Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synthesis and physicochemical, DFT, thermal and DNA-binding analysis of a new pentadentate N3S2 Schiff base ligand and its [CuN3S2]2+ complexes
Ismail Warad *a,
Hadeel Subohb,
Nabil Al-Zaqri
*a,
Hadeel Subohb,
Nabil Al-Zaqri cd,
Ali Alsalme
cd,
Ali Alsalme c,
Fahad A. Alharthi
c,
Fahad A. Alharthi c,
Meshari M. Aljohanie and
Abdelkader Zarroukf
c,
Meshari M. Aljohanie and
Abdelkader Zarroukf
aDepartment of Chemistry and Earth Sciences, Qatar University, PO Box 2713, Doha, Qatar. E-mail: ismail.warad@qu.edu.qa
bDepartment of Chemistry, Science College, An-Najah National University, P.O. Box 7, Nablus, Palestine
cDepartment of Chemistry, College of Science, King Saud University, P.O. Box 2455, Riyadh 11451, Saudi Arabia
dDepartment of Chemistry, College of Science, Ibb University, P.O. Box 70270, Ibb, Yemen
eDepartment of Chemistry, Faculty of Science, University of Tabuk, Tabuk-71491, Saudi Arabia
fLaboratory of Materials, Nanotechnology and Environment, Faculty of Sciences, Mohammed V University, Av. Ibn Battouta, Box 1014, Agdal-Rabat, Morocco
First published on 10th June 2020
Abstract
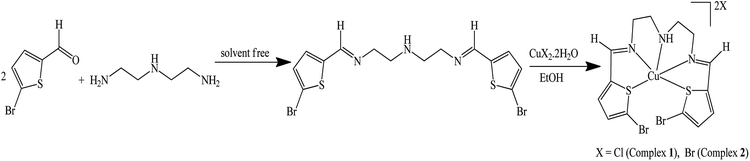
A new N3S2 pentadentate Schiff base ligand derived from 5-bromothiophene-2-carbaldehyde, (E)-N1-((5-bromothiophen-2-yl)methylene)-N2-(2-((E)-((5-bromothiophen-2-yl)-methylene amino) ethyl ethane-1,2-diamine, is prepared. The ligand and its complexes are subjected to extensive physical and theoretical analyses and the results are consistent with their predicted compositions. Dicationic Cu(II) complexes ([CuN3S2]X2) with a coordination number of 5 are proposed on the basis of the spectral data with N3S2 serving as a pentadentate ligand. The prepared complexes display a square pyramidal geometry around the Cu(II) center. TG shows different thermal behavior for the N3S2 ligand and its complexes. Solvatochromism of the complexes is promoted by the polarity of the solvent used. A one-electron transfer Cu(II)/Cu(I) reversible redox reaction is promoted by CV. SEM and EDS of the free ligand and its complexes support the morphology and composition changes observed upon the complexation of Cu(II). As an outstanding goal to develop anticancer new metal chemotherapy, preliminary studies of the binding of the desired complexes with DNA were carried out, as it is through judging the strength of interactions that a future drug can be designed and synthesized. The viscosity and absorption results obtained for complex 1 indicated its enhanced CT-DNA binding properties as compared to those of complex 2 with Kb values of 3.2 × 105 and 2.5 × 105 M−1, respectively.
1. Background
Multidentate ligands are frequently used in organometallic chemistry because of their poly-bonding ability to coordinate with several metal ions.1–3 Consequently, systems containing multidentate ligands are critical towards the success of coordination studies.3 One of the most famous multidentate ligands are Schiff bases, which have attracted the attention of researchers due to their ability to coordinate to metals via several sites, which can stabilize novel structures around the metal center.3–8 In general, Schiff base-transition metal complexes that have been extensively investigated because the Schiff base ligand can coordinate with metal ion centers via one or more sites leading to the synthesis of several types of complex with different metal centers and stereochemistry, and a broad range of applications.8–10 Recently, several reports have shown that these ligands and their complexes can be used toward the design of new drug candidates exhibiting anti-cancer, enzyme inhibition, anti-malarial, antifungal, antibacterial, and anti-inflammatory activity.8–16Copper is essential in the human body and plays a critical role in biological processes involving electron transfer reactions. In fact, Cu(II) complexes bearing {S, O, N} donor chelating ligands are excellent anti-cancer agents because of their strong DNA binding ability.4–10 Due to the highly selective permeability of copper(II) ions through the cell membrane of cancer cells, copper is considered to be one of the most effective anti-tumor agents with low cost and few side effects.7–16 Thus, various complexes bearing several types of ligand have been prepared and evaluated against cancer cells.12–24 Pentadentate Schiff base ligands have received less attention as compared to mono-, di-, tri-, and tetradentate ligands due to the difficulty in their synthesis and unexpected multimode coordination behavior.15–24 In view of the several coordination modes exhibited by pentadentate N3S2 ligands derived from thiophene, their interesting structures, and the CT-DNA binding affinity of their Cu(II) complexes, we herein report two mononuclear copper(II) complexes obtained using a new pentadentate N3S2 ligand. The CT-DNA binding affinity of their corresponding Cu(II) complexes is also evaluated.
2. Experimental
2.1. Material and instrumentation
All chemicals and solvents were purchased from Sigma and used without any further purification. TLC was performed to evaluate the purity of the as-synthesized compounds when needed. Elemental analysis was carried out on an Elementar-Vario EL analyzer. TG/DTG curves were recorded on a Perkin-Elmer thermogravimetric analyzer. FT-IR spectroscopy was recorded on a Perkin-Elmer Spectrum 1000 FT-IR spectrometer. UV-Vis spectroscopy was recorded on a TU-1901double-beam UV-visible spectrophotometer. All electrochemical experiments were carried out at room temperature under an argon atmosphere using a three-electrode cell Voltalab 80 potentio-state PGZ402 equipped with a Pt-electrode (Metrohm, A = 0.0064 cm2) used as the working electrode and platinum wire spiral (£1 mm) with a diameter of 7 mm used as the counter electrode.2.2. Synthesis of N-[(1E)-(5-bromothien-2-yl)methylene]-N′-(2-{[(1E)-(5-bromothien-2-yl)methylene]-amino}ethyl)ethane-1,2-diamine [N3S2]
5-Bromothiophene-2-carbaldehyde (0.026 mol) was added to diethylenetriamine (0.013 mol) in the absence of solvent and the resulting mixture was stirred for 30 min at RT until a viscous oil was formed. The temperature increased and the viscosity of the reaction mixture ensured the condensation reaction occurred. Dichloromethane (10 mL) was added to the reaction mixture and the resulting solution was sealed and stirred for 1 h. A colorless oily product was obtained after evaporation of the solvent.Yield: 85%. The product was a colorless oil at RT. Molecular formula: C14H15Br2N3S2. 1H NMR (250 MHz, CDCl3) δ (ppm): 2.4 (t, 4H, –HNC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH2N
2CH2N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–), 3.9 (t, 4H, –HNCH2C
CH–), 3.9 (t, 4H, –HNCH2C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2N
2N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–), 4.3 (broad s, 1H, HN only observed in free dissolved sample), 6.7 (d, 2H, thiophene), 7.6 (d, 2H, thiophene), 8.2 (s, 2H, –HC
CH–), 4.3 (broad s, 1H, HN only observed in free dissolved sample), 6.7 (d, 2H, thiophene), 7.6 (d, 2H, thiophene), 8.2 (s, 2H, –HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–). 13C NMR δ (ppm): 15.8 (s, 2C, –HN
N–). 13C NMR δ (ppm): 15.8 (s, 2C, –HN![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2CH2N
H2CH2N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–), 61.1 (s, 2C, –HNCH2
CH–), 61.1 (s, 2C, –HNCH2![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2N
H2N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–), 125.6, 130.0, 140.0, 143.0 (d, 8C, thiophene), 156.1 (s, 2C, –HC
CH–), 125.6, 130.0, 140.0, 143.0 (d, 8C, thiophene), 156.1 (s, 2C, –HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–). MS: m/z = 446.2 [M+]. UV-Vis (EtOH) (nm): 240 (sh), 280. IR (cm−1): ν = 3320 (N–H), 3020 (C–H thiophene), 2960–2770 (C–H aliphatic), 1675 (C
N–). MS: m/z = 446.2 [M+]. UV-Vis (EtOH) (nm): 240 (sh), 280. IR (cm−1): ν = 3320 (N–H), 3020 (C–H thiophene), 2960–2770 (C–H aliphatic), 1675 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N).
N).
2.3. Synthesis of complexes 1–2
A solution of N3S2 (0.18 mmol) in EtOH (5 mL) was added to the Cu(II) salt (0.17 mmol) dissolved in freshly distilled EtOH (20 mL). The color of the reaction mixture changed from brown to blue and the temperature increased upon the addition of the ligand solution. The product complex is poorly soluble in EtOH and precipitates from the reaction mixture. A reduction in the volume of the reaction mixture under vacuo led to most of the blue Cu(II) complex being precipitated, which was then filtered and washed with cooled EtOH and ethyl acetate.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1590 (N–H), 1150 (N–C), 520 (Cu–N). UV-Vis (water, nm): 250 (2.2 × 104 M−1 L−1) and 615 (2.8 × 102 M−1 L−1).
N), 1590 (N–H), 1150 (N–C), 520 (Cu–N). UV-Vis (water, nm): 250 (2.2 × 104 M−1 L−1) and 615 (2.8 × 102 M−1 L−1).![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1580 (N–H), 1180 (C–N), 510 (Cu–N). UV-Vis (water, nm): 260 (2.3 × 104 M−1 L−1) and 625 (2.2 × 102 M−1 L−1).
N), 1580 (N–H), 1180 (C–N), 510 (Cu–N). UV-Vis (water, nm): 260 (2.3 × 104 M−1 L−1) and 625 (2.2 × 102 M−1 L−1).2.4. DNA binding experiments
The experimental titration absorption spectra were recorded in Tris–HCl buffer (5 mM Tris–HCl/50 mM NaCl buffer at pH 7.2) using a Cu(II) complex concentration of 5.0 × 10−5 M (complex 1) and 1.0 × 10−5 M (complex 2) throughout the experiment. The CT-DNA concentrations were varied between 0 and 5.0 × 10−5 M (complex 1) and 0 and 1.0 ×10−3 M (complex 2), maintaining the total mixture volume constant at 10.0 mL. The resulting mixed solution of Cu(II)/CT-DNA was allowed to equilibrate for 10 min at RT for each experiment prior to carrying out the absorption measurement.35–392.5. Viscosity experiments
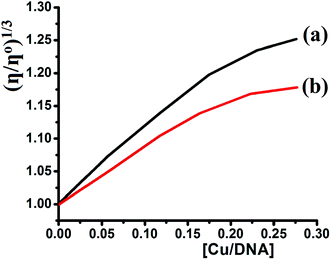
Viscosity experiments were performed on a Ubbelodhe viscometer at 25.0 (±0.1) °C. The flow time was measured using a stopwatch with different concentrations of the complexes (0, 2.5 × 10−5, 6.25 × 10−5, 8.75 × 10−4, 1.12 × 10−4, and 1.37 × 10−4 M) and a fixed concentration of DNA = 5.0 × 10−4 M in Tris–HCl buffer. Each sample was measured in triplicate and the average flow time was calculated. Data are presented as (η/ηo)1/3 versus the binding ratio [Cu]/[DNA],35–41 where η is the viscosity of DNA in the presence of the complex and ηo is the viscosity of the pure DNA solution.3. Results and discussion
3.1. Synthesis
A solvent-free condensation reaction using a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio of 5-bromothiophene-2-carbaldehyde and diethylenetriamine under an air atmosphere rapidly affords a new pentadentate Schiff base ligand, (E)-N1-((5-bromothiophen-2-yl)methylene)-N2-(2-((E)-((5-bromothiophen-2-yl)methylene)amino)ethyl)ethane-1,2-diamine (N3S2), in good yield (Scheme 1).
1 molar ratio of 5-bromothiophene-2-carbaldehyde and diethylenetriamine under an air atmosphere rapidly affords a new pentadentate Schiff base ligand, (E)-N1-((5-bromothiophen-2-yl)methylene)-N2-(2-((E)-((5-bromothiophen-2-yl)methylene)amino)ethyl)ethane-1,2-diamine (N3S2), in good yield (Scheme 1).
The desired Cu(II) complexes were prepared by mixing an equivalent amount of N3S2 with the hydrated CuX2 salt in EtOH at RT under an atmosphere. The preparation of complexes 1 and 2 was confirmed by the color and temperature changes observed in the reaction. The addition of the colorless ligand solution to the copper salt solution was accompanied by a distinct color change from brown to blue and the isolated complexes were characterized using spectroscopic, electrochemical, and thermal analysis.
The complexes are very soluble in coordinating-solvents such as water, DMSO, and DMF, and poorly soluble in alcohols including ethanol, which strongly indicates the complexes are ionic. The molar conductivity of the aqueous complex solutions was 185–210 μS cm−1, which is within the range observed for a [1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1] electrolyte.
1] electrolyte.
The structures of the N3S2 ligand and its complexes were analyzed using EA, MS, FT-IR, CV, NMR, UV-Vis, SEM, EDS, and TG/DTG. DFT calculations on the free ligand were performed using Gaussian 09 software. The analysis of the complexes revealed the construction of [Cu:N3S2]X2-type square pyramid complexes. The mass spectra, conductance, and water solubility of the as-prepared complexes supported their dicationic mononuclear salt nature, as illustrated in Scheme 1.
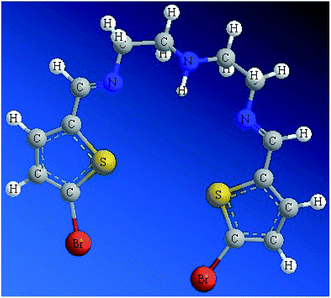
3.2. Optimized structure of N3S2
Since the ligand is an oil at RT, it was not possible to collect a stable and suitable crystal for X-ray analysis. Subsequently, the structure of the N2S2 ligand was optimized at the DFT-B3LYP level of theory, as depicted in Fig. 1. The optimized molecular structure of the N2S2 ligand revealed the (E,E)-isomer of both C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N groups was the kinetically favored isomer with the least internal strict repulsion effect, which forces the S-heterocyclic rings to be in the same plane creating a semi-vacant site center suitable to be occupied by metal ions. The presence of two aromatic rings conjugated to the two C
N groups was the kinetically favored isomer with the least internal strict repulsion effect, which forces the S-heterocyclic rings to be in the same plane creating a semi-vacant site center suitable to be occupied by metal ions. The presence of two aromatic rings conjugated to the two C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N groups results in the increased acidity of the N–H group in the ligand.4–10
N groups results in the increased acidity of the N–H group in the ligand.4–10
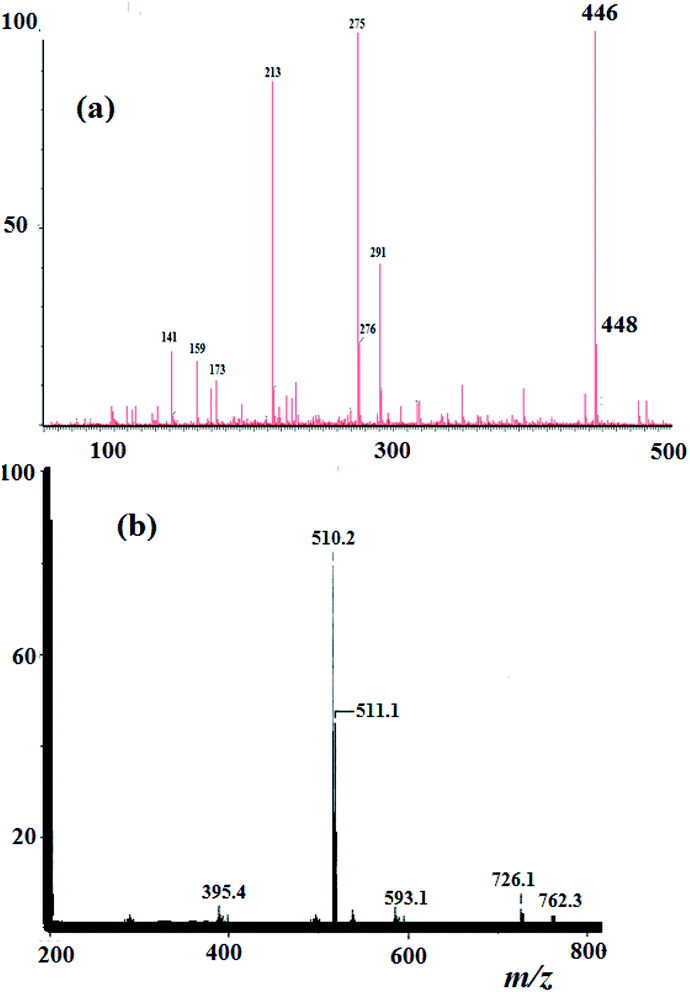
3.3. MS and elemental analysis
Elemental analysis result of the N3S2 ligand and its Cu(ii) complexes was in agreement with their proposed molecular formulae. For the N3S2 ligand: C14H15Br2N3S2, calcd. C, 37.43; H, 3.37%; found: C, 37.25; H, 3.21%. EI-MS of the ligand was in accordance with its assigned structure: m/z = 446.2 [M+] and 448.2 [M+ + 2], (theoretical m/z = 446.9), as shown in Fig. 2a.The ESI-MS data obtained for the complexes are consistent with their proposed formula and support their monomeric dicationic structure. The theoretical m/z value of complex 2 is 509.2 [M+] and was observed experimentally with molecular ion peaks at m/z = 510.2 [M+ + 1] and 511.1 [M+ + 2], which confirm its dicationic mononuclear structure and molecular formula, as shown in Fig. 2b.
3.4. 1H and 13C-NMR spectra of the N3S2 ligand
The experimental 1H-NMR spectrum of the N3S2 ligand was recorded in CDCl3 and shown in Fig. 3a; the theoretical spectrum is depicted in Fig. 3b and a comparison between the theoretical and experimental 1H NMR spectra is shown in Fig. 3c. The 1H NMR spectrum shows two sharp triplet signals at δ = 2.4 and 2.9 ppm corresponding to![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NCH2C
NCH2C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2NHC
2NHC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH2N
2CH2N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) and
and ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NC
NC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH2NHCH2C
2CH2NHCH2C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2N
2N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) , respectively. No signal for the NH proton was detected due to the rapid D/H exchange parallel to the CDCl3/CHCl3 singlet observed at δ = 7.2 ppm (the NH proton appears as a broad singlet peak at δ = 4.3 ppm in the freshly prepared NMR solution of the ligand), which is in consistent with the calculated acidity of the NH group. The thiophene protons were observed as two multiplet peaks at δ = 6.7 and 7.6 ppm, and the azomethine proton (N
, respectively. No signal for the NH proton was detected due to the rapid D/H exchange parallel to the CDCl3/CHCl3 singlet observed at δ = 7.2 ppm (the NH proton appears as a broad singlet peak at δ = 4.3 ppm in the freshly prepared NMR solution of the ligand), which is in consistent with the calculated acidity of the NH group. The thiophene protons were observed as two multiplet peaks at δ = 6.7 and 7.6 ppm, and the azomethine proton (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH) was detected as a singlet at δ = 8.2 ppm.
CH) was detected as a singlet at δ = 8.2 ppm.
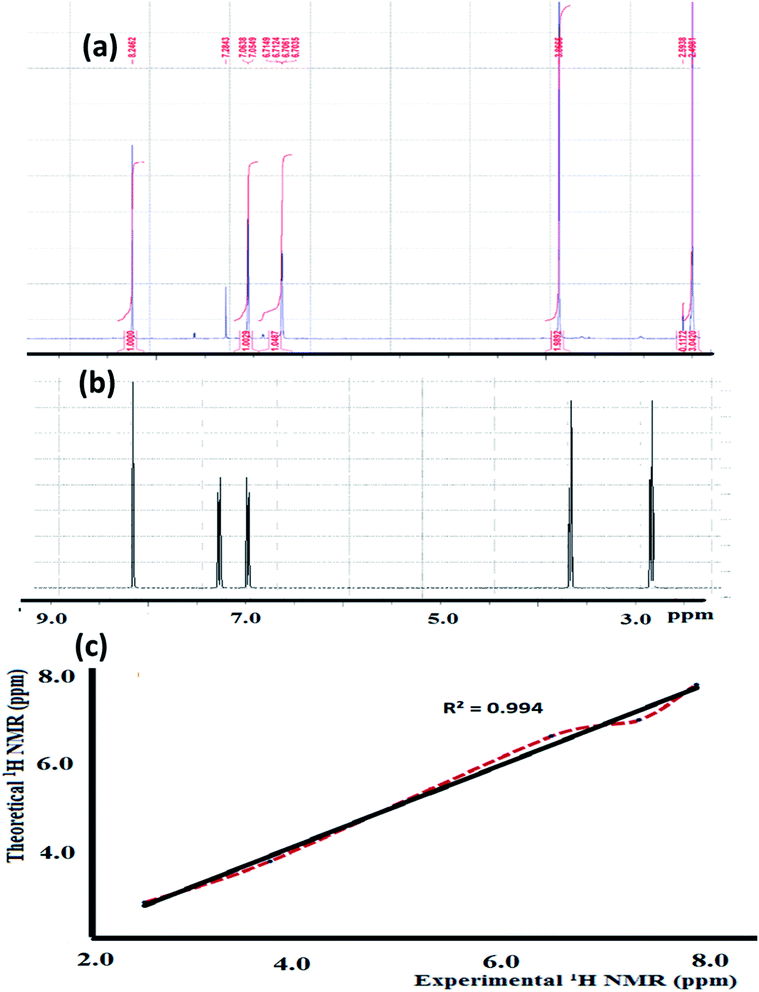 | ||
| Fig. 3 (a) Experimental and (b) theoretical 1H NMR spectrum of the N3S2 ligand recorded in CDCl3 at RT and (c) comparison of the experimental and theoretical spectra. | ||
The theoretical 1H-NMR spectrum shows several peaks belonging to the aliphatic, thiophene, and azomethine protons in the N3S2 ligand, which are consistent with the experimental spectrum. For comparison, the theoretical 1H-NMR spectrum was plotted against the experimental one, as depicted in Fig. 3c. Fig. 3c shows the matched linear relationship between the theoretical and experimental 1H-NMR spectra, which reflects the excellent degree of agreement.
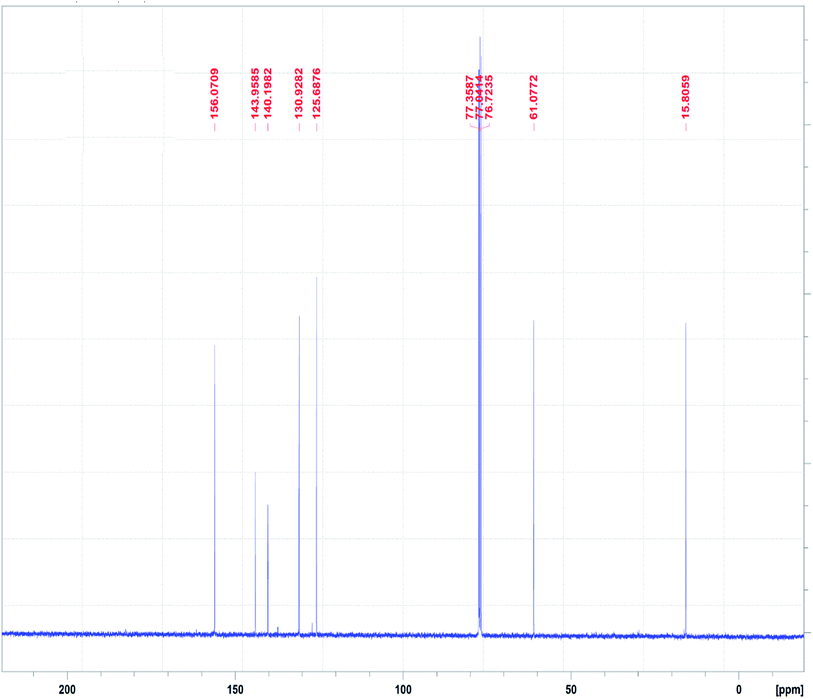
The 13C NMR spectrum of the N3S2 ligand is shown in Fig. 4. The 13C NMR spectrum exhibits two singlet peaks at δ = 15.0 (CH2NH) and 61.1 (CH2N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ) ppm. The four aromatic carbon signals are observed at δ = 125.6, 130.0, 140.0, and 143.0 ppm, and the N
) ppm. The four aromatic carbon signals are observed at δ = 125.6, 130.0, 140.0, and 143.0 ppm, and the N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH signal was observed at δ =156.0 ppm.
CH signal was observed at δ =156.0 ppm.
3.5. FT-IR and DFT-IR spectroscopy
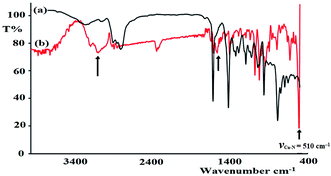
FT-IR spectroscopy was utilized to monitor the condensation reaction during the ligand formation step. The FT-IR spectra of the starting materials, 5-bromothiophene-2-carbaldehyde and N1-(2-aminoethyl)ethane-1,2-diamine, before and after the condensation reaction were recorded (Fig. 5). The formation of the ligand was confirmed by two major changes: (1) the primary N–H stretching vibration in diethylenetriamine observed at 3340 and 3270 cm−1 (Fig. 5a) is reduced to one single peak at 3240 cm−1 due to the formation of the secondary amine in the ligand (Fig. 5b); and (2) the stretching vibration belonging to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group in carbaldehyde observed at 1742 cm−1 is shifted by ∼60 cm−1 due to the formation of the C
O group in carbaldehyde observed at 1742 cm−1 is shifted by ∼60 cm−1 due to the formation of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N– group (1688 cm−1) in the ligand (Fig. 5c).
N– group (1688 cm−1) in the ligand (Fig. 5c).
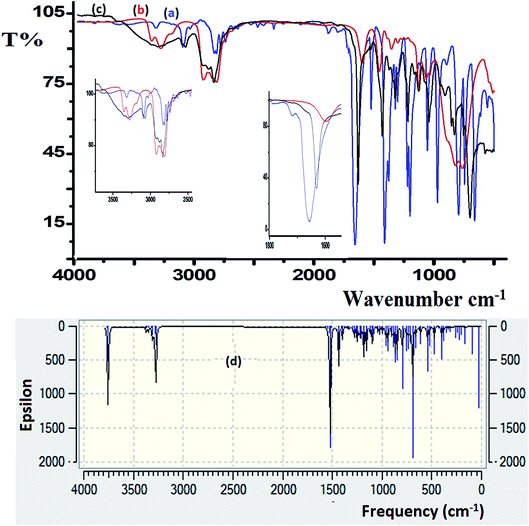 | ||
| Fig. 5 FT-IR spectra recorded for (a) 5-bromothiophene-2-carbaldehyde, (b) diethylenetriamine, and (c) the N3S2 ligand. (d) DFT-IR theoretical spectrum of the N3S2 ligand. | ||
DFT-IR calculations were also carried out for the free ligand, as shown in Fig. 5d. The experimental result was expected to be lower than the theoretically calculated value because the DFT-IR calculations were performed for a free molecule in the gaseous state, while the experimental spectrum was recorded in the solid state.25–27
The theoretical and experimental FT-IR spectra are illustrated in Fig. 5c and d for comparison, in which the vibrational frequencies and intensities were in agreement with each other.
The FT-IR spectra of the as-synthesized complexes are similar to that observed for the free ligand with slight shifts in the peak positions (Fig. 6). Fig. 6 illustrates the differences observed in the FT-IR spectra recorded for the ligand and complex 2. In complex 2, the water peak vibrations are observed at ∼3425 (ν(O–H)) and 1422 (ν(bend)) cm−1 indicating the presence of uncoordinated water molecules in the lattice of the complex and not in the ligand, since the complex is water soluble, but the ligand is not. The ν(N–H) band observed at 3250 cm−1 in the complex was shifted to a lower wavenumber with high intensity compared to that of the N3S2 ligand (3320 cm−1), which may be attributed to the coordination of the NH group to the copper metal center. In addition, the ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) vibration peak of the complex was shifted by ∼23 cm−1 (from 1688 to 1665 cm−1) due to the formation of the C
N) vibration peak of the complex was shifted by ∼23 cm−1 (from 1688 to 1665 cm−1) due to the formation of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N → Cu(II) bond. The most important observation in this FT-IR study was the presence of a sharp peak at 510 cm−1 in the complex spectrum due to the ν(Cu–N) vibrations, which support the direct formation of the new N → Cu(II) bonds.
N → Cu(II) bond. The most important observation in this FT-IR study was the presence of a sharp peak at 510 cm−1 in the complex spectrum due to the ν(Cu–N) vibrations, which support the direct formation of the new N → Cu(II) bonds.
3.6. Frontier molecular orbital calculations
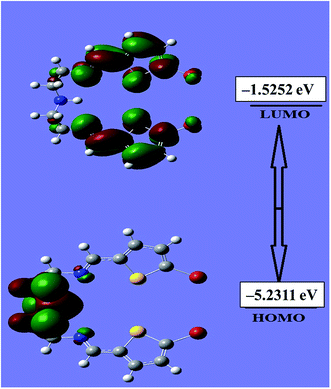
An evaluation of the HOMO/LUMO energies is beneficial toward estimating the chemical behavior of the N3S2 ligand. The HOMO/LUMO energy gap controls many chemical reactivity descriptors, such as hardness, electrophilicity, quantum chemistry terms, chemical potential, electronegativity, and local reactivity.25–28 For example, the nucleophilicity of the ligand can be evaluated by its ability to donate electrons, which is associated with the HOMO energy level, while the electron affinity is characterized by the LUMO. Fig. 7 shows a schematic representation of the HOMO/LUMO orbitals in the gaseous phase. The HOMO is located at −5.2311 eV, while the LUMO is located at −1.5252 eV with an energy gap of 3.7059 eV. The calculated energy gap value reveals the ease of HOMO to LUMO electron excitation, which is reflected in the HOMO being the predominant molecular orbital and consistent with overall nature of the pentadentate ligand as a strong electron donor with a high degree of nucleophilicity. It is very easy for electrons to be excited from the ground to excited state with such a small energy gap. The HOMO/LUMO gap is related to the chemical reactivity or kinetic stability, since both have negative values. Consequently, the HOMO and LUMO decide the chemical stability of the ligand.22–31 Several parameters related to the HOMO/LUMO energy gap value have been theoretically calculated and are illustrated Table 1.| Basis set | B3LYP/3-21G |
|---|---|
| EHOMO | −5.2311 |
| ELUMO | −1.5252 |
| Chemical potential (μ) | −6.7563 |
| Dipole moment | 2.49140 |
| Chemical hardness (η) | 1.85255 |
| Electronegativity (X) | 6.75632 |
3.7. UV-Vis spectroscopy
The electronic absorption behavior of the N3S2 ligand and its complexes were measured at RT in ethanol and water, respectively. The absorption bands observed for the ligand were also assigned using theoretical calculations at the TD-DFT/B3LYP/3-21 level of theory. The UV-Vis spectrum of the N3S2 ligand shows highly intense transitions at λmax = 240 (sharp) and 280 nm (3.0 × 104 M−1 L−1), which correspond to the π–π* transitions, as shown in Fig. 8a. The absorption maxima in the time-dependent DFT UV-Vis spectra were observed at 275 (sharp) and ∼1100 nm (broad, out of range >800 nm), as shown in Fig. 8b. The theoretical UV-Vis calculations of the molecular orbital geometry revealed the visible absorption maximum of the N3S2 ligand corresponds to the electron transition between the HOMO and LUMO. A good agreement between the theoretical-TD-DFT results and experimental UV-Vis spectra was observed and the slight ∼5 nm shift was attributed to the solvent effect.26–30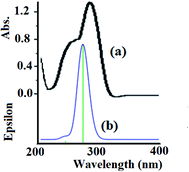 | ||
| Fig. 8 (a) Experimental UV-Vis spectrum of the N3S2 ligand recorded in ethanol at RT and (b) the TD DFT/B3LYP/3-311 theoretical spectrum calculated in the gaseous phase. | ||
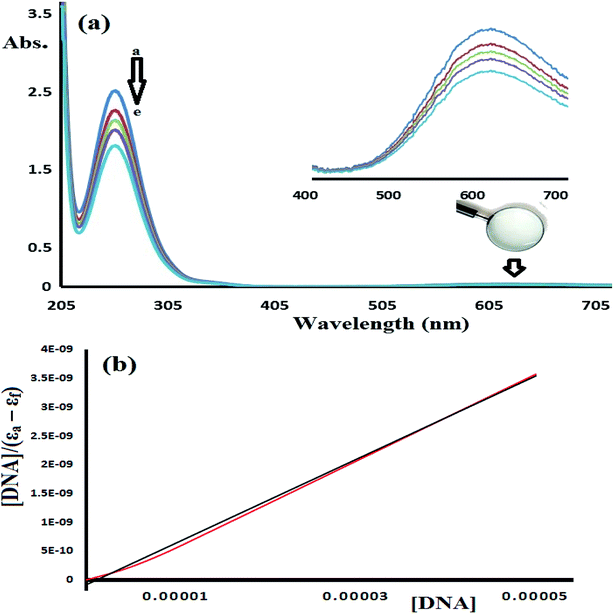
In water, complex 1 and 2 exhibit similar electronic behavior. The signals corresponding to the π–π* electron transition were shifted from 280 (free ligand) to ∼250 nm in the complexes due to the coordination of the ligand to the Cu(II) center. In addition, broad bands in the visible region of 600–640 nm were observed upon complexation with the Cu(II) center, which were not exhibited by the free ligand or CuX2 starting material. These bands were attributed to the blue color of the resulting N–Cu(II) complexes. The blue color absorption was assigned to the d–d electron transition in the square pyramid geometry around the Cu(II) complex center, as shown in Fig. 9 for complex 1.
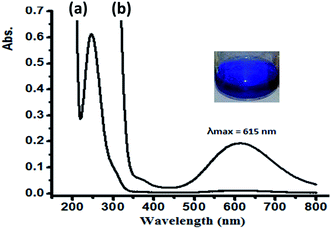 | ||
| Fig. 9 UV-Vis spectra recorded for complex 1 dissolved in water at RT at a concentration of (a) 5 × 10−5 M and (b) 5 × 10−4 M. | ||
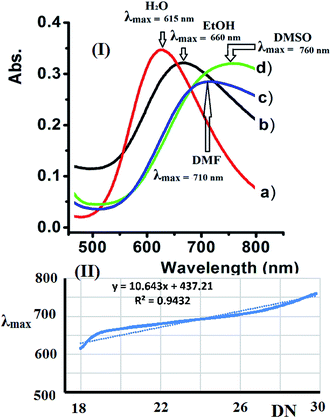
3.8. Solvatochromism of complex 2
Water, EtOH, DMF, and DMSO were used to evaluate the solvatochromism phenomena observed due to the solubility of the dicationic complexes. The UV-Vis spectra of complex 2 recorded in the selected solvents exhibit a broad band at 600–800 nm. The complexes exhibited a significant positive λmax shift upon increasing the polarity of the solvent due to the expected Jahn–Teller effect observed at the Cu(II) center with a d9 electronic configuration and coordination number of 5.The UV-Vis spectra of complex 2 shift depending on the solvent's donor number polarity, as shown in Fig. 10a. Bathochromic shifts were recorded due to the direct coordination of the polar solvent molecules to the vacant sites of the five coordination Cu(II) center with different strengths, which is in accordance with the mechanism of solvatochromism reported for this type of complex.32–34
Accordingly, the λmax values observed for complex 2 in the different solvents studied increases linearly when Gutmann's donor number (DN) of the selected solvent increased. The linear trend in λmax observed for complex 2 verses DN is shown in Fig. 10b.
3.9. Thermal analysis
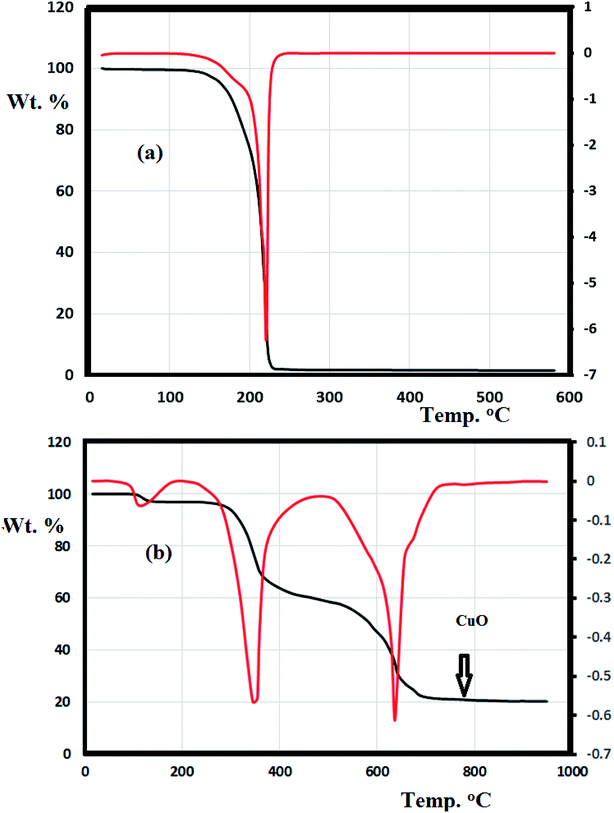
The thermal behavior of the N3S2 ligand and complex 2 were investigated using TG/DTG under an open air atmosphere over a temperature range of 0–900 °C at a heating rate of 10 °C min−1 (Fig. 11).Fig. 11a shows the TG curve obtained for the N3S2 ligand, which displays noticeable thermal stability up to 140 °C. Decomposition begins after 140 °C and ends at ∼260 °C. The ligand was totally decomposed into light gases including SO2, NO2, and CO2 in a broad step with ∼100% weight loss. No intermediate degradation steps, physical phonemes, or residues were recorded and the compound exhibited a simple one-step thermal decomposition mechanism.
Complexes 1 and 2 exhibit similar thermogravimetric behavior. The TG/DTG curve obtained for complex 2 shows three main steps (Fig. 11b). The first step (<100 °C) corresponds to the loss of uncoordinated water molecules in accordance with the FT-IR results. The second decomposition step at 280–450 °C (40% weight loss) was attributed to the decomposition of the ligand from the backbone of complex 2 to give CuBr2 as the final product. The third step starts from 460 °C and ends at 750 °C, which was attributed to the reaction between CuBr2 and O2 with the elimination of bromide ions in one broad step to form copper oxide (Cu![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, 18%) as the final product.
O, 18%) as the final product.
3.10. Electrochemistry of complex 2
As a representative example, the electron-transfer conductance of complex 2 in acetonitrile was investigated using cyclic voltammetry, as shown in Fig. 12. The N3S2 ligand is electroinactive from 0 to −1.5 V, while complex 2 exhibits a one electron redox transfer process in this range. The electrochemical behavior observed at the Pt working electrode was E1/2 = −0.760 V, ipa/ipc = 0.92, and ΔEp =130 mV, and the plot of ipc vs. v1/2 was linear with slope = 0.991. All these parameters suggest that the Cu(I)/Cu(II) redox process becomes quasi-reversible with responses at −650 and −780 mV (vs. Ag/AgNO3).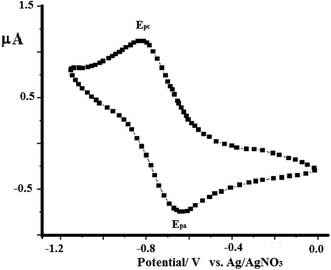 | ||
| Fig. 12 CV curve obtained for complex 1 dissolved in acetonitrile (1 × 10−4 M) at RT in the presence of 0.1 M TBAHF at a scan rate of 0.10 V s−1 using Ag/AgNO3 as reference electrode. | ||
3.11. SEM and EDS
The surface morphologies of both the free N3S2 ligand and complex 2 were subjected to SEM and EDS. The SEM image of the free ligand displays a semi-square single phase with a block over block morphology with unequal boundaries and various micrometer volumes (Fig. 13a). The SEM image of complex 2 shows a different morphology with a smooth, homogeneous, and uniform rod-like morphology with various pore sizes (Fig. 13b).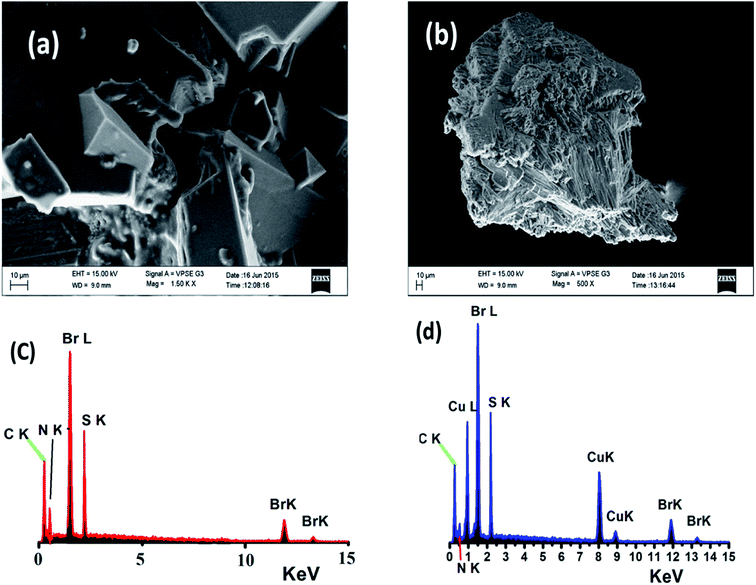 | ||
| Fig. 13 SEM images of (a) the N3S2 ligand and (b) complex 2, and EDS images of (c) the N3S2 ligand and (d) complex 2. | ||
Since the SEM image of the surface of the free N3S2 ligand was different than that of its corresponding complex, the change in the morphology of the ligand before and after its coordination to Cu(II) confirmed the formation of the L–M complex and allowed us to differentiate the chemical composition of the N3S2 ligand and its complex. Therefore, the composition of the N3S2 ligand and its complex were determined using EDS, as shown in Fig. 13c and d, respectively. Comparison of the spectra indicated that the N3S2 ligand only contains C, N, S, and Br, while its complex contains C, N, S, Br, and Cu, which again confirmed the formation of the copper complex. The absence of O atoms in the ligand and complex indicates the stability of the compounds toward atmospheric O2. The absence of unknown peaks reflects the high purity of both the ligand and its complex.
3.12. CT-DNA binding affinity of complexes
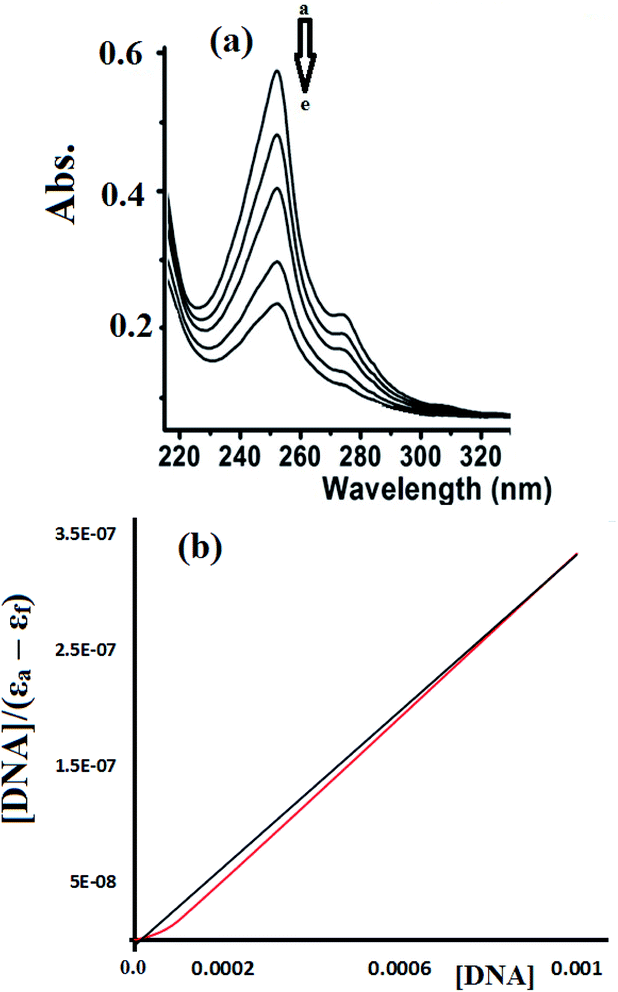
To estimate the binding ability of the Cu(II) complexes, the intrinsic binding constant (Kb) was evaluated by monitoring the changes in the absorbance spectra verses the CT-DNA concentration using the following equation.35–38
| [DNA]/(εa − εf) = [DNA]/(εb − εf) + 1/(Kb (εb − εf) |
Fig. 14 shows the UV-Vis spectra of CT-DNA-1. A high concentration of complex 1 (5 × 10−5 M) was used in this experiment in order to monitor the complex absorption behavior in both the UV and visible light regions during the DNA titration experiment. The two characteristic absorption peaks observed at 250 and 625 nm decrease in intensity upon the addition of CT-DNA at different concentrations, as shown in Fig. 14.
Fig. 15 shows the UV-Vis spectra recorded during the complex 2-CT-DNA binding titration experiment at 255 nm using a lower concentration of complex 2 (1 × 10−5 M).
The UV-abs. spectral titrations behavior of DNA with the Cu(II) complexes were used to make the comparison between the desired complexes with the recent synthesized Schiff bases/Cu(II) complexes. The small shift in Abs. wavelength and the intercalating binding constant (Kb) values were used as a criterion in judging the [Cu(II):DNA] binding strength as seen in Table 2.
| No. | Complex no. | Shift in wavelength | Kb | Ref. |
|---|---|---|---|---|
| 1 | Complexes 1–6 | Bathochromic | 2.41 × 104–1.60 × 105 | 41 |
| 2 | CuCl2(SB)2 | Hyperchromic | 7.85 × 103 | 42 |
| 3 | Complex 2 | Hyperchromic | 3.14 × 103 | 43 |
| 4 | Complex 3 | Hyperchromic | 1.06 × 105 | 44 |
| 5 | CuLB | No shift | 6.09 × 105 | 45 |
| 6 | Complex 1 | Hyperchromic | 3.34 × 104 | 46 |
| 7 | Cu(4a) | Bathochromic | 9.05 × 105 | 47 |
| 8 | Complexes 1 and 2 | Hyperchromic | 3.20 × 105 and 2.51 × 105 | This study |
Depending on the results illustrated in Table 1, one can say that complexes 1 and 2 prepared in this study are classified as a good binder among their peers' complexes, as their Kb values are higher than all the complexes except complexes in entries 5 and 7. Such high activities can be attributed to the pentadentate coordination mode of N3S2 Schiff base ligand, which gave their Cu(II) desired complexes an additional stability and opportunities to bind the DNA strings in different types and more binding modes, these features are equivalent to those previously observed for Cu(II) complexes.38–40
4. Conclusions
A new Schiff base, (E)-N1-((5-bromothiophen-2-yl)methylene)-N2-(2-((E)-((5-bromothiophen-2-yl)methylene)amino)ethyl)ethane-1,2-diamine, was synthesized via the condensation of 5-bromothiophene-2-carbaldehyde and diethylenetriamine. The N3S2 ligand was characterized using spectroscopic and theoretical analyses, and the condensation reaction used in its synthesis was monitored through FT-IR spectroscopy. Water soluble square pyramid dicationic complexes with the general formula [Cu(N3S2)]X2 were formed because the N3S2 ligand acts as a pentadentate ligand. The TG results demonstrated the different thermal behavior observed between the free ligand and its complexes. The polarity of the solvent used plays a critical role in controlling the solvatochromatic behavior of the complexes. SEM and EDS supported the complexation of the N3S2 ligand to the Cu(II) metal center. The complexes exhibited a one electron redox transfer process with negative voltages. Both the viscosity and absorption spectra showed that complex 1 is a better CT-DNA binder than complex 2 with Kb values of 3.20 × 105 and 2.51 × 105 M−1, respectively. The hypochromism interactions indicated and intercalative binding between the aromatic-DNA base pairs and the complexes theophine aromatic ring via π–π stacking interactions. The preliminary complex:DNA binding results observed in this study reflected the desired complexes as very good binder, which means that such compounds can be used as new chemotherapy.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through research group no (RG-1440-141).Notes and references
- A. Golcu, M. Tumer, H. Demirelli and R. A. Wheatley, Cd (II) and Cu (II) complexes of polydentate Schiff base ligands: synthesis, characterization, properties and biological activity, Inorg. Chim. Acta, 2005, 358(6), 1785–1797 CrossRef CAS.
- A. T. Chaviara, P. C. Christidis, A. Papageorgiou, E. Chrysogelou, D. J. Hadjipavlou-Litina and C. A. Bolos, In vivo anticancer, anti-inflammatory, and toxicity studies of mixed-ligand Cu (II) complexes of dien and its Schiff dibases with heterocyclic aldehydes and 2-amino-2-thiazoline. Crystal structure of [Cu (dien)(Br)(2a-2tzn)](Br)(H2O), J. Inorg. Biochem., 2005, 99(11), 2102–2109 CrossRef CAS PubMed.
- A. T. Chaviara, P. J. Cox, K. H. Repana, A. A. Pantazaki, K. T. Papazisis and A. H. Kortsaris, et al., The unexpected formation of biologically active Cu(II) Schiff mono-base complexes with 2-thiophene-carboxaldehyde and dipropylenetriamine: crystal and molecular structure of CudptaSCl2, J. Inorg. Biochem., 2005, 99(2), 467–476 CrossRef CAS PubMed.
- A. T. Chaviara, P. J. Cox, K. H. Repana, R. M. Papi, K. T. Papazisis and D. Zambouli, et al., Copper (II) Schiff base coordination compounds of dien with heterocyclic aldehydes and 2-amino-5-methyl-thiazole: synthesis, characterization, antiproliferative and antibacterial studies. Crystal structure of CudienOOCl2, J. Inorg. Biochem., 2004, 98(8), 1271–1283 CrossRef CAS PubMed.
- I. Warad, A. A. Khan, M. Azam, S. I. Al-Resayes and S. F. Haddad, Design and structural studies of diimine/CdX2 (X = Cl, I) complexes based on 2,2-dimethyl-1,3-diaminopropane ligand, J. Mol. Struct., 2014, 1062, 167–173 CrossRef CAS.
- I. Warad, M. Azam, S. I. Al-Resayes, M. S. Khan, P. Ahmad and M. Al-Nuri, et al., Structural studies on Cd(II) complexes incorporating di-2-pyridyl ligand and the X-ray crystal structure of the chloroform solvated DPMNPH/CdI2 complex, Inorg. Chem. Commun., 2014, 43, 155–161 CrossRef CAS.
- E. Pontiki, D. Hadjipavlou-Litina and A. T. Chaviara, Evaluation of anti-inflammatory and antioxidant activities of copper (II) Schiff mono-base and copper (II) Schiff base coordination compounds of dien with heterocyclic aldehydes and 2-amino-5-methyl-thiazole, J. Enzyme Inhib. Med. Chem., 2008, 23(6), 1011–1017 CrossRef CAS PubMed.
- Z. H. Chohan, M. S. Iqbal, S. K. Aftab and A. Rauf, Antibacterial dimeric copper (II) complexes with chromone-derived compounds, J. Enzyme Inhib. Med. Chem., 2012, 27(2), 223–231 CrossRef CAS PubMed.
- C. T. Dillon, T. W. Hambley, B. J. Kennedy, P. A. Lay, J. E. Weder and Q. Zhou, Copper and zinc complexes as antiinflammatory drugs, Met. Ions Biol. Syst., 2004, 41, 253 CAS.
- K. Nakajima, Y. Ando, H. Mano and M. Kojima, Photosubstitution reactivity, crystal structures, and electrochemistry of ruthenium(II)(III) complexes containing tetradentate (O2N2, S2N2, and P2N2) Schiff base ligands, Inorg. Chim. Acta, 1998, 274(2), 184–191 CrossRef CAS.
- N. Al-Zaqri, T. Khatib, A. Alsalme, F. Alharthi, Z. Sarouks and I. Warad, Synthesis and amide ⇔ imidic prototropic tautomerization in thiophene-2-carbohydrazide: XRD, DFT/HSA-computation, DNA-docking, TG and isoconversional kinetics via FWO and KAS models, RSC Adv., 2020, 10, 2037–2048 RSC.
- M.-H. Shih and F.-Y. Ke, Syntheses and evaluation of antioxidant activity of sydnonyl substituted thiazolidinone and thiazoline derivatives, Bioorg. Med. Chem., 2004, 12(17), 4633–4643 CrossRef CAS PubMed.
- K. Singh, M. S. Barwa and P. Tyagi, Synthesis, characterization and biological studies of Co(II), Ni(II), Cu(II) and Zn(II) complexes with bidentate Schiff bases derived by heterocyclic ketone, Eur. J. Med. Chem., 2006, 41(1), 147–153 CrossRef CAS PubMed.
- M. Pellei, G. G. Lobbia, C. Santini, R. Spagna, M. Camalli and D. Fedeli, et al., Synthesis, characterization and antioxidant activity of new copper (I) complexes of scorpionate and water soluble phosphane ligands, Dalton Trans., 2004, 17, 2822–2828 RSC.
- J. Vančo, O. Švajlenová, E. Račanská, J. Muselík and J. Valentová, Antiradical activity of different copper (II) Schiff base complexes and their effect on alloxan-induced diabetes, J. Trace Elem. Med. Biol., 2004, 18(2), 155–161 CrossRef PubMed.
- Z. H. Chohan, H. Pervez, A. Rauf, A. Scozzafava and C. T. Supuran, Antibacterial Co (II), Cu (II), Ni (II) and Zn (II) complexes of thiadiazole derived furanyl, thiophenyl and pyrrolyl Schiff bases, J. Enzyme Inhib. Med. Chem., 2002, 17(2), 117–122 CrossRef CAS PubMed.
- E. Pontiki, D. Hadjipavlou-Litina, A. T. Chaviara and C. A. Bolos, Evaluation of anti-inflammatory and antioxidant activities of mixed-ligand Cu (II) complexes of dien and its Schiff dibases with heterocyclic aldehydes and 2-amino-2-thiazoline, Bioorg. Med. Chem. Lett., 2006, 16(8), 2234–2237 CrossRef CAS PubMed.
- C. Santini, M. Pellei, G. G. Lobbia, D. Fedeli and G. Falcioni, Synthesis and characterization of new copper (I) complexes containing 4-(diphenylphosphane) benzoic acid and “scorpionate” ligands with “in vitro” superoxide scavenging activity, J. Inorg. Biochem., 2003, 94(4), 348–354 CrossRef CAS PubMed.
- S. U. Rehman, Z. H. Chohan, F. Gulnaz and C. T. Supuran, In vitro antibacterial, antifungal and cytotoxic activities of some coumarins and their metal complexes, J. Enzyme Inhib. Med. Chem., 2005, 20(4), 333–340 CrossRef CAS PubMed.
- S. H. Sumrra and Z. H. Chohan, In vitro antibacterial, antifungal and cytotoxic activities of some triazole Schiff bases and their oxovanadium(IV) complexes, J. Enzyme Inhib. Med. Chem., 2013, 28(6), 1291–1299 CrossRef CAS PubMed.
- Z. H. Chohan and M. Hanif, Design, synthesis, and biological properties of triazole derived compounds and their transition metal complexes, J. Enzyme Inhib. Med. Chem., 2010, 25(5), 737–749 CrossRef CAS PubMed.
- Z. H. Chohan and S. H. Sumrra, Synthesis, characterization and biological properties of thienyl derived triazole Schiff bases and their oxovanadium(IV) complexes, J. Enzyme Inhib. Med. Chem., 2012, 27(2), 187–193 CrossRef CAS PubMed.
- M. AL-Noaimi, M. I. Choudhary, F. F. Awwadi, W. H. Talib, T. B. Hadda and I. Warad, et al., Characterization and biological activities of two copper(II) complexes with dipropylenetriamine and diamine as ligands, Spectrochim. Acta, Part A, 2014, 127, 225–230 CrossRef CAS PubMed.
- I. Warad, O. H. Abd-Elkader, A. Boshaala, N. Al-Zaqri, B. Hammouti and T. B. Hadda, Synthesis, spectral, thermal, and a crystalline structure of complexes containing [MeC(CH2PPh2)3Cu(I)], Res. Chem. Intermed., 2013, 39(2), 721–732 CrossRef CAS.
- J. Aihara, Reduced HOMO–LUMO gap as an index of kinetic stability for polycyclic aromatic hydrocarbons, J. Phys. Chem. A, 1999, 103(37), 7487–7495 CrossRef CAS.
- K. Fukui, Role of frontier orbitals in chemical reactions, Science, 1982, 218(4574), 747–754 CrossRef CAS PubMed.
- P. Udhayakala, T. V. Rajendiran, S. Seshadri and S. Gunasekaran, Quantum chemical vibrational study, molecular property and HOMO-LUMO energies of 3-bromoacetophenone for Pharmaceutical application, J. Chem. Pharm. Res., 2011, 3(3), 610–625 CAS.
- S. Sebastian, S. Sylvestre, J. Jayabharathi, S. Ayyapan, M. Amalanathan and K. Oudayakumar, et al., Study on conformational stability, molecular structure, vibrational spectra, NBO, TD-DFT, HOMO and LUMO analysis of 3, 5-dinitrosalicylic acid by DFT techniques, Spectrochim. Acta, Part A, 2015, 136, 1107–1118 CrossRef CAS PubMed.
- V. L. Furer, A. E. Vandyukov, C. Padie, J. P. Majoral, A. M. Caminade and V. I. Kovalenko, FT-Raman, FT-IR spectroscopic and DFT studies of hexaphenoxycyclotriphosphazene, J. Mol. Struct., 2016, 1115, 124–135 CrossRef CAS.
- S. Tarchouna, I. Chaabane and A. B. Rahaiem, FT-IR and Raman spectra and vibrational investigation of bis (4-acetylanilinium) hexachlorostannate using DFT (B3LYP) calculation, J. Chem. Pharm. Res., 2016, 83, 186–194 CAS.
- V. L. Furer, A. E. Vandyukov, J. P. Majoral, A. M. Caminade and V. I. Kovalenko, Structure, IR and Raman spectra of phosphotrihydrazide studied by DFT, Spectrochim. Acta, Part A, 2016, 166, 19–24 CrossRef CAS PubMed.
- H. Golchoubian, H. Ghorbanpour and E. Rezaee, Dinuclear copper (II) complexes with bridging oximato group: Synthesis, crystal structure and solvatochromism property, Inorg. Chim. Acta, 2016, 442, 30–36 CrossRef CAS.
- H. Golchoubian, G. Moayyedi, E. Rezaee and G. Bruno, Synthesis, characterization and solvatochromism study of mixed-chelate copper (II) complexes: a combined experimental and density functional theoretical study, Polyhedron, 2015, 96, 71–78 CrossRef CAS.
- H. Golchoubian and E. Rezaee, Synthesis, characterization and solvatochromism studies of two new mixed-chelate copper (II) complexes containing β-ketoamine and diamine ligands, Polyhedron, 2013, 55, 162–168 CrossRef CAS.
- X.-Q. Zhou, Y. Li, D.-Y. Zhang, Y. Nie, Z.-J. Li and W. Gu, et al., Copper complexes based on chiral Schiff-base ligands: DNA/BSA binding ability, DNA cleavage activity, cytotoxicity and mechanism of apoptosis, Eur. J. Med. Chem., 2016, 114, 244–256 CrossRef CAS PubMed.
- P. R. Inamdar, R. Chauhan, J. Abraham and A. Sheela, DNA interaction and cytotoxic activity of copper complex based on tridentate hydrazone derived ligand and nitrogen donor heterocycle, Inorg. Chem. Commun., 2016, 67, 67–71 CrossRef CAS.
- Z. Shokohi-Pour, H. Chiniforoshan, A. A. Momtazi-Borojeni and B. Notash, A novel Schiff base derived from the gabapentin drug and copper (II) complex: Synthesis, characterization, interaction with DNA/protein and cytotoxic activity, J. Photochem. Photobiol., B, 2016, 162, 34–44 CrossRef CAS PubMed.
- R. Pradhan, M. Banik, D. B. Cordes, A. M. Z. Slawin and N. C. Saha, Synthesis, characterization, X-ray crystallography and DNA binding activities of Co (III) and Cu (II) complexes with a pyrimidine-based Schiff base ligand, Inorg. Chim. Acta, 2016, 442, 70–80 CrossRef CAS.
- L. H. Abdel-Rahman, A. M. Abu-Dief, M. Ismael, M. A. A. Mohamed and N. A. Hashem, Synthesis, structure elucidation, biological screening, molecular modeling and DNA binding of some Cu (II) chelates incorporating imines derived from amino acids, J. Mol. Struct., 2016, 1103, 232–244 CrossRef CAS.
- L. Jia, J. Xu, X. Zhao, S. Shen, T. Zhou and Z. Xu, et al., Synthesis, characterization, and antitumor activity of three ternary dinuclear copper(II) complexes with a reduced Schiff base ligand and diimine coligands in vitro and in vivo, J. Inorg. Biochem., 2016, 159, 107–119 CrossRef CAS PubMed.
- M. Al-Noaimi, M. Suleiman, H. W. Darwish, A. H. Bakheit, M. Abdoh and I. Warad, et al., DNA Binding Test, X-Ray Crystal Structure, Spectral Studies, TG-DTA, and Electrochemistry of [CoX2 (dmdphphen)](Dmdphphen Is 2,9-Dimethyl-4,7-diphenyl-1, 10-phenanthroline, X = Cl, and NCS) Complexes, Bioinorg. Chem. Appl., 2014, 2014, 1–8 CrossRef PubMed.
- G. Ramesh, S. Daravath, N. Ganji, A. Rambabu and K. Venkateswarlu, Facile synthesis, structural characterization, DNA binding, incision evaluation, antioxidant and antimicrobial activity studies of Cobalt (II), Nickle (II) and Copper (II) complexes of 3-amino-5-(4-fluorophenyl) isoxazole derivatives, J. Mol. Struct., 2020, 1202, 127338–127354 CrossRef CAS.
- R. Kalarani, M. Sankarganesh, G. G. V. Kumar and M. Kalanithi, Synthesis, spectral, DFT calculation, sensor, antimicrobial and DNA binding studies of Co (II), Cu (II) and Zn (II) metal complexes with 2-amino benzimidazole Schiff base, J. Mol. Struct., 2020, 127725–127732 CrossRef CAS.
- Z. Afsan, T. Roisnel, S. Tabassum and F. Arjmand, Structure elucidation {spectroscopic, single crystal X-ray diffraction and computational DFT studies} of new tailored benzenesulfonamide derived Schiff base copper (II) intercalating complexes: Comprehensive biological profile {DNA binding, pBR322 DNA cle, Bioorg. Chem., 2020, 94, 103427–103438 CrossRef CAS PubMed.
- S. K. Maiti, M. Kalita, A. Singh, J. Deka and P. Barman, Investigation of DNA binding and bioactivities of thioether containing Schiff base copper(II), cobalt(II) and palladium(II) complexes: synthesis, characterization, spectrochemical study, viscosity measurement, Polyhedron, 2020, 184, 114559–114566 CrossRef CAS.
- S. Hemalatha, J. Dharmaraja, S. Shobana, P. Subbaraj, T. Esakkidurai and N. Raman, Chemical and pharmacological aspects of novel hetero MLB complexes derived from NO2 type Schiff base and N2 type 1,10-phenanthroline ligands, J. Saudi Chem. Soc., 2020, 24(1), 61–80 CrossRef.
- N. N. Rao, E. kishan, K. Gopichand, R. Nagaraju, A. M. Ganai and P. V. Rao, Design, synthesis, spectral characterization, DNA binding, photo cleavage and antibacterial studies of transition metal complexes of benzothiazole Schiff base, Chem. Data Collect., 2020, 27, 100368–1003576 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2020 |