 Open Access Article
Open Access ArticleRapid characterization of chemical constituents of Shaoyao Gancao decoction using UHPLC coupled with Fourier transform ion cyclotron resonance mass spectrometry†
Lin Sun‡
a,
Min Zhao‡ a,
Yanhui Zhao
a,
Yanhui Zhao a,
Xue Jianga,
Miao Wang*b,
Yixin Zhanga and
Chunjie Zhao
a,
Xue Jianga,
Miao Wang*b,
Yixin Zhanga and
Chunjie Zhao *a
*a
aSchool of Pharmacy, Shenyang Pharmaceutical University, Wenhua Road 103, Shenyang, Liaoning Province, China. E-mail: lab433@163.com
bSchool of Life Science and Biopharmaceutics, Shenyang Pharmaceutical University, Wenhua Road 103, Shenyang, Liaoning Province, China
First published on 11th August 2020
Abstract
Shaoyao Gancao decoction (SGD), a well-known Chinese herbal formula, has been used to treat liver injury for a long time. In this study, chemical profiles of SGD were identified using ultra high-performance liquid chromatography combined with Fourier transform ion cyclotron resonance mass spectrometry (UHPLC-FT-ICR-MS/MS). Liquid chromatography was performed on a C18 column (150 mm × 2.1 mm, 1.8 μm); the mobile phase comprised 0.1% formic acid (A) and acetonitrile (B). We then characterized 73 chemical compounds; the primary constituents in SGD included phenols and monoterpenes (in Paeoniae Radix Alba), triterpene saponins, and flavonoids (in Glycyrrhizae Radix et Rhizoma Praeparata Cum Melle). Thus, this study provides a basis for further study on SGD and is expected to be useful for rapidly characterizing constituents in other traditional Chinese herbal formulations.
1 Introduction
In China, Traditional Chinese Medicine (TCM) and its formulas have an extended history for treating diseases. Their integrated and synergistic effects on multiple targets have been extensively praised.1 However, it is difficult for researchers to explain the component that plays a major role in the efficacy of the materials because of their massive chemical composition, which is an obstacle for TCM in the international market.2 In reaction to this phenomenon, multiple studies have focused on examining the chemical components of TCM. However, TCM is numerous in quantity and complicated in composition, so the condition of current research is far from enough. Because of the continuous development in science and technology, a rapid method for identifying chemical components of TCM is necessary, which will then act as the basis for TCM's pharmacology research and clinical applications.Initially, Shaoyao Gancao decoction (SGD) was described in Shang Han Lun, a clinical TCM book written by Zhang Zhongjing in the Eastern Han Dynasty.3 It contains two herbs: Paeoniae Radix Alba and Glycyrrhizae Radix et Rhizoma Praeparata Cum Melle. The SGD was a classical formula of TCM and extensively used for treating febrile diseases such as relief of nourishing liver, relaxing spasm, and relieving pain.4
At present, few studies have focused on the chemical components of SGD.5 To improve the detection range and sensitivity of previous method, researchers are increasingly using UHPLC-FT-ICR-MS, which is a type of powerful qualitative screening platform with a high mass resolving power that demonstrates powerful separation and can generate accurate molecular measurements. For example, using this method, Wang et al. characterized 33 chemical compounds in Cortex Fraxini and Guan et al. characterized 120 chemical compounds in Sijunzi decoction.6,7
In our work, we selected UHPLC-FT-ICR-MS to systematically characterize the chemical profiles of SGD. This study is thus able to provide a substantial base and provide considerable information for SGD-related pharmacological research.
2 Materials and methods
2.1 Chemicals and materials
Paeoniae Radix Alba (batch number: 18061201, source: Anhui China) and Glycyrrhizae Radix et Rhizoma Praeparata Cum Melle (batch number: 180518; source: Neimenggu China), which were authenticated by Professor Jingming Jia (Department of TCM, Shenyang Pharmaceutical University, Shenyang, China), were purchased from Guoda pharmacy (Shenyang, China). The primary source of reference compounds (purity > 98%), including benzoyl paeoniflorin, albiflorin, ononin, and glycyrrhizic acid, was Shanghai Yuanye Bio-Technology Co., Ltd (Shanghai, China), while gallic acid, liquiritin, paeoniflorin, and liquiritin apioside were obtained from the National Institute for the Control of Pharmaceutical and Biological Products (Beijing, China). Moreover, acetonitrile of HPLC grade and formic acid of LC-MS grade were obtained from Fisher Scientific (Fair Lawn, NJ, USA); purified water was then purchased from Wahaha (Hangzhou, China).2.2 Preparation of SGD for analysis
As per SGD's original composition, two constituting herbs, Paeoniae Radix Alba (250 g) and Glycyrrhizae Radix et Rhizoma Praeparata (250 g), were mixed and macerated in purified water (5 L) for 0.5 h, then boiled at 100 °C for 1.5 h, and then the extracted solution was filtered through five layer gauzes. The residue was decocted twice with boiling water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8, v/v) for 1 h each and the extracted solution was filtered using five layer gauzes. These three extractions were then combined and dried using lyophilization. Before analysis, dried powder (0.5 g) was dissolved in water (10 mL), and then vortexed for 1 min for complete dissolution.
8, v/v) for 1 h each and the extracted solution was filtered using five layer gauzes. These three extractions were then combined and dried using lyophilization. Before analysis, dried powder (0.5 g) was dissolved in water (10 mL), and then vortexed for 1 min for complete dissolution.
2.3 Instrument and analytical conditions
Chromatographic analysis was performed using an Agilent 1260 UHPLC system (USA) using a universal XB C18 column (150 mm × 2.1 mm, 1.8 μm; Kromat, USA) at the column temperature of 35 °C. The mobile phase comprised 0.1% formic acid (A) and acetonitrile (B), and the gradient elution program was carried out for chromatographic separation as follows: 2–10% (B) from 0 to 12 min, 10–25% (B) from 12 to 32 min, 25–62% (B) from 32 to 52 min, and 62–65% (B) from 52 to 55 min. The flow rate was 0.20 mL min−1, and the injection volume was 2 μL.Mass spectra analysis was conducted on a Bruker Solarix 7.0 T FT-ICR-MS system (Bruker, Germany) and a Bruker Compass-Hystar workstation (Bruker, Germany) using both positive and negative electrospray ionization (ESI) modes, followed by optimized conditions: nebulizer gas pressure of 4.0 bar; dry gas flow rate of 8 L min−1; dry gas temperature of 200 °C; ion accumulation time of 0.15 s; time of flight of 0.6 ms; capillary voltage of 4.5 kV; and endplate offset of 500 V. The recording of the full-scan mass spectrum data was performed between m/z 100 and 3000. In respect to the auto MS/MS mode, the selection of both MS/MS boost and MS/MS isolation was made; moreover, the range of collision power was maintained between 10 and 30 eV for MS/MS experimentations.
3 Results and discussion
Fig. 1 shows the base peak ion chromatograms (BPC) of SGD and the reference compounds. The extracted ion chromatograms (EIC) for each molecular weight, which are shown in the ESI (Fig. S1 and S2),† were correspondingly obtained for detecting the associated compound. Among the identified compounds, the accurate identification of eight compounds was performed by comparing the retention time (tR) and the MS/MS data associated with the reference compounds in the positive ion mode. The other compounds were determined by their retention times, as well as the molecular weight and the MS/MS fragments. Bruker workstation was used for computing the molecular formulas of the compounds by comparing the known molecular weights with the measured molecular weights, followed by limiting the acceptable error values to <3.0 ppm. Using MS/MS data, additional speculations of the layouts of the compounds were conducted. In aggregate, we reported 73 compounds and their layouts are presented in Fig. S3 and S4.† Fig. S5† shows the presentation of MS/MS spectra of the typical compounds while displaying their possible fragmentation pathways in Fig. 2. The inferences of each ingredient were carried out with the help of the molecular formulas and fragmentation pathways, followed by additional confirmation with reference to the previous literatures.8–16 Table 1 lists the retention time, formula, molecular weight, calculated m/z, detected m/z, error value and MS/MS data of ingredients. A concrete illustration of the ingredients' characterization was performed as hereunder.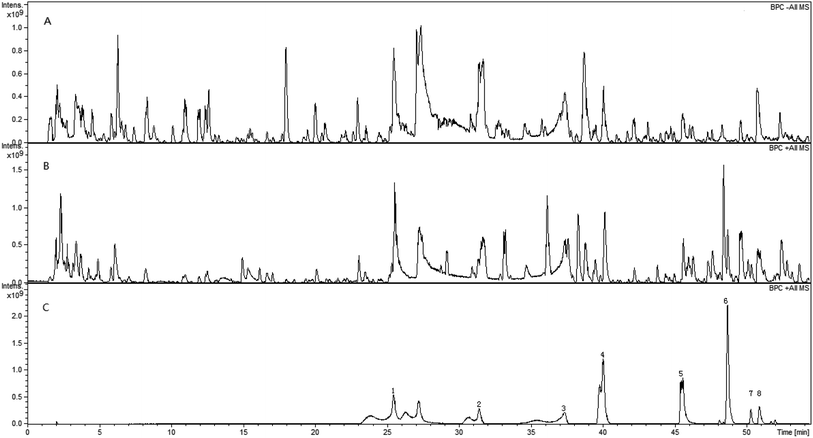 | ||
| Fig. 1 The base peak ion chromatograms (BPC) of SGD in both positive (A) and negative (B) ion modes and the corresponding compounds (C). | ||
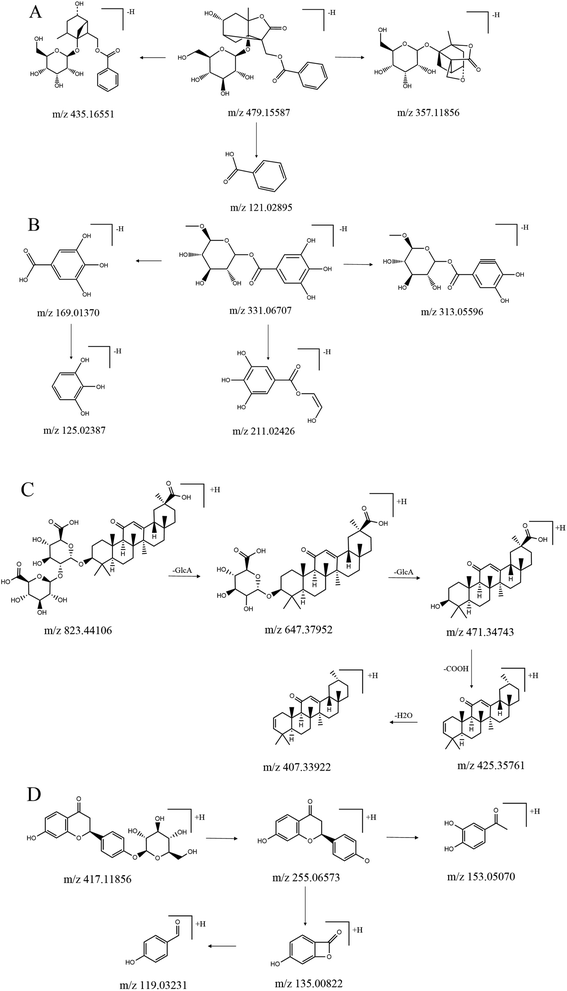 | ||
| Fig. 2 The possible fragmentation pathways of the typical compounds. (A) Albiflorin, (B) glucogallin, (C) glycyrrhizic acid, (D) liquiritin. | ||
| No. | tR (min) | Identification | Formula | Molecular weight | Ion mode | MS (m/z) | ppm | MS/MS (m/z) |
|---|---|---|---|---|---|---|---|---|
| a Ps: 1–30 from Paeoniae Radix Alba; 31–73 from Glycyrrhizae Radix et Rhizoma Praeparata. | ||||||||
| 1 | 3.43 | Citric acid | C6H8O7 | 192.0270 | [M + H]+ | 193.03428 | −0.55 | 191.05401; 111.00913 |
| [M − H]− | 191.01973 | 0.44 | 133.01330 | |||||
| 2 | 6.2 | Gallic acid | C7H6O5 | 170.0215 | [M + H]+ | 171.0288 | 5.15 | 126.02387 |
| [M − H]− | 169.01425 | 0.54 | 125.02453; 108.02271 | |||||
| 3 | 6.72 | Debenzoyl paeoniflorin | C16H24O10 | 376.1369 | [M + H]+ | 377.14422 | 1.62 | 375.12803; 345.11810; 195.06531; 139.07233 |
| 4 | 7.43 | 1-O-β-D-Glucopyranosyl-paeonisuffrone | C16H24O9 | 360.1420 | [M + H]+ | 361.14931 | 1.49 | 181.08418; 163.01784; 127.01413 |
| 5 | 11.93 | Glucogallin | C13H16O10 | 332.0743 | [M + H]+ | 333.08162 | 1.37 | 207.05048; 125.02387 |
| [M − H]− | 331.06707 | −0.11 | 313.05596; 211.02426; 169.01370; 125.02387 | |||||
| 6 | 12.97 | 6-O-β-D-Glucopyranosyl lactinolide | C16H26O9 | 362.1576 | [M − H]− | 361.15041 | 0.28 | 185.11777; 163.06065; 113.06025 |
| 7 | 15.66 | Ethyl gallic acid | C9H10O5 | 198.0528 | [M + H]+ | 199.0601 | 3.32 | 125.10300 |
| 8 | 17.64 | Mudanpioside F | C16H24O8 | 344.1471 | [M − H]− | 343.13981 | 1.1 | 179.05556; 165.09115 |
| 9 | 24.29 | Galloylpaeoniflorin | C30H32O15 | 632.1741 | [M + H]+ | 633.1814 | 1.00 | 631.16613; 613.15570; 509.12952; 491.11895; 463.12404 |
| 10 | 24.48 | 1′-O-Benzoylsucrose | C19H26O12 | 446.1424 | [M − H]− | 445.13515 | 0.07 | 179.14800; 132.04226; 121.02895 |
| 11 | 24.87 | Isomaltopaeoniflorin | C29H38O16 | 642.2159 | [M + H]+ | 643.22326 | 1.49 | 643.22326; 191.11500 |
| 12 | 25.22 | Paeonol | C9H10O3 | 166.0630 | [M − H]− | 165.05572 | 0.59 | 165.05572 |
| 13 | 25.54 | Paeonilactone B | C10H12O4 | 196.0735 | [M + H]+ | 197.08084 | 0.43 | 133.0662; 105.0688; 103.0545 |
| 14 | 25.56 | Paeonilactone C | C17H18O6 | 318.1103 | [M + H]+ | 319.11761 | 0.34 | 183.06573; 135.04460 |
| 15 | 26.92 | Oxypaeoniflorin | C23H28O12 | 496.1581 | [M + H]+ | 497.16535 | 1.07 | 267.08286; 180.07864; 163.06065; 137.02837 |
| 16 | 28.75 | Schaftoside | C26H28O14 | 564.1479 | [M + H]+ | 565.15518 | 1.41 | 565.15518; 501.13969; 163.03952 |
| [M − H]− | 563.14063 | 1.16 | 563.14063; 499.1404 | |||||
| 17 | 31.1 | Palmitic acid | C16H32O2 | 256.2402 | [M + H]+ | 257.08084 | 1.19 | 143.07120; 113.11303 |
| 18 | 31.33 | Paeoniflorigenone | C17H18O6 | 318.1103 | [M + H]+ | 319.11761 | 1.29 | 137.05818; 133.06662; 105.03324 |
| 19 | 31.35 | Albiflorin | C23H28O11 | 480.1631 | [M + H]+ | 481.17044 | 0.53 | 197.08113; 151.07255; 133.02649; 105.01342 |
| [M − H]− | 479.15587 | 0.67 | 435.16551; 357.11856; 121.02895 | |||||
| 20 | 31.85 | Kaempferitrin | C27H30O14 | 578.1635 | [M + H]+ | 579.17083 | 0.49 | 623.15923; 315.05121; 314.04118; 299.01050 |
| 21 | 38.83 | Lactiflorin | C23H26O10 | 462.1526 | [M + H]+ | 463.15987 | 1.59 | 179.07100; 151.07186; 135.08121 |
| [M − H]− | 461.14532 | 0.45 | 461.14532; 285.06104; 121.08956 | |||||
| 22 | 37.45 | Paeonisuffrone C | C10H14O4 | 198.0892 | [M − H]− | 197.08084 | 0.6 | 197.08084 |
| 23 | 37.53 | Paeoniflorin | C23H28O11 | 480.1631 | [M + H]+ | 481.17044 | 0.8 | 481.17044; 451.16042; 375.12972; 329.12364; 123.04460 |
| 24 | 40.58 | Benzoyloxypaeoniflorin | C30H32O13 | 600.1842 | [M − H]− | 599.17701 | 0.7 | 599.17618; 509.19525; 491.23538; 293.21011; 137.10284 |
| 25 | 40.59 | Mudanpioside D | C24H30O12 | 510.1737 | [M − H]− | 509.16645 | 0.81 | 509.16645; 463.15486; 121.02994 |
| 26 | 44.4 | Hederagenin | C30H48O4 | 472.3553 | [M + H]+ | 473.36254 | 1.45 | 426.31340; 251.20111; 168.11503 |
| 27 | 46.06 | Benzoylpaeoniflorin | C30H32O12 | 584.1893 | [M + H]+ | 585.19665 | 1.33 | 585.19665; 463.16042; 433.14986; |
| 28 | 49.25 | Benzoyl paeonifloride | C30H32O12 | 584.1893 | [M + H]+ | 585.19665 | 0.99 | 585.19665; 567.18664; 463.45900 |
| 29 | 50.79 | Astrantiagenin D | C30H46O4 | 470.3396 | [M + H]+ | 471.34689 | 1.04 | 234.16198; 209.45415 |
| 30 | 52.15 | Oleanolic acid | C30H48O3 | 456.3604 | [M + H]+ | 457.36762 | 1.17 | 411.28992; 203.16068; 153.15942; |
| 31 | 2.01 | Gentiobiose | C12H22O11 | 342.1161 | [M − H]− | 341.10894 | 0.29 | 341.10894; 221.06613; 179.05556; 161.04500 |
| 32 | 23.43 | Liquiritigenin-7,4-diglucoside | C27H32O14 | 580.1791 | [M − H]− | 579.17193 | 1.66 | 579.17193; 417.11856; 253.05008 |
| 33 | 23.59 | Liquiritin | C21H22O9 | 418.1263 | [M − H]− | 417.11911 | 0.26 | 255.06573; 153.05070; 135.00822; 119.03231 |
| 34 | 26.07 | Vicenin-2 | C27H30O15 | 594.1584 | [M + H]+ | 595.16575 | 0.85 | 595.16575; 451.14517 |
| 25.97 | [M − H]− | 593.15119 | 1.31 | 593.15119; 449.12952; 363.12912 | ||||
| 35 | 28.75 | Schaftoside | C26H28O14 | 564.1479 | [M + H]+ | 565.15518 | 1.41 | 446.11564; 431.10298; 401.09589 |
| [M − H]− | 563.14063 | 1.16 | 403.10291; 271.05008 | |||||
| 36 | 30.22 | Choerospondin | C21H22O10 | 434.1213 | [M − H]− | 433.11249 | −0.12 | 282.11643; 271.06593; 152.01479 |
| 37 | 31.7 | Pinocembrin | C15H12O4 | 256.0735 | [M + H]+ | 257.08084 | 1.96 | 257.08084; 108.02113 |
| [M − H]− | 255.06628 | 0.16 | 255.06628; 150.03169; 106.04186 | |||||
| 38 | 30.88 | Glucoliquiritin apioside | C32H40O18 | 712.2214 | [M + H]+ | 713.22874 | 0.94 | 551.17647; 459.70586; 255.13625 |
| 39 | 31.12 | Licoagroside A | C23H24O12 | 492.1268 | [M − H]− | 491.11950 | 0.88 | 327.07693; 164.03532; 148.03643 |
| 40 | 31.66 | Liquiritin apioside | C26H30O13 | 550.1686 | [M + H]+ | 551.17592 | 0.92 | 551.17592; 257.09195; 137.02387 |
| [M − H]− | 549.16136 | 1.24 | 549.16136; 417.17138; 255.13727; 135.00822 | |||||
| 41 | 30.95 | Liquiritigenin | C15H12O4 | 256.0735 | [M + H]+ | 257.08084 | 1.19 | 257.08084; 135.09800 |
| 42 | 31.10 | Isoliquiritigenin | C15H12O4 | 256.0735 | [M + H]+ | 257.08084 | 1.19 | 163.06592; 150.03169; 106.12400 |
| 43 | 31.24 | Trifolirhizin | C22H22O10 | 446.1213 | [M + H]+ | 447.12857 | 1.15 | 285.07128; 229.08474; 149.02177 |
| 44 | 31.53 | Neoliquiritin | C21H22O9 | 418.1263 | [M + H]+ | 419.13366 | 0.33 | 419.13366; 257.08138 |
| [M − H]− | 417.1186 | 0.13 | 417.11856; 255.06573 | |||||
| 45 | 31.85 | Violanthin | C27H30O14 | 578.1635 | [M + H]+ | 579.17083 | 0.49 | 579.17083; 549.16082; 495.12912 |
| 46 | 36.34 | Naringenin-7-O-glucoside | C21H22O10 | 434.1212 | [M − H]− | 433.11402 | 1.9 | 433.11402; 271.06065 |
| 47 | 37.53 | Albiflorin | C23H28O11 | 480.1631 | [M + H]+ | 481.17044 | 0.82 | 481.17044; 451.44800; 359.31451; 329.12364 |
| 48 | 40.13 | Ononin | C22H22O9 | 430.1264 | [M + H]+ | 431.13366 | 0.46 | 323.07669; 179.05556; 144.02113; 107.04969 |
| 49 | 40.16 | Pallidiflorin | C16H12O4 | 268.0735 | [M + H]+ | 269.08084 | 0.66 | 269.08084; 254.05791; 241.05008; 181.06534 |
| [M − H]− | 267.06628 | 0.22 | 267.06628; 252.04226; 223.03952 | |||||
| 50 | 40.25 | Isoliquiritin apioside | C26H30O13 | 550.1686 | [M + H]+ | 551.17592 | 0.92 | 419.13421; 255.06572; 137.04460 |
| [M − H]− | 549.16136 | 1.24 | 549.16082; 431.11895; 415.16042 | |||||
| 51 | 40.98 | 5,7-Dihydroxyflavone | C15H12O4 | 256.0735 | [M − H]− | 255.06628 | 0.46 | 255.06628; 135.03954; 119.04960 |
| 52 | 41.06 | Licochalcone B | C16H14O5 | 286.08412 | [M − H]− | 285.07685 | 0.15 | 255.07891; 193.05761; 165.06538 |
| 53 | 43.22 | Licorice-saponin O4 | C54H84O24 | 1116.5352 | [M + H]+ | 1117.5425 | 0.57 | 516.34509; 327.32421; 192.02700; 189.16433 |
| 54 | 44.05 | Echinatin | C16H14O4 | 270.08921 | [M + H]+ | 271.09649 | 1.14 | 239.07549; 149.06349; 121.03782 |
| 55 | 44.23 | Uralsaponin T | C48H74O19 | 954.48240 | [M + H]+ | 955.48899 | 0.75 | 779.44623; 458.35522; 179.04616 |
| 56 | 44.46 | Uralsaponin P | C42H64O16 | 824.41944 | [M + H]+ | 825.42671 | 1.04 | 663.36548; 487.33574; 165.06255 |
| 57 | 45.46 | Licorice-saponin M3 | C48H74O19 | 954.4824 | [M + H]+ | 955.48971 | 1.32 | 955.48971; 517.23599; 366.04062; 163.06065 |
| 58 | 45.84 | Uralsaponin F | C44H64O19 | 896.4041 | [M + H]+ | 897.41146 | 0.7 | 721.14563; 545.33269; 527.88076; 467.33254; 421.11257; 497.88210; 375.33245 |
| [M − H]− | 895.3969 | 1.81 | 719.36098; 543.11527; 525.35432; 465.88908; 419.44671; 495.54490; 373.32157 | |||||
| 59 | 47.3 | 22-Acetoxyl-glycyrrhizin | C44H64O18 | 880.4092 | [M + H]+ | 881.4165 | 1.53 | 705.13564; 529.11253; 518.00490; 451.33235; 405.44267 |
| 60 | 47.6 | Licorice-saponin G2 | C42H62O17 | 838.3986 | [M + H]+ | 839.40598 | 0.57 | 839.40598; 663.35370; 487.37913 |
| [M − H]− | 837.39142 | 0.6 | 837.39142; 661.12531; 485.90786; 351.11236 | |||||
| 61 | 48.42 | Licorice-saponin A3 | C48H72O21 | 984.4565 | [M + H]+ | 985.46389 | 0.23 | 985.46389; 823.88097; 647.32446 |
| [M − H]− | 983.44933 | 0.3 | 983.44933; 821.57765; 645.33542; 351.11676 | |||||
| 62 | 48.49 | Uralsaponin N | C42H62O17 | 838.3987 | [M + H]+ | 839.40598 | 0.57 | 663.37644; 487.32988; 179.05516 |
| 63 | 48.91 | Licorice-saponin B2 | C42H64O15 | 808.4244 | [M + H]+ | 809.43180 | 1.13 | 809.43180; 633.40026; 439.39439 |
| 64 | 49.28 | Formononetin | C16H12O4 | 268.0735 | [M − H]− | 267.06628 | 0.54 | 267.06628; 252.04226; 195.04460 |
| 65 | 50.14 | 22-β-Acetoxylglyrrhaldehyde | C44H64O17 | 864.4142 | [M + H]+ | 865.42163 | −0.33 | 689.37723; 513.34358; 179.04966 |
| [M − H]− | 863.40707 | 2.76 | 481.33178; 353.07200; 193.03483 | |||||
| 66 | 50.76 | Glycyrrhizic acid | C42H62O16 | 822.4037 | [M + H]+ | 823.41106 | 1.57 | 647.37952; 471.34743; 425.35761; 407.33922 |
| 67 | 50.79 | Glycyrrhetinic acid | C30H46O4 | 470.3396 | [M + H]+ | 471.34689 | 1.04 | 339.26538; 189.16722; 137.13835 |
| 68 | 54.41 | Licorice-saponin K2 | C42H62O16 | 822.4037 | [M + H]+ | 823.41106 | 1.08 | 823.41106; 647.82600; 471.70200 |
| [M − H]− | 821.39651 | 1.37 | 821.39651; 646.55342 | |||||
| 69 | 52.84 | 3′-Methoxyglabridin | C21H22O5 | 354.1467 | [M − H]− | 353.13945 | 0.27 | 353.13945; 338.15542; 147.04734 |
| 70 | 53.16 | Licorice-saponin H2 | C42H62O16 | 822.4037 | [M + H]+ | 823.41106 | 1.08 | 823.41106; 647.37952; 471.34743 |
| 71 | 54.23 | Licorice-saponin J2 | C42H64O16 | 824.4194 | [M + H]+ | 825.42671 | 1.53 | 825.42671; 649.39517; 455.40456 |
| [M − H]− | 823.41216 | 0.59 | 823.41216; 647.37952; 193.03483 | |||||
| 72 | 53.65 | Uralsaponin C | C42H64O16 | 824.4194 | [M + H]+ | 825.42671 | 1.53 | 649.39517; 473.36309; 455.35252; 437.34196 |
| 73 | 53.96 | Glycycoumarin | C21H20O6 | 368.1259 | [M − H]− | 367.11871 | 0.3 | 367.11817; 296.27800; 369.13811; 313.07121; 285.07630 |
3.1 Characterization of the constituents in Paeoniae Radix Alba
Monoterpenes and several phenols were the primary active ingredients in Paeoniae Radix Alba with majority of them being monoterpenes. In this study, a tentative characterization of 30 compounds of Paeoniae Radix Alba in SGD was performed, followed by the identification of four of them. Peaks 1, 2, 3 and 6 in Fig. 1C can be attributed to gallic acid, albiflorin, paeoniflorin, and benzoyl paeoniflorin, respectively. Albiflorin was used as an illustration for demonstrating the fragmentation pathways of monoterpenes in Paeoniae Radix Alba. In the negative mode, the ion at m/z 479.15587 was inferred to be the adduct ion ([M − H]−), followed by the calculation of the formula as C23H28O11. In the MS/MS spectrum, the key fragment ions found were at m/z 435.16551, 357.11856, and 121.02895, which suggested the loss of CO2 (44 Da), C7H5O2 (122 Da) and C16H22O9 (358 Da) from the precursor ion, respectively. Glucogallin was selected as an illustration for demonstrating the fragmentation pathways of phenol. In respect to the negative mode, the ion at m/z 331.06707 was deducted to be the adduct ion ([M − H]−) and the calculated formula was C13H16O10. The important fragment ions found in the MS/MS spectrum were at m/z 313.05596, 211.02426, 169.01370 and 125.02387. The ion at m/z 313.05596 can be attributed to the loss of OH (17 Da) from the precursor ion, whereas the ion at m/z 211.02426 can be attributed to the loss of C4H8O4 (120 Da) from the precursor ion. The ions at m/z 169.01370 and 125.02387 represented C7H6O5 and C6H6O3, respectively, and the characterization of other compounds in Paeoniae Radix Alba was performed based on fragmentation patterns and related literature.10,12,133.2 Characterization of constituents in Glycyrrhizae Radix et Rhizoma Praeparata Cum Melle
Triterpene saponins and flavonoids were the primary active constituents in Glycyrrhizae Radix et Rhizoma Praeparata Cum Melle.14 In this research, tentative characterization of 43 ingredients of Glycyrrhizae Radix et Rhizoma Praeparata Cum Melle in SGD was performed, followed by the precise identification of four among them. Peaks 4, 5, 7 and 8 in Fig. 1C represented liquiritin, ononin, isoliquiritigenin, and glycyrrhizic acid, respectively. Glycyrrhizic acid was used as a common triterpene saponins composition of Glycyrrhizae Radix et Rhizoma Praeparata Cum Melle as an illustration for demonstrating the fragmentation pathways. In the positive mode, the ion at m/z 823.44106 was inferred to be the adduct ion ([M + H]+), followed by the calculation of the formula as C42H62O16. The key fragment ions found in the MS/MS spectrum were at m/z 647.37952, 471.34743, 425.35761 and 407.33922. The ion at m/z 647.37952 suggested the loss of C6H8O6 (176 Da) from the precursor ion, that at m/z 471.34743 revealed the loss of C6H8O6 (176 Da) from the m/z 647.37952, that at m/z 425.35761 suggested the loss of CHO2 (46 Da) from the m/z 471.34743, and that at m/z 407.33922 revealed the loss of H2O (18 Da) from the m/z 425.35761. Liquiritin was used as an example for demonstrating the fragmentation pathways of flavonoids in Glycyrrhizae Radix et Rhizoma Praeparata Cum Melle. In respect to the negative mode, the ion at m/z 417.11856 was confirmed to be the adduct ion ([M − H]−), followed by the calculation of the formula as C21H22O9. The key fragment ions found in the MS/MS spectrum were at m/z 255.06573, 153.05070, 135.00822 and 119.03231. The ion at m/z 255.06573 denoted the loss of C6H11O5 (178 Da) from the precursor ion; that at m/z 153.05070 denoted the loss of C7H3O (102 Da) from the m/z 255.06573; that at m/z 135.00822 denoted the loss of C8H7O (120 Da) from the m/z 255.06573; and that at m/z 119.03231 denoted the loss of O (16 Da) from the m/z 135.00822. Characterization of the other ingredients in Glycyrrhizae Radix et Rhizoma was performed based on the fragmentation patterns and related literature.14,16SGD is a classical formula of traditional Chinese medicine that is extensively used in the clinic due to its anti-inflammatory, immunoregulatory, analgesic, antidepression, hepatoprotective and neuroprotective effects.12 Moreover, there is a wealth of study on the pharmacological effects of certain active components in the Paeoniae Radix Alba and Glycyrrhizae Radix et Rhizoma Praeparata Cum Melle. This study revealed that monoterpenes and several phenols (in Paeoniae Radix Alba) and the triterpene saponins and flavonoids (in Glycyrrhizae Radix et Rhizoma Praeparata Cum Melle) constituted the key ingredients in SGD. Some of these chemical components have been reported to be the active ingredients in SGD.8,10,13,17,18 For example, paeoniflorin was reported to have anti-inflammatory, hepatoprotective and neuroprotective effects.19,20 Albiflorin was shown to be both anti-inflammatory and antioxidant.9,21 Polyphenol was reported to play a role in antioxidant and antiviral activity. Pentagalloylglucose was shown to have anti-inflammatory, anti-allergic, antitumor, antiviral and antibacterial effects. Paeonol was reported to have anti-inflammatory, antitumor, anti-allergic, antioxidant activities, along with cardiovascular and neuroprotective effects.22 Liquiritin had antidepressive and neuroprotective effects.23,24 Liquiritigenin had been reported to exhibit anti-inflammatory effect.25 Saponins from liquorice demonstrated anti-inflammatory, antiarrhythmia and hepatoprotective effects.26,27 To better understand the major functional compounds and the mechanism of SGD, additional research is required. This study provides a good basis for identifying the prototype components and metabolites in SGD, which can better illustrate its medicinal value.
4 Conclusions
A rapid method was performed to systematically characterize 73 chemical constituents of SGD in total with the help of UHPLC-FT-ICR-MS. Experimental results reveal that phenols and monoterpenes (in Paeoniae Radix Alba), triterpene saponins and flavonoids (in Glycyrrhizae Radix et Rhizoma Praeparata Cum Melle) are the primary components in SGD. Moreover, it provides more information about the compounds in SGD than the previous literature. Therefore, the results of this study can be used to evaluate the quality of SGD and provide a basis for subsequent in vivo studies of SGD. Furthermore, this work provides a method for rapid identification of other TCMs. However, additional studies are required to overcome the limitation of identifying only known compounds using this method.Abbreviations
| BPC | Base peak ion chromatograms |
| EIC | Extracted ion chromatograms |
| SGD | Shaoyao Gancao decoction |
| TCM | Traditional Chinese medicine |
| UHPLC-FT-ICR-MS/MS | Ultra high-performance liquid chromatography coupled with Fourier transform ion cyclotron resonance mass spectrometry |
Conflicts of interest
The authors declare that there are no conflicts of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC: 81973284). This work was supported by Natural Science Foundation of Liaoning Province 2018 (Grant No. 20180551259).References
- P. Dong, L. Zhang, L. Zhan and Y. Liu, J. Sep. Sci., 2016, 39, 595–602 CrossRef CAS PubMed.
- Y. Jiang, R. Liu, J. Chen, M. Liu, M. Liu, B. Liu, L. Yi and S. Liu, J. Chromatogr. A, 2019, 1600, 197–208 CrossRef CAS PubMed.
- Z. J. Zhang, Treatise on Febrile Diseases (Shang Han Lun), The Eastern Han Dynasty Search PubMed.
- C.-H. Xu, P. Wang, Y. Wang, Y. Yang, D.-H. Li, H.-F. Li, S.-Q. Sun and X.-Z. Wu, J. Ethnopharmacol., 2013, 149, 443–452 CrossRef CAS PubMed.
- Q. Yin, P. Wang, A. Zhang, H. Sun, X. Wu and X. Wang, J. Sep. Sci., 2013, 36, 1238–1246 CrossRef CAS PubMed.
- Y. Wang, F. Han, A. Song, M. Wang, M. Zhao and C. Zhao, J. Sep. Sci., 2016, 39, 4325–4334 CrossRef CAS PubMed.
- Z. Guan, M. Wang, Y. Cai, H. Yang, M. Zhao and C. Zhao, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2018, 1086, 11–22 CrossRef CAS PubMed.
- Z. Li, H. Gao, J. Li and Y. Zhang, J. Sep. Sci., 2017, 40, 2558–2564 CrossRef CAS PubMed.
- Y. Yan, C.-Z. Chai, D.-W. Wang, J. Wu, H.-H. Xiao, L.-X. Huo, D.-N. Zhu and B.-Y. Yu, J. Pharm. Biomed. Anal., 2014, 95, 76–84 CrossRef CAS PubMed.
- B. Yan, M. Shen, J. Fang, D. Wei and L. Qin, J. Pharm. Biomed. Anal., 2018, 160, 276–288 CrossRef CAS PubMed.
- K. Pei, Y. Duan, H. Cai, S. Tu, F. Qiao, X. Song, X. Liu, G. Cao, K. Fan and B. Cai, Anal. Methods, 2015, 7, 9442–9451 RSC.
- S.-L. Li, J.-Z. Song, F. F. K. Choi, C.-F. Qiao, Y. Zhou, Q.-B. Han and H.-X. Xu, J. Pharm. Biomed. Anal., 2009, 49, 253–266 CrossRef CAS PubMed.
- L. Shan, N. Yang, Y. Zhao, X. Sheng, S. Yang and Y. Li, J. Sep. Sci., 2018, 41, 3791–3805 CrossRef CAS PubMed.
- Y. Cui, T. Liu, Y. Zhang, R. Wang, X. Liu, Q. Zhang, P. Yu, Y. Zhao and Z. Yu, J. Pharm. Biomed. Anal., 2018, 159, 318–325 CrossRef CAS PubMed.
- Y. Xu, H. Cai, G. Cao, Y. Duan, K. Pei, S. Tu, J. Zhou, L. Xie, D. Sun, J. Zhao, J. Liu, X. Wang and L. Shen, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2018, 1083, 110–123 CrossRef CAS PubMed.
- C. Schmid, C. Dawid, V. Peters and T. Hofmann, J. Nat. Prod., 2018, 81, 1734–1744 CrossRef CAS PubMed.
- N. Y. Tang, C. H. Liu, C. T. Hsieh and C. L. Hsieh, Am. J. Chin. Med., 2010, 38, 51–64 CrossRef CAS.
- C. Feng, M. Liu, X. W. Shi, W. Yang, D. Z. Kong, K. P. Duan and Q. Wang, J. Ethnopharmacol., 2010, 130, 407–413 CrossRef CAS PubMed.
- X. Li, J. Shen, Z. Zhong, J. Peng, H. Wen, J. Li, Q. Luo and W. Wei, Parasitology, 2010, 137, 1213–1225 CrossRef CAS PubMed.
- A. Manayi, S. Omidpanah, D. Barreca, S. Ficarra, M. Daglia, S. F. Nabavi and S. M. Nabavi, Phytochem. Rev., 2017, 16, 1173–1181 CrossRef CAS.
- K. S. Suh, E. M. Choi, Y. S. Lee and Y. S. Kim, Fitoterapia, 2013, 89, 33–41 CrossRef CAS PubMed.
- S. Parker, B. May, C. Zhang, A. L. Zhang, C. J. Lu and C. C. Xue, Phytother Res., 2016, 30, 1445–1473 CrossRef PubMed.
- W. Wang, X. Hu, Z. Zhao, P. Liu, Y. C. Hu, J. P. Zhou, D. F. Zhou, Z. B. Wang, D. Guo and H. Z. Guo, Prog. Neuro-Psychopharmacol. Biol. Psychiatry, 2008, 32, 1179–1184 CrossRef CAS.
- L. S. Teng, Q. F. Meng, J. H. Lu, J. Xie and Z. Z. Wang, Mol. Med. Rep., 2014, 10, 818–824 CrossRef CAS PubMed.
- J. E. Mersereau, N. Levy, R. E. Staub, S. Baggett, T. Zogric, S. Chow, W. A. Ricke, M. Tagliaferri, I. Cohen and L. F. Bjeldanes, Mol. Cell. Endocrinol., 2008, 283, 49–57 CrossRef CAS PubMed.
- H. Hosseinzadeh and M. Nassiri-Asl, Phytother Res., 2015, 29, 1868–1886 CrossRef PubMed.
- B. Liang, X. L. Guo, J. Jin, Y. C. Ma and Z. Q. Feng, World J. Gastroenterol., 2015, 21, 5271–5280 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra04701e |
| ‡ Co-first authors. |
| This journal is © The Royal Society of Chemistry 2020 |
