation potential; Bacillus cereus, Microbacterium laevaniformans, and Sphingomonas sp. showed medium biofilm formation potential; and Acidovorax defluvii had weak biofilm formation potential. In contrast, the order of chlorine resistance was Bacillus cereus > Sphingomonas sp. > Microbacterium laevaniformans > Acidovorax defluvii > Acinetobacter sp. The chlorine resistance of single-species biofilms has little relation with biofilm formation potential. The single-species biofilm with the strongest biofilm formation potential was highly sensitive to disinfectant. The single-species biofilm associated with spore formation exhibited the strongest chlorine resistance. The ability of a single bacterial biofilm to change physiological state, such as by forming spores or secreting EPS, can protect the biofilm against a disinfectant and reduce the biofilm's sensitivity to the disinfectant.Conflicts of interest
The authors declare that there are no conflicts of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (51608198, 61872141), the support from Science and Technology Research Project of Education Department of Jiangxi Province (Grant No. GJJ180342), and the Natural Science Foundation of Jiangxi Province (Grant No. 20161BAB216138). We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.References
- G. Liu, Y. Zhang, E. van der Mark, A. Magic-Knezev, A. Pinto, B. van den Bogert, W. Liu, W. van der Meer and G. Medema, Water Res., 2018, 138, 86–96 CrossRef CAS PubMed.
- I. B. Gomes, L. C. Simoes and M. Simoes, RSC Adv., 2019, 9, 32184–32196 RSC.
- G. Liu, G. L. Bakker, S. Li, J. H. G. Vreeburg, J. Q. J. C. Verberk, G. J. Medema, W. T. Liu and J. C. Van Dijk, Environ. Sci. Technol., 2014, 48, 5467–5476 CrossRef CAS PubMed.
- F. Bimakr, M. P. Ginige, A. H. Kaksonen, D. C. Sutton, G. J. Puzon and K. Y. Cheng, Sens. Actuators, B, 2018, 277, 526–534 CrossRef CAS.
- K. C. Makris, S. S. Andra and G. Botsaris, Crit. Rev. Environ. Sci. Technol., 2014, 44, 1477–1523 CrossRef CAS.
- Z. Xue and Y. Seo, Environ. Sci. Technol., 2013, 47, 1365–1372 CrossRef CAS PubMed.
- S. Liu, C. Gunawan, N. Barraud, S. A. Rice, E. J. Harry and R. Amal, Environ. Sci. Technol., 2016, 50, 8954–8976 CrossRef CAS PubMed.
- I. Douterelo, B. E. Dutilh, K. Arkhipova, C. Calero and S. Husband, Water Res., 2020, 173, 115586 CrossRef CAS PubMed.
- S. Bae, C. Lyons and N. Onstad, Water Res. X, 2019, 2, 100026 CrossRef PubMed.
- Y. Perrin, D. Bouchon, V. Delafont, L. Moulin and Y. Héchard, Water Res., 2019, 149, 375–385 CrossRef CAS PubMed.
- I. Douterelo, M. Jackson, C. Solomon and J. Boxall, Sci. Total Environ., 2017, 581–582, 277–288 CrossRef CAS PubMed.
- J. Liu, H. Ren, X. Ye, W. Wang, Y. Liu, L. Lou, D. Cheng, X. He, X. Zhou, S. Qiu, L. Fu and B. Hu, Appl. Microbiol. Biotechnol., 2016, 101, 749–759 CrossRef PubMed.
- R. A. Li, J. A. McDonald, A. Sathasivan and S. J. Khan, Water Res., 2019, 153, 335–348 CrossRef CAS PubMed.
- T. E. Cloete, Int. Biodeterior. Biodegrad., 2003, 51, 277–282 CrossRef CAS.
- C. Bertelli, S. Courtois, M. Rosikiewicz, P. Piriou, S. Aeby, S. Robert, J. F. Loret and G. Greub, Front. Microbiol., 2018, 9, 2520 CrossRef PubMed.
- T. B. Afonso, L. C. Simões and N. Lima, Biofouling, 2019, 35, 1041–1054 CrossRef CAS PubMed.
- H. L. Roder, S. J. Sorensen and M. Burmolle, Trends Microbiol., 2016, 24, 503–513 CrossRef PubMed.
- A. F. Thompson, E. L. English, A. M. Nock, G. G. Willsey, K. Eckstrom, B. Cairns, M. Bavelock, S. W. Tighe, A. Foote, H. Shulman, A. Pericleous, S. Gupta, D. E. Kadouri and M. J. Wargo, Microbiology, 2020, 166, 34–43 CrossRef CAS PubMed.
- W. Sun, W. Liu, L. Cui, M. Zhang and B. Wang, Sci. Total Environ., 2013, 458–460, 169–175 CrossRef CAS PubMed.
- M. L. Zhang, W. J. Liu, X. B. Nie, C. P. Li, J. N. Gu and C. Zhang, Microbes Environ., 2012, 27, 443–448 CrossRef PubMed.
- J. G. Szabo, G. Meiners, L. Heckman, E. W. Rice and J. Hall, J. Environ. Manage., 2017, 187, 1–7 CrossRef CAS PubMed.
- C. H. Tan, K. W. Lee, M. Burmolle, S. Kjelleberg and S. A. Rice, Environ. Microbiol., 2017, 19, 42–53 CrossRef PubMed.
- D. Dai, L. Raskin and C. Xi, J. Appl. Microbiol., 2017, 123, 1614–1627 CrossRef CAS PubMed.
- H. L. Roder, N. M. C. Olsen, M. Whiteley and M. Burmolle, Environ. Microbiol., 2020, 22, 5–16 CrossRef PubMed.
- D. Zhong, W. Ma, X. Jiang, Y. Yuan, H. Li and Y. Li, Desalin. Water Treat., 2018, 133, 191–198 CrossRef CAS.
- L. C. Simoes, M. Simoes and M. J. Vieira, Biofouling, 2011, 27, 685–699 CrossRef CAS PubMed.
- L. C. Simoes, M. Simoes and M. J. Vieira, Appl. Environ. Microbiol., 2010, 76, 6673–6679 CrossRef CAS PubMed.
- V. J. Pereira, R. Marques, M. Marques, M. J. Benoliel and M. T. Barreto Crespo, Water Res., 2013, 47, 517–523 CrossRef CAS PubMed.
- S. Stepanovic, D. Vukovic, I. Dakic, B. Savic and M. Svabic-Vlahovic, J. Microbiol. Methods, 2000, 40, 175–179 CrossRef CAS PubMed.
- M. Schwering, J. Song, M. Louie, R. J. Turner and H. Ceri, Biofouling, 2013, 29, 917–928 CrossRef CAS PubMed.
- P. Gulati and M. Ghosh, J. Water Health, 2017, 15, 942–954 CrossRef PubMed.
- L. C. Simoes, M. Simoes and M. J. Vieira, Appl. Environ. Microbiol., 2007, 73, 6192–6200 CrossRef CAS PubMed.
- W. Ding, W. Jin, S. Cao, X. Zhou, C. Wang, Q. Jiang, H. Huang, R. Tu, S. F. Han and Q. Wang, Water Res., 2019, 160, 339–349 CrossRef CAS PubMed.
- J. G. Szabo, N. Muhammad, L. Heckman, E. W. Rice and J. Hall, Appl. Environ. Microbiol., 2012, 78, 2449–2451 CrossRef CAS PubMed.
- I. Vaz-Moreira, C. Egas, O. C. Nunes and C. M. Manaia, FEMS Microbiol. Ecol., 2013, 83, 361–374 CrossRef CAS PubMed.
- S. L. Gora, K. D. Rauch, C. C. Ontiveros, A. K. Stoddart and G. A. Gagnon, Water Res., 2019, 151, 193–202 CrossRef CAS PubMed.
- I. B. Gomes, M. Simoes and L. C. Simoes, Sci. Total Environ., 2016, 565, 40–48 CrossRef CAS PubMed.
- S. Maes, T. Vackier, S. Nguyen Huu, M. Heyndrickx, H. Steenackers, I. Sampers, K. Raes, A. Verplaetse and K. De Reu, BMC Microbiol., 2019, 19, 77 CrossRef PubMed.
- Y. Furuichi, S. Yoshimoto, T. Inaba, N. Nomura and K. Hori, Environ. Sci. Technol., 2020, 54, 2520–2529 CrossRef CAS PubMed.
- L. C. Simoes and M. Simoes, RSC Adv., 2013, 3, 2520–2533 RSC.
- I. Vaz-Moreira, O. C. Nunes and C. M. Manaia, Sci. Total Environ., 2017, 586, 1141–1149 CrossRef CAS PubMed.
- J. Mir, J. Morato and F. Ribas, J. Appl. Microbiol., 1997, 82, 7–18 CrossRef CAS PubMed.
- Z. Xue, V. R. Sendamangalam, C. L. Gruden and Y. Seo, Environ. Sci. Technol., 2012, 46, 13212–13219 CrossRef CAS PubMed.
- Y. H. Wang, Y. H. Wu, T. Yu, X. H. Zhao, X. Tong, Y. Bai, Z. Y. Huo and H. Y. Hu, Sci. Total Environ., 2019, 693, 133579 CrossRef CAS PubMed.
- J. Tang, Y. Wu, S. Esquivel-Elizondo, S. J. Sorensen and B. E. Rittmann, Trends Biotechnol., 2018, 36, 1171–1182 CrossRef CAS PubMed.
- J. Szabo and S. Minamyer, Environ. Int., 2014, 72, 124–128 CrossRef CAS PubMed.
- G.-Q. Li, Z.-Y. Huo, Q.-Y. Wu, Y. Lu and H.-Y. Hu, Sci. Total Environ., 2018, 639, 1233–1240 CrossRef CAS PubMed.
- F. Zeng, S. Cao, W. Jin, X. Zhou, W. Ding, R. Tu, S.-F. Han, C. Wang, Q. Jiang, H. Huang and F. Ding, J. Cleaner Prod., 2020, 243, 118666 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra04985a |
| This journal is © The Royal Society of Chemistry 2020 |