Open Access Article
Open Access ArticleAn aptasensor for the label-free detection of thrombin based on turn-on fluorescent DNA-templated Cu/Ag nanoclusters†
Baozhu Zhanga and
Chunying Wei *b
*b
aCollege of Chemistry and Chemical Engineering, Jinzhong University, Yuci 030619, P. R. China
bKey Laboratory of Chemical Biology and Molecular Engineering of Ministry of Education, Institute of Molecular Science, Shanxi University, Taiyuan 030006, P. R. China. E-mail: weichuny@sxu.edu.cn
First published on 24th September 2020
Abstract
A highly sensitive thrombin aptasensor was constructed based on the alteration of the aptamer conformation induced by the target recognition and the turn-on fluorescence due to the proximity of two darkish DNA-templated copper/silver nanoclusters (DNA-Cu/Ag NCs). Two DNA templates were designed as the functional structures consisting of the Cu/Ag NC-nucleation segment located at two termini or one terminus and the aptamer segment in the middle of a DNA template. Two darkish DNA-Cu/Ag NCs came close to each other when the aptamer combined with the target due to the conformational alteration of the aptamer structure, resulting in an increased fluorescence signal readout. Thrombin was sensitively determined as low as 1.6 nM in the range of 1.6–8.0 nM with a high selectivity. Finally, this sensor succeeded in detecting thrombin in a real fetal bovine serum.
1 Introduction
Thrombin, as a specific serine endoprotease, has attracted increasing attention due to not only playing important roles in numerous pathological and physiological processes including angiogenesis, thrombosis, blood coagulation cascade and tumor growth haemostasis,1–3 but also regulating platelet aggregation, endothelial cell activation, and as a hormone other important responses in vascular biology.4–6 Therefore, different analysis methods for the detection of thrombin have been reported over the past few years, for instance, colorimetry,7 fluorescence,8–10 surface plasmon resonance (SPR),11 surface enhanced Raman spectroscopy (SERS),12 electrochemistry,13–16 and electrochemiluminescence (ECL).17 Although these assays have many advantages including sensitivity, specificity and accuracy, these processes are time consuming and inevitably suffer from expensive instrumentation and some of them need expensive labeling substances. Therefore, it is essential to develop a label-free, simple, rapid, convenient and low-price method to detect thrombin selectively with high sensitivity.Aptamers possess great promise for the biosensing of macromolecules or low-molecular-weight substrates owing to their good stability, high specificity, cost-effectiveness, and relative ease of isolation and modification.18–20 Various optical,21–23 electrochemical,24–26 and atomic force microscopy (AFM)27 aptasensors have been developed in the past decade. Among these aptasensors, some require labeling the aptamers using dye molecules for achieving high sensitivity, or functionalizing the aptamers with various functional groups (such as –NH2 and –SH) to fabricate the sensing interface. However, such labeling processes suffer from complicated experimental preparations, expensiveness, and affect the binding affinity between the aptamers and their targets to a certain degree. On the other hand, some of available label-free optical aptasensors usually employ organic fluorescent dyes as a signal transduction medium. However, most of these organic fluorescent dyes are poisonous. Therefore, the fabrication of an aptasensing platform possessing the advantages of high sensitivity, low toxicity, and label-free is still a challenge.
In recent years, DNA-stabilized metal nanoclusters (DNA-NCs), as an emerging alternative to organic fluorophores, have been extensively exploited in fluorescent sensors for the assay of various chemical or biological analytes.28 In contrast with organic fluorophores or quantum dots, DNA-NCs possess a series of advantages, including excellent stability, good biocompatibility, low toxicity, ultra-small size, large Stokes shift, and a strong fluorescence emission.29–32 Moreover, the quantum yield and fluorescence emission of DNA-NCs can be altered by adjusting the length and sequence of DNA. Hence, DNA-NCs can be designed as fluorescence probes for the assay of metal ions,33,34 various biomolecules34,35 and in vitro or in vivo imaging.5,36
It was reported that the fluorescence of non-emissive DNA-templated Ag NCs (DNA-Ag NCs) can significantly enhance in proximity to each other or by interacting with different enhancer sequences.37,38 Motivated by this view, an interesting aptasensor was constructed based on DNA-Cu/Ag NCs for thrombin detection. As shown in Scheme 1, DNA templates consist of a Cu/Ag NC-nucleation segment located at the two termini and an aptamer segment of thrombin is in the middle. The aptamer combines with the target in the presence of thrombin, and two dark coloured DNA-Cu/Ag NCs get close each other due to the conformational alteration of the aptamer, resulting in an increase in fluorescence intensity of DNA-Cu/Ag NCs. Therefore, the proposed DNA-Cu/Ag NC sensing system provides a turn-on and label-free strategy for detecting thrombin.
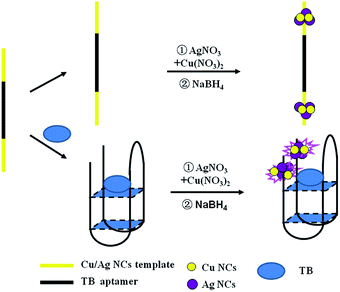 | ||
| Scheme 1 Schematic of the sensing strategy for the detection of thrombin based on ssDNA-templated copper/silver nanoclusters combined with a thrombin aptamer. | ||
2 Experimental
2.1 Reagents and apparatus
Oligonucleotides employed in this assay were purchased from Sangon Biotechnology Inc. (Shanghai, China), and the sequences and names of DNA are displayed in ESI Table S1.† Thrombin (TB), immunoglobulin G (IgG), bovine serum albumin (BSA) and human serum albumin (HSA) were purchased from Sigma Aldrich (China). Fetal bovine serum was bought from Beijing Solarbio Biotechnology Co. Ltd. (China). Cysteine (Cys), glutamic acid (Glu), AgNO3, Cu(NO3)2 and NaBH4 were obtained from Aladdin Bio-chem technology Co. Ltd. (Shanghai, China). All chemical reagents mentioned above were of analytical grade and directly used as received without further purification. Tris-HAc buffer (10 mM, pH 7.0) was used as the working buffer in all of the experiments. Milli-Q water (18.2 MΩ cm) was utilized in all solutions.Fluorescence measurements were obtained using a Hitachi F-4600 fluorescence spectrophotometer at room temperature, and the excitation and emission slit widths were 5 nm and 10 nm, respectively. UV-vis absorption spectra were recorded using a Cary 50 Bio spectrophotometer (Varian Inc., CA) at room temperature. Time-resolved fluorescence measurements were performed using an FL920 fluorescent lifetime spectrometer (Edinburgh Instruments, Livingston, UK), operating in the time-correlated single photon counting (TCSPC) mode using a semiconductor laser (405 nm) as the excitation source. A commercial software by Edinburgh Instruments was used for data analyses. When 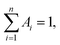 the average excited-state lifetime is expressed by the equation
the average excited-state lifetime is expressed by the equation 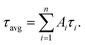 The average size and morphological measurements of TBA1-Cu/Ag NCs were performed on a JEOL JEM-2100 transmission electron microscope at an acceleration voltage of 200 kV. Circular dichroism (CD) spectra were obtained on a Chirascan Circular Dichroism Spectrometer (Applied Photophysics Ltd., Surrey, UK) at room temperature, and the spectra were recorded within the range of 220–320 nm at 1 nm intervals employing a 1 mm optical path-length quartz cell at an instrument scanning speed of 120 nm min−1. The FTIR spectrum of TBA1-Cu/Ag NCs was recorded in the range 4000–500 cm−1 on a Bruker ALPHA spectrometer. X-ray photoelectron spectroscopy (XPS) (ESCAL-ab 220i-XL, VG Scientific, England) was performed using a monochromic Al Ka as source at 1486.6 eV.
The average size and morphological measurements of TBA1-Cu/Ag NCs were performed on a JEOL JEM-2100 transmission electron microscope at an acceleration voltage of 200 kV. Circular dichroism (CD) spectra were obtained on a Chirascan Circular Dichroism Spectrometer (Applied Photophysics Ltd., Surrey, UK) at room temperature, and the spectra were recorded within the range of 220–320 nm at 1 nm intervals employing a 1 mm optical path-length quartz cell at an instrument scanning speed of 120 nm min−1. The FTIR spectrum of TBA1-Cu/Ag NCs was recorded in the range 4000–500 cm−1 on a Bruker ALPHA spectrometer. X-ray photoelectron spectroscopy (XPS) (ESCAL-ab 220i-XL, VG Scientific, England) was performed using a monochromic Al Ka as source at 1486.6 eV.
2.2 Synthesis of DNA-Cu/Ag NCs and determination of thrombin
DNA-Cu/Ag NCs were synthesized according to a previously published protocol with a minor modulation.39 Briefly, DNA oligonucleotides (1.5 μM) with different concentrations (0–11.2 nM) of thrombin in the Tris-HAc buffer (10 mM, pH 7.0) were first mixed and incubated for 0.5 h. AgNO3 and Cu(NO3)2 were sequentially added, and the mixture was kept in dark for 20 min at 4 °C. Then, Ag+ and Cu2+ were reduced by the freshly prepared NaBH4 and the solution was shaken vigorously for 1 min. The molar ratio of the final concentrations of DNA, Cu(NO3)2, AgNO3, and NaBH4 was equal 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8
8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20. The final mixture solution was kept away from light for 1 h at 4 °C. Furthermore, the fluorescence spectrum of each sample was recorded at ambient temperature.
20. The final mixture solution was kept away from light for 1 h at 4 °C. Furthermore, the fluorescence spectrum of each sample was recorded at ambient temperature.
2.3 Specificity of thrombin
To confirm whether different agents could interfere with the assay of thrombin, 5 interfering agents such as HSA, BSA, IgG, Cys, and Glu were used, and their concentration used was 16 nM, while the concentration of thrombin is 8.0 nM. The assay method was same as that of just thrombin.2.4 Circular dichroism measurements
DNA-Cu/Ag NCs were prepared in terms of the method described above (see section about the Detection of thrombin). The concentration of DNA oligonucleotides was 10 μM, and the ratio of the concentration of DNA, Cu(NO3)2, AgNO3, and NaBH4 was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8
8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20. CD spectra of DNA-Cu/Ag NCs without and with 8 nM thrombin were measured, respectively.
20. CD spectra of DNA-Cu/Ag NCs without and with 8 nM thrombin were measured, respectively.
2.5 Application of the proposed sensor
To demonstrate the practical application of the TBA1-Cu/Ag NC detection system, thrombin in a fetal bovine serum solution were detected using this sensing system. The samples were spiked with thrombin at different concentration levels, and the actual samples were measured using the same approach as described above.3 Results and discussion
3.1 Optical characterization of DNA-Cu/Ag NCs
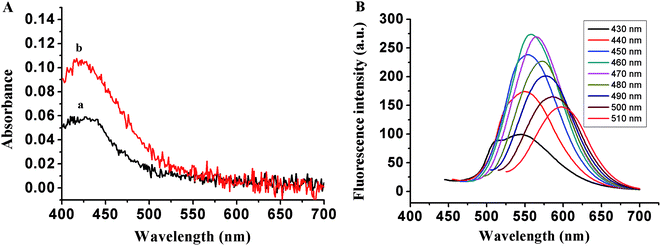
To fabricate the DNA-Cu/AgNC probe for the detection of thrombin, two different DNA templates, namely TBA1 and TBA2, were designed according to our previous research work,40 and their names are (ESI Table S1†). Among these the middle segment of the template (italic) is the aptamer of thrombin, while the 5′ and/or 3′ ends (bold) are the C-rich segments stabilized by DNA-Cu/AgNCs, and the TTTTT linker (underline) is inserted between the aptamer and C-rich sequences. As shown in Fig. 1A, only an obvious peak at 430 nm appears in UV-Vis spectra of TBA1-Cu/Ag NCs alone (curve a) and with 8.0 nM thrombin (curve b) due to the characteristic plasmon absorption band of nanoparticles.41,42 In addition, the absorption intensity of the latter is far stronger than that of the former at 430 nm, and the spectrum of the latter blue shifts a little owing to the binding of the aptamer to thrombin. Fig. 1B demonstrates that the fluorescence emission spectra of TBA1-Cu/Ag NCs depends on their excitation wavelengths, and the maximum excitation and emission wavelengths are 460 and 560 nm, respectively. In addition, the optical characterizations of TBA2-Cu/Ag NCs were also measured (EMS Fig. S1†). Its absorption and fluorescence spectra are similar to those of TBA1-Cu/Ag NCs, while the fluorescence intensity of TBA2-Cu/Ag NCs is stronger than that of TBA1-Cu/AgNCs.The FTIR spectrum of TBA1-Cu/Ag NCs was recorded in the range of 4000–500 cm−1 on a Bruker ALPHA spectrometer. As shown in ESI Fig. S2,† the bands at 3421.8, 1629.8, 1047.3 cm−1 are ascribed to the stretching vibrations of N–H (O–H), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, and C–O bonds in G, C, T, and A bases of DNA (TBA1) (no O–H bond in A base), respectively. Additionally, the absorption peak at 2941.5 and 1386.8 cm−1 can be assigned to the stretching and bending vibrations of C–H (methyl) in the T base of DNA (TBA1), respectively. Therefore, indicating that N–H, O–H, C
C, and C–O bonds in G, C, T, and A bases of DNA (TBA1) (no O–H bond in A base), respectively. Additionally, the absorption peak at 2941.5 and 1386.8 cm−1 can be assigned to the stretching and bending vibrations of C–H (methyl) in the T base of DNA (TBA1), respectively. Therefore, indicating that N–H, O–H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C–O and C–H bonds exist in the DNA (TBA1) template of DNA-Cu/Ag NCs.
C, C–O and C–H bonds exist in the DNA (TBA1) template of DNA-Cu/Ag NCs.
The stability of the probe has a great influence on the detection property. Therefore, the changes in their fluorescence intensities against the storage time were measured. As shown in ESI Fig. S3,† TBA1-Cu/Ag NCs needed about 20 min to reach the maximum emission intensity and maintain that for about 60 min, while the fluorescence of TBA2-Cu/Ag NCs became stable after 30 min and the stability time was only 30 min. Hence, the stability of TBA1-Cu/Ag NCs was better than that of TBA2-Cu/Ag NCs.
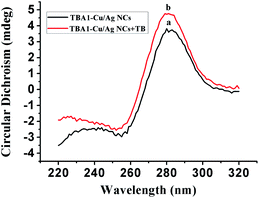
To confirm the conformation of TBA1-Cu/Ag NCs, the CD spectra of TBA1 and TBA1-Cu/Ag NCs were recorded. As shown in Fig. 2, both the CD spectra of TBA1 alone (curve a) and TBA1-Cu/Ag NCs alone (curve b) present a negative band at 255 nm and a broad positive band centered at 280 nm. However, the CD spectrum of TBA1-Cu/Ag NCs shifted about 1 mdeg, and the intensity also became a little stronger owing to the formation of Cu/Ag NCs at the C-rich sequences of the TBA1 template, indicating that TBA1-Cu/Ag NCs suffered from a slight conformation change in TBA1.
3.2 Optimization of the experimental conditions for thrombin determination
3.3 TEM of TBA1-Cu/Ag NCs
TEM images were obtained to characterize the size of TBA1-Cu/Ag NCs alone and with 8.0 nM thrombin. As shown in Fig. 3, the distribution of Cu/Ag NCs is uniform and the average size is about 2 nm (the inset of Fig. 3A and B), which meets the requirement of the core size of metal NCs being below 2.0 nm.45 Compared with the size distribution histogram displayed in the insets, the number of TBA1-Cu/Ag NCs with 8.0 nM thrombin in the range of 1–2 nm and 2–3 nm is a little more than that of TBA1-Cu/Ag NCs alone, indicating that the size of TBA1-Cu/Ag NCs may have altered slightly owing to the change in the microenvironment in the presence of thrombin.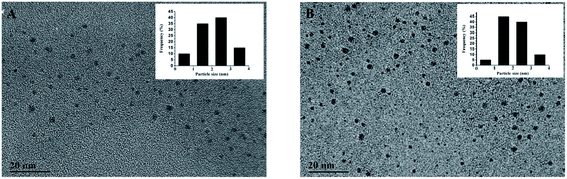 | ||
| Fig. 3 TEM images of TBA1-Cu/Ag NCs alone (A) and with 8.0 nM thrombin (B). The inset shows the size distribution histogram. | ||
3.4 XPS of TBA1-Cu/Ag NCs
The presence of P, B, C, Ag, N, Na, O and Cu atoms in the TBA1-Cu/Ag NCs were revealed by the XPS spectrum (Fig. 4A). As shown in Fig. 4B, the binding energy values at 368.2 eV and 374.3 eV belong to Ag 3d5/2 and Ag 3d3/2 in the expanded spectrum of Ag 3d, and is attributed to Ag (0) in the TBA1-Cu/Ag NCs, respectively.46,47 In addition, two peaks are observed in the magnified spectrum of Cu 2p (Fig. 4C), and the binding energy peak at 932.6 eV for Cu 2p3/2 suggests the presence of Cu(I) and/or Cu(0) in the TBA1-Cu/Ag NCs.48,49 Cu(I) and Cu(0) species cannot be differentiated by Cu 2p3/2 XPS due to a small separation of 0.1 eV.49 The peak at 952.7 eV corresponds to the literature value of 952.2 eV (Cu 2p1/2),43,48 which speculates the presence of Cu(0) in TBA1-Cu/Ag NCs.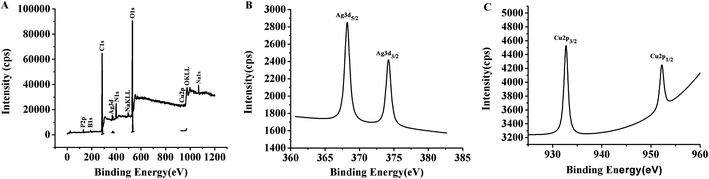 | ||
| Fig. 4 (A) XPS spectra of TBA1-Cu/Ag NCs. (B) Ag 3d region of XPS spectrum of TBA1-Cu/Ag NCs. (C) Cu 2p region of XPS spectrum of TBA1-Cu/Ag NCs. | ||
3.5 Detection of thrombin
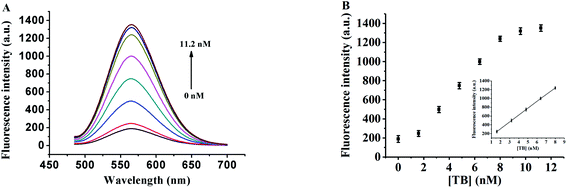
In order to evaluate the detection capability of the proposed sensor, different concentrations of thrombin were separately added into TBA1 and incubated for 0.5 h, and then TBA1-Cu/Ag NCs were synthesized (see section about the Detection of thrombin) and the fluorescence emission spectrum of each of these sample was collected under the optimal experimental conditions. Fig. 5 shows that the fluorescence intensity of TBA1-Cu/Ag NCs increases with the increase in the thrombin concentration. It is seen from the calibration plot present in the inset of Fig. 5B that the fluorescence intensity linearly depends on the thrombin concentration from 1.6 to 8.0 nM. Its regression equation is F = 0.1222 + 124.3Cthrombin (R = 0.9999). The limit of detection (LOD) is as low as 1.6 nM. As shown in ESI Table S2,† the lowest LOD is 30 pM for other methods,9,10,50–53 wherein the detection is carried out using exonuclease I-assisted target recycling and SYBR Green I-aided signal amplification,9 while the other method with the LOD of 31.3 pM is based on the dye-labeled aptamer-assembled graphene.10 These methods improve the sensitivity; however, SYBR Green I is a toxic organic dye while labeling an aptamer is expensive and time consuming. The LOD of the proposed method is 1.6 nM, which is lower than that observed from UV-Vis absorption in ESI Table S2,† demonstrating the immense potential of implemented DNA-Cu/Ag NCs as a fluorescence biosensing platform in biological systems.Furthermore, the fluorescence lifetimes of TBA1-Cu/Ag NCs alone and with different concentrations of thrombin were investigated at 560 nm of the emission wavelength of TBA1-Cu/Ag NCs (ESI Fig. S6†). The fluorescence transients of TBA1-Cu/Ag NCs display the tri-exponential time constants (ESI Table S3†) with an average lifetime of 3.8 ns. The fluorescence lifetimes of TBA1-Cu/Ag NCs remain almost unchanged with the addition of thrombin; therefore, the results indicate that the interaction between thrombin and Cu/Ag NCs is a static process.
3.6 Specificity of the DNA-Cu/Ag NCs towards thrombin
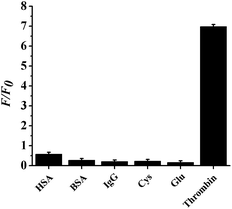
The specificity of the probe depends on the high specific binding affinity between the aptamer and the target.54 For the evaluation of the specificity of the proposed method, thrombin and the effect of 5 interfering agents, including HSA, BSA, IgG, Cys, and Glu were investigated while carrying out the assay of thrombin, and the results are exhibited in Fig. 6. Only thrombin causes an obvious enhancement in the relative fluorescent intensity (F/F0), while the interfering agents hardly cause any change. Hence, the interfering agents almost have no effect on the detection of thrombin, indicating that the proposed probe has high specificity.3.7 Application of the proposed sensor
To verify the accuracy and practical utility of the proposed probe in actual samples, 1.6, 3.2, 4.8, and 6.4 nM thrombin were separately spiked in a 100-fold diluted fetal bovine, serum and the fluorescence intensity was determined by the proposed sensor. Table 1 shows that the recoveries vary in the range of 98.3% to 105.0%, and the relative standard deviations are within the range of 1.22–6.19%. This indicates that the proposed probe can be exploited in actual samples to detect thrombin reliably and accurately and has high precision. Furthermore, the results can be compared with those of the previous reports.52,554 Conclusions
In summary, an interesting “fluorescence turn-on” and label-free aptasensor for the detection of thrombin was constructed based on DNA-Cu/Ag NCs. Not only is the assay very simple in operation, but also the proposed sensor is inexpensive and convenient, and avoids chemical modification, enzymatic reaction, organic co-solvents and sophisticated instrumentals. It presents a high sensitivity and selectivity, with an LOD of 1.6 nM in the range of 1.6–8.0 nM. In addition, the proposed probe has succeeded in detecting thrombin in the fetal bovine serum. Therefore, this sensor has great potential application in the fields of clinical diagnosis and biochemical research.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21171108), the Innovation Team Construction Plan of “1331 Project” of Jinzhong University (jzxycxtd2019010 and jzxycxtd2019007), the Startup Foundation of Doctors of Jinzhong University, and the Transformation of Scientific and Technological Achievements Programs of Higher Education Institutions in Shanxi (2019KJ008).Notes and references
- Z. Chen, Y. Tan, C. Zhang, L. Yin, H. Ma, N. Ye, H. Qiang and Y. Lin, Biosens. Bioelectron., 2014, 56, 46–50 CrossRef CAS.
- Q. Zhao and J. Gao, Chem. Commun., 2013, 49, 7720–7722 RSC.
- L. Yang, J. Zhu, Y. Xu, W. Yun, R. Zhang, P. He and Y. Fang, Electroanalysis, 2011, 23, 1007–1012 CrossRef CAS.
- Y. Xu, W. Zhou, M. Zhou, Y. Xiang, R. Yuan and Y. Chai, Biosens. Bioelectron., 2015, 64, 306–310 CrossRef CAS.
- J. Qiao, X. Mu, L. Qi, J. Deng and L. Mao, Chem. Commun., 2013, 49, 8030–8032 RSC.
- Y. Kitamoto, E. Nakamura, S. Kudo, H. Tokunaga, E. Murakami, K. Noguchi and T. Imamura, Clin. Chim. Acta, 2008, 398, 159–160 CrossRef CAS.
- Y. Huang, J. Chen, S. Zhao, M. Shi, Z. F. Chen and H. Liang, Anal. Chem., 2013, 85, 4423–4430 CrossRef CAS.
- J. Liu, J. Li, Y. Jiang, S. Yang, W. Tan and R. Yang, Chem. Commun., 2011, 47, 11321–11323 RSC.
- X. Chen, T. Li, X. Tu and L. Luo, Sens. Actuators, B, 2018, 265, 98–103 CrossRef CAS.
- H. Chang, L. Tang, Y. Wang, J. Jiang and J. Li, Anal. Chem., 2010, 82, 2341–2346 CrossRef CAS.
- L. Wang, C. Zhu, L. Han, L. Jin, M. Zhou and S. Dong, Chem. Commun., 2011, 47, 7794–7796 RSC.
- H. Cho, B. R. Baker, S. Wachsmann-Hogiu, C. V. Pagba, T. A. Laurence, S. M. Lane, L. P. Lee and J. B. H. Tok, Nano Lett., 2008, 8, 4386–4390 CrossRef CAS.
- Y. Li, L. Liu, X. Fang, J. Bao, M. Han and Z. Dai, Electrochim. Acta, 2012, 65, 1–6 CrossRef CAS.
- Y. Wu, W. Xu, L. Bai, Y. Yuan, H. Yi, Y. Chai and R. Yuan, Biosens. Bioelectron., 2013, 50, 50–56 CrossRef CAS.
- J. Li, J. Wang, X. Guo, Q. Zheng, J. Peng, H. Tang and S. Yao, Anal. Chem., 2015, 87, 7610–7617 CrossRef CAS.
- Y.-P. Dong, T.-T. Gao, Y. Zhou and J.-J. Zhu, Anal. Chem., 2014, 86, 11373–11379 CrossRef CAS.
- Y. Xiao, B. D. Piorek, K. W. Plaxco and A. J. Heeger, J. Am. Chem. Soc., 2005, 127, 17990–17991 CrossRef CAS.
- S. P. Song, L. H. Wang, J. Li, J. L. Zhao and C. H. Fan, TrAC, Trends Anal. Chem., 2008, 27, 108–117 CrossRef CAS.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS.
- A. B. Iliuk, L. H. Hu and W. A. Tao, Anal. Chem., 2011, 83, 4440–4452 CrossRef CAS.
- D. Tang, D. L. Liao, Q. K. Zhu, F. Y. Wang, H. P. Jiao, Y. J. Zhang and C. Yu, Chem. Commun., 2011, 47, 5485–5487 RSC.
- Z. W. Tang, P. Mallikaratchy, R. H. Yang, Y. M. Kim, Z. Zhu, H. Wang and W. H. Tan, J. Am. Chem. Soc., 2008, 130, 11268–11269 CrossRef CAS.
- R. Z. Nutiu and Y. F. Li, J. Am. Chem. Soc., 2003, 125, 4771–4778 CrossRef CAS.
- Y. Liu, N. Tuleouva, E. Ramanculov and A. Revzin, Anal. Chem., 2010, 82, 8131–8136 CrossRef CAS.
- A. A. Rowe, E. A. Miller and K. W. Plaxco, Anal. Chem., 2010, 82, 7090–7095 CrossRef CAS.
- Y. L. Zhang, Y. Huang, J. H. Jiang, G. L. Shen and R. Q. Yu, J. Am. Chem. Soc., 2007, 129, 15448–15449 CrossRef CAS.
- B. Basnar, R. Elnathan and I. Willner, Anal. Chem., 2006, 78, 3638–3642 CrossRef CAS.
- L. Zhang, R.-P. Liang, S.-J. Xiao, J.-M. Bai, L.-L. Zheng, L. Zhan, X.-J. Zhao, J.-D. Qiu and C.-Z. Huang, Talanta, 2014, 118, 339–347 CrossRef CAS.
- N. Enkin, E. Sharon, E. Golub and I. Willner, Nano Lett., 2014, 14, 4918–4922 CrossRef CAS.
- I. Diez, M. I. Kanyuk, A. P. Demchenko, A. Walther, H. Jiang, O. Ikkala and R. H. A. Ras, Nanoscale, 2012, 4, 4434–4437 RSC.
- K. S. Park and H. G. Park, Curr. Opin. Biotechnol., 2014, 28, 17–24 CrossRef CAS.
- C. Lu, Q. Su and X. Yang, Nanoscale, 2019, 11, 16036–16042 RSC.
- H. Ding, C. Liang, K. Sun, H. Wang, J. K. Hiltunen, Z. Chen and J. Shen, Biosens. Bioelectron., 2014, 59, 216–220 CrossRef CAS.
- Y. Qiao, Y. Zhang, C. Zhang, L. Shi, G. Zhang, S. Shuang, C. Dong and H. Ma, Sens. Actuators, B, 2016, 224, 458–464 CrossRef CAS.
- S. N. Ding, C. M. Li and N. Bao, Biosens. Bioelectron., 2015, 64, 333–337 CrossRef CAS.
- X. L. Hu, X. M. Wu, X. Fang, Z. J. Li and G. L. Wang, Biosens. Bioelectron., 2016, 77, 666–672 CrossRef CAS.
- B.-C. Yin, J.-L. Ma, H.-N. Le, S. Wang, Z. Xu and B.-C. Ye, Chem. Commun., 2014, 50, 15991–15994 RSC.
- H.-C. Yeh, J. Sharma, I.-M. Shih, D. M. Vu, J. S. Martinez and J. H. Werner, J. Am. Chem. Soc., 2012, 134, 11550–11558 CrossRef CAS.
- Y.-T. Su, G.-Y. Lan, W.-Y. Chen and H.-T. Chang, Anal. Chem., 2010, 82, 8566–8572 CrossRef CAS.
- B. Zhang and C. Wei, Anal. Bioanal. Chem., 2020, 412, 2529–2536 CrossRef CAS.
- X. Liu, R. Hu, Z. Gao and N. Shao, Langmuir, 2015, 31, 5859–5867 CrossRef CAS.
- E. Shaviv, O. Schubert, M. Alves-Santos, G. Goldoni, R. Di Felice, F. Vallée, N. Del Fatti, U. Banin and C. Sönnichsen, ACS Nano, 2011, 5, 4712–4719 CrossRef CAS.
- K. Loo, N. Degtyareva, J. Park, B. Sengupta, M. Reddish, C. C. Rogers, A. Bryant and J. T. Petty, J. Phys. Chem. B, 2010, 114, 4320–4326 CrossRef CAS.
- E. G. Gwinn, P. O'Neill, A. J. Guerrero, D. Bouwmeester and D. K. Fygenson, Adv. Mater., 2008, 20, 279–283 CrossRef CAS.
- X.-R. Song, N. Goswami, H.-H. Yang and J. Xie, Analyst, 2016, 114, 3126–3140 RSC.
- Q. Zhou, Y. Lin, M. Xu, Z. Gao, H. Yang and D. Tang, Anal. Chem., 2016, 88, 8886–8892 CrossRef CAS.
- R. B. Platzman, H. Haick and R. Tannenbaum, J. Phys. Chem. C, 2008, 112, 1101–1108 CrossRef.
- K. L. Deutsch and B. H. Shanks, J. Catal., 2012, 285, 235–241 CrossRef CAS.
- P. Liu and E. J. Hensen, J. Am. Chem. Soc., 2013, 135, 14032–14035 CrossRef CAS.
- Y. Bai, F. Feng, L. Zhao, C. Wang, H. Wang, M. Tian, J. Qin, Y. Duan and X. He, Biosens. Bioelectron., 2013, 47, 265–270 CrossRef CAS.
- Y. Wang, L. Bao, Z. Liu and D.-W. Pang, Anal. Chem., 2011, 83, 8130–8137 CrossRef CAS.
- J. Feng, C. Zhuo, X. Ma, S. Li and Y. Zhang, Analyst, 2016, 141, 4481–4487 RSC.
- G. Liang, S. Cai, P. Zhang, Y. Peng, H. Chen, S. Zhang and J. Kong, Anal. Chim. Acta, 2011, 689, 243–249 CrossRef CAS.
- C.-H. Lu, F. Wang and I. Willner, Chem. Sci., 2012, 3, 2616–2622 RSC.
- Y. Zhu, X. C. Hu, S. Shi, R. R. Gao, H. L. Huang, Y. Y. Zhu, X. Y. Lv and T. M. Yao, Biosens. Bioelectron., 2016, 79, 205–212 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra04609d |
| This journal is © The Royal Society of Chemistry 2020 |