Open Access Article
Open Access ArticleA 3D mixing-based portable magnetic device for fully automatic immunofluorescence staining of γ-H2AX in UVC-irradiated CD4+ cells†
Runtao Zhong a,
Liangsheng Houb,
Yingbo Zhaoa,
Tianle Wang‡
a,
Shaohua Wanga,
Mengyu Wanga,
Dan Xua and
Yeqing Sun*a
a,
Liangsheng Houb,
Yingbo Zhaoa,
Tianle Wang‡
a,
Shaohua Wanga,
Mengyu Wanga,
Dan Xua and
Yeqing Sun*a
aInstitute of Environmental Systems Biology, Dalian Maritime University, 1 Linghai Road, Dalian 116026, China. E-mail: yqsun@dlmu.edu.cn
bCollege of Marine Engineering, Dalian Maritime University, Dalian, 1 Linghai Road, Dalian 116026, China
First published on 7th August 2020
Abstract
Immunofluorescence (IF) is a common method used in cell biology. The conventional protocol for IF staining is time and labor-intensive, operator dependent and reagent-consuming. Magnetic Bead (MB)-based microdevices are frequently utilized in cellular assays, but integration of simple and efficient mixing with downstream multi-step manipulation of MBs for automatic IF staining is still challenging. We herein present a portable, inexpensive and integratable device for MB-based automatic IF staining. First, a front-end cell capture step is performed using a 3D-mixing module, which is built upon a novel mechanism named ec-2MagRotors and generates periodically changing 3D magnetic fields. A 5-fold enhancement of cell capture efficiency was attained even with a low bead-to-cell concentration ratio (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), when conducting magnetic 3D mixing. Second, a 1D-moving module is employed downstream to automatically manipulate MB–cell complexes for IF staining. Further, a simplified protocol for staining of γ-H2AX, a biomarker widely used in evaluation of cell radiation damage, is presented for proof-of-principle study of the magnetic device. Using UVC-irradiated CD4+ cells as samples, our device achieved fully automatic γ-H2AX staining within 40 minutes at room temperature and showed a linear dose–response relationship. The developed portable magnetic device is automatic, efficient, cost-effective and simple-to-use, holding great potential for applications in different IF assays.
1), when conducting magnetic 3D mixing. Second, a 1D-moving module is employed downstream to automatically manipulate MB–cell complexes for IF staining. Further, a simplified protocol for staining of γ-H2AX, a biomarker widely used in evaluation of cell radiation damage, is presented for proof-of-principle study of the magnetic device. Using UVC-irradiated CD4+ cells as samples, our device achieved fully automatic γ-H2AX staining within 40 minutes at room temperature and showed a linear dose–response relationship. The developed portable magnetic device is automatic, efficient, cost-effective and simple-to-use, holding great potential for applications in different IF assays.
Introduction
Immunofluorescence (IF) is a highly sensitive and specific method that has been widely used in cell biology to study differential expression, localization and distribution of proteins at the tissue, cellular, and subcellular level.1–3 It has been regarded as a routine technique in basic biomedical research and an effective approach for biomarker diagnostics.4–6 The nucleosomal histone protein H2AX is specifically phosphorylated (γ-H2AX) adjacent to DNA double-strand breaks (DSBs) and is used for quantifying DSBs.7 Many chemotherapies and ionizing radiation (IR) used in cancer treatment result in DSBs. Therefore, γ-H2AX has significant potential as a biomarker in evaluating patient sensitivity and responsiveness to IR and chemotherapy.8,9 Peripheral Blood Monocytic Cell (PBMCs) analysis is, by far, the method most widely used to examine γ-H2AX, mainly due to the easy sampling of venous blood from patients.10 Generally, the blood sample-based intracellular IF assay involves isolation, fixation and permeabilization of target cells for antibody accessibility, blocking, and staining with fluorophore-conjugated antibodies before examination. However, macroscale methods to isolate and staining cells of interest require long, laborious, and expensive procedures that typically result in difficulties in point-of-care (POC) IF assays, which require automatic and rapid IF staining.11–14Microfluidics (lab on a chip) is a science and technology characterized by manipulation of fluids in a micrometer-scale space.15,16 Microfluidic technology has shown promising potentials to overcome limitations of macroscale IF staining protocol, most remarkably by miniaturization and integration of operational units,17,18 thus enabling portability, automaticity, rapidness and low cost.19,20 A lot of studies related to microfluidic IF staining have been reported,21–24 demonstrating the feasibilities of automatic and rapid IF assays. Superparamagnetic microparticles (known as magnetic beads, MBs) are widely used in biochemical assays for decades, owing to their high surface-to-volume ratio, surface tunability and easy control by means of external magnetic fields.25,26 The versatility in sizes, magnetic properties and surface functionalizations, as well as commercial availability of MBs have been promoting their use in lab-on-a-chip devices,25 thus lowering complexity in chip design and fluid control (free of pump and valve), and improving flexibility and automation of the assays.26,27 Researches on MB-based on-chip IF staining have shown the possibilities of magnetic POC IF assays.11,28–31
As for MB-based on-chip IF staining, a front-end MBs mixing step allows efficient capture of target cells,31 which is determined by the extent of interactions between MBs and cells. These kind of interactions are not controlled by diffusion because of the large size of cells.32 Therefore, the collisions and binding events in the stop-flow chip strongly depend on the movement of MBs.30 However, ensembles of MBs are known to aggregate into reversible concentration of beads in a confined space due to the action of an external magnetic field.33 The multi-particle clustering reduces the accessibility of the biorecognition molecules on the surface of MBs and thereby decreases the extent of interactions between MBs and cells, resulting in insufficient capture of target cells. In this circumstance, some approaches to conduct mixing of MBs and cells off-chip28,30,31 or on-chip using additional sophisticated instruments have been performed,29 which significantly lower the integration and automation of the assays; while a few methods to control and mix MBs on-chip using relatively complex actuators and specific mechanisms have been proposed to enhance the interactions between MBs and cells.33–35 Alternatively, a rather high bead-to-cell concentration ratio (5.4 × 103![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was used under the application of a 1D-moving magnet to get a proper cell capture efficiency (68 ± 4%),11 which would inevitably increase the consumption of expensive functionalized MBs, thus the cost of tests.
1) was used under the application of a 1D-moving magnet to get a proper cell capture efficiency (68 ± 4%),11 which would inevitably increase the consumption of expensive functionalized MBs, thus the cost of tests.
While magnetic microdevices have been implemented in quite a few applications,25,26 their mixing and handling of MBs on-chip is normally either not optimal with aggregation of the MBs under regular magnetic fields, or requires rather complex magnetic actuators. The ability to integrate simple, highly efficient and flexible on-chip mixing with downstream multi-step manipulation of MBs for automatic cell capture and IF staining will enable the development of inexpensive micro total analysis systems (μ-TAS) for POC IF assays, which has rarely been completely achieved.
We have previously demonstrated rapid detection of IF stained γ-H2AX in CD4+ T lymphocytes (CD4+ cells) on a microfluidic cytometer, but the cultured CD4+ cells were intracellularly stained using time-, labor-, and reagent-intensive in-tube protocol.14 In order to automate and speed up the IF staining of γ-H2AX to obtain rapid and accurate detection of γ-H2AX level to assess the radiation damage, we herein introduce a portable, cost-effective and integratable magnetic device to perform on-chip efficient cell capture and automatic intracellular IF staining. Key features include: a novel mechanism of on-chip magnetic 3D mixing (ec-2MagRotors) for on-chip thorough mixing of MBs and cells, a portable magnetic device for automatic multi-step manipulation of MB–cell complexes, and a simplified protocol for IF staining of γ-H2AX in CD4+ cells. We simulated the generation of a periodically changing 3D magnetic field based on the mechanism of ec-2MagRotors and characterized the 3D mixing-based capture efficiency of CD4+ cells. As proof-of-principle analysis, we conducted on-chip CD4+ cell capture and IF staining of γ-H2AX in UVC-irradiated CD4+ cells automatically and obtained similar linearity for radiation dose–response relationship in contrast to that of in-tube staining protocol. We believe that the 3D mixing-based portable magnetic device has the potential to be beneficial for automatic and rapid IF staining in many applications and enabling further integration with detection modules to establish μ-TAS for POC IF assays.
Materials and methods
Fluidic chip design and fabrication
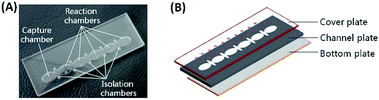
The IF-staining fluidic chip (76.2 mm × 25.4 mm in size) contains a channel with six large elliptical chambers (9 mm × 5.4 mm for major and minor axes, respectively) and five small isolation chambers (7.4 mm × 2 mm for major and minor axes, respectively) between neighboring large chambers (Fig. 1A). The large chambers are used for storage of sample/reagent to perform cell capture and downstream multi-step IF staining. The small chambers are designed to fill the isolation medium (such as mineral oil) to separate different reagents during the traverses of MB–cell complexes. The disposable chip is composed of three layers, i.e. the cover plate with injection ports (0.8 mm in diameter), the channel plate with chambers and the bottom plate without any structures (Fig. 1B), having the thickness of 1 mm, 2 mm and 0.5 mm, respectively. To assemble the three layers, poly(methyl methacrylate) (PMMA) and double-sided adhesive film (Adhesives Research, Inc., Glen Rock, PA) were cut using a CO2-laser cutter (YoungChip, Inc., China). Two layers of double-sided adhesive films between PMMA layers were cut with the same design as the middle channel plate. After alignment and assembly, the chip was firmly bonded by using an air press to obtain a uniform seal.Portable magnetic device construction
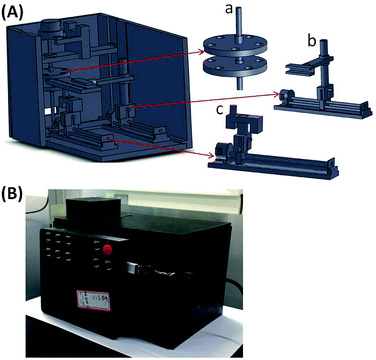
To automate MB-based IF staining on-chip, a portable magnetic device which consists of a 3D-mixing module and a 1D-moving module (Fig. 2), was developed. The 3D-mixing module is used to perform on-chip thorough mixing of MBs to achieve efficient cell capture on functionalized MBs, which is built upon a novel mechanism of ec-2MagRotors, i.e. eccentrically-rotating upper and bottom circular plates embedded with staggeredly arranged magnets. Each of the two circular plates measures a diameter of 40 mm and a thickness of 3 mm, containing five slots evenly arranged along the periphery for holding of magnets and an eccentric hole about 3 mm away from the center of the plate. A rotation axis runs through the upper and the lower circular plates from their eccentric holes and installs the two plates to align them and mutually stagger the magnet slots vertically. The two circular plates are embedded with ten cylindrical NdFeB magnets (each with a diameter of 5 mm and a height of 6 mm) and driven by a rotary motor to generate a periodically changing 3D magnetic field between the two plates. Combining with the 3D-mixing module, we further integrated a 1D-moving module into a single portable device to conduct multi-step transmission and reaction of MB–cell complexes. The 1D-moving module includes a chip moving unit and a magnet moving unit, each containing a moving motor to enable positioning of the fluidic chip and programmed movement of a cylindrical magnet, respectively. The light-tight plastic housing and support components were three-dimensionally printed in the laboratory. The assembled magnetic device features a size of 192 × 123 × 122 mm3 (Fig. 3), a weight about 0.666 kg and a power consumption less than 10 W, with a built-in Single Chip Microcomputer (SCM, STC12C2052AD) for control of three stepper motors (one rotary motor and two moving motors) to fully automate the mixing and movement of MBs (Fig. S1 and S2†).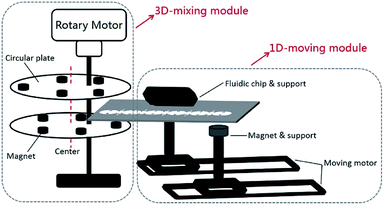 | ||
| Fig. 2 Schematic diagram of the portable magnetic device including a 3D-mixing module (based on the novel mechanism of ec-2MagRotors) and a 1D-moving module. | ||
CD4+ cell culture and UVC irradiation
Human CD4+ T lymphocytes (CD4+ cells), purchased from Shanghai Meixuan Biotechnology Co., Ltd., were suspended in the RPMI1640 medium (Jiangsu Kaiji Biological Co., Ltd., China) containing 10% inactivated fetal bovine serum (Life Technologies, Inc., USA), 1% penicillin and streptomycin (GE Healthcare Hyclone™, USA), and cultured in an incubator with 5% CO2 at 37 °C. Culture medium was replaced for an average of two days, and cell passage was conducted for an average of four days at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.
4.
After cultured for two days, CD4+ cells in exponential growth phase were collected in a 15 mL centrifuge tube, and the supernatant was removed by centrifugation at 1000 revolutions per minute (rpm) for 5 min. The cell concentration was then adjusted to about 107 cells mL−1 by adding an appropriate amount of pre-incubated (37 °C) culture medium and pipetting 10 times to mix well. About 60 μL of this cell suspension and 940 μL of the culture medium were added into a 60 mm culture dish and irradiation in a UVC radiation box (Fig. S3†). The irradiation time was 0, 0.625, 1.25, 2.5, 5 and 10 min, corresponding to radiation doses of 0, 4, 8, 16, 32, 64 J m−2, respectively. After irradiation, the culture dish was covered by the lid and transferred into the 5% CO2 incubator to incubate at 37 °C for 30 min. The processed CD4+ cells were collected in a 1.5 mL centrifuge tube, and the supernatant was removed by centrifugation at 2000 rpm for 5 min. Finally, 60 μL of PBS solution was added into the 1.5 mL tube and pipetted 10 times to mix well. The CD4+ cell sample could be used immediately or stored at 4 °C until further use.
MB-based on-chip IF staining of γ-H2AX in CD4+ cells
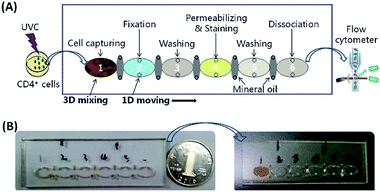
Two Dynabeads® CD4 kits (Life technologies, Inc., USA), i.e. Dynabeads® FlowComp™ Human CD4 kit (11361D, with 2.8 μm diameter FlowComp™ Dynabeads®) and Dynabeads® CD4 Positive Isolation Kit (11331D, with 4.5 μm diameter Dynabeads® CD4), were tested for on-chip CD4+ cell capture and staining. H2A.X Phosphorylation Assay Kit (Flow Cytometry) (17-344, Upstate Biotechnology, Inc., USA) was employed to stain and determine the levels of γ-H2AX in CD4+ cells. For the Dynabeads® FlowComp™ Human CD4 kit, 5 μL of the FlowComp™ Human CD4 Antibody was added to the 60 μL of UVC-irradiated CD4+ cell sample and incubated for 10–20 min on ice with gentle mixing before sample loading. To load the fluidic chip, 60 μL of the prepared CD4+ cell sample and 30 μL of the pre-washed MBs with desired concentration were firstly pipetted into chamber 1 (Fig. 4A). Next, about 90 μL of the 1× fixation solution (20-257, Upstate Biotechnology, USA), 1× PBS solution, 1× wash solution (20-258, Upstate Biotechnology, USA) and bead dissociation solution (Life technologies, Inc., USA) were loaded into chambers 2, 3, 5 and 6, respectively. A mixture (90 μL in volume) of 1× permeabilization solution (20-259, Upstate Biotechnology, USA) and appropriate volume of Anti-phospho-Histone H2A.X (Ser139), FITC conjugate (16-202, Upstate Biotechnology, USA) was then injected into chamber 4. The large elliptical chambers (1–6) were filled up with the sample/reagents without leaking into adjacent small isolation chambers because of high surface tension from the interface between the aqueous solutions and the inner surface and the shape of the chambers. Lastly, about 25 μL of mineral oil (Sigma-Aldrich, St. Louis, MO) was loaded into each of the isolation chamber without introducing any air bubbles.The loaded chip (Fig. 4B) was fixed on the chip support and positioned to the initial position between the two circular plates of the 3D-mixing module (Fig. 2 and 3A), then the lid on the top of the device was closed to protect FITC from light (Fig. 3B). After pressed the “AUTO” button on the front panel (the red button in Fig. 3B), the instrument started to run via an in-house developed program. Magnetic force was utilized to actuate MBs for 3D mixing and programmable transmission, enabling automatic γ-H2AX staining on-chip in 40 min.
Upon beginning of the protocol, a rotary motor actuated the two eccentric circular plates with magnets at a spinning speed of 6 rpm for 15 min under a 3 V DC power supply. A periodically changing 3D magnetic field between the two plates was generated to result in 3D mixing of MBs and CD4+ cells in chamber 1, thus increasing the binding of MBs to CD4+ cells via molecular interaction. After 3D mixing, the chip was transported by the chip moving motor to the position where chamber 1 was directly above the magnet in the magnet moving unit. The MBs carrying the bound CD4+ cells were then transferred from chamber 1 to separate with the aqueous solution, crossed the oil barrier in the isolation chamber, and moved into chamber 2, via actuation of the magnet moving motor. In chamber 2, cell fixation was performed by moving the CD4+ cell-conjugated beads back and forth five cycles at a translational speed of 0.25 mm s−1. The back-and-forth motion of the magnet resulted in adequate mixing and collection of the MBs within the chamber. By controlling the chamber-to-chamber transmission and back-to-forth motion within reaction chambers using the magnet moving unit, the MB–CD4+ cell complexes were sequentially moved from chamber 3 to chamber 6, conducting washing, cell permeabilization and γ-H2AX IF staining, another washing to remove unbound, non-specifically attached antibody–FITC conjugate, and bead dissociation, respectively. The entire on-chip γ-H2AX staining process was fully automated and completed in 40 min under room temperature. Following release of MBs, the stained CD4+ cells were aspirated from chamber 6 to a centrifuge tube, washed and resuspended in PBS buffer for fluorescence detection using flow cytometry.
Alternatively, for the Dynabeads® FlowComp™ Human CD4 kit, bead dissociation could also be conducted off-chip by extracting the stained CD4+ cells from chamber 6 and then adding 1 mL of FlowComp™ Release Buffer to incubate for 10–20 min at room temperature under rolling and tilting, according to the manufacturer's instructions.
CD4+ cell detection and analysis
CD4+ cells were counted using cell-count boards under an inverted microscope. For assessing the capture efficiency of CD4+ cells to MBs, it was calculated based on the following equation,
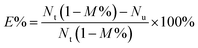 | (1) |
Results and discussion
Working principle of on-chip magnetic 3D mixing
Alonso et al. described a magnetic actuator containing a single layer of circular plate with eccentrically embedded magnets on the circumference to enhance the mixing of a MB-based immunoassay.30 Rotation of the actuator triggers the back-and-forth movement of the MBs within a reaction chamber to increase the effective surface area of the MBs (creating a loose cloud of MBs following the magnets) and make the reagents from the entire reaction chamber available to the MBs, leading to a 2.7-fold sensitivity enhancement. To further increase the chances of MBs collisions and binding events to enhance cell capture efficiency, this work proposed the mechanism of ec-2MagRotors, which employs two eccentrically rotating plates embedded with staggeredly arranged magnets. Based on the structure of the 3D-mixing module (Fig. S4A†), the distances of the embedded magnets to the eccentric rotation axis are different and periodically distributed. One revolution of the circular plates represents one cycle of MBs movement, i.e. from one end to the other end of the capture chamber and back to the starting point. When rotating, the magnets pass alternately under or above the capture chamber to induce the movement of the MBs, producing a loose and movable cloud of MBs following the magnets (Fig. S4B†), and the relative positions of the magnets and the capture chamber change periodically (top right in Fig. S4A† shows 1/2 cycle). Therefore, the 3D-mixing module developed herein provides a periodically changing 3D magnetic field and the MBs are actuated to perform periodic 3D motion within the capture chamber. To further understand the actuation of MBs under the control of the 3D-mixing module, a computational model was developed to simulate the magnetic field intensity and relative magnetic force on MBs in the generated magnetic field (Fig. S5†). Results show that a periodically changing magnetic field in three spatial dimensions of X, Y, and Z directions is generated between the two magnetic plates when the two plates rotate eccentrically. MBs in the capture chamber are subjected to periodically changing magnetic forces in a rotating 3D magnetic field (Fig. 5). Continuous rotation of the two circular plates causes uninterrupted movements of the MBs in three-dimensional directions, thus significantly increasing the bead/cell solution interface and the chances of interaction between MBs and cells from the entire capture chamber, as well as decreasing aggregation of MBs (Fig. S4†). It also can be seen from the simulation results that the ratio of the motion period in the Z-direction to that in the XY-plane is 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. S5†).
1 (Fig. S5†).
 | ||
| Fig. 5 Movement of MBs under control of the rotating 3D magnetic field: (A) in the capture chamber, (B) the trajectory. | ||
Meanwhile, it would be rather easy to further integrate the 3D-mixing module with other magnetic modules to perform downstream complex actuations of MBs, as the 1D-moving module described in this work, or conduct parallel applications by using the chip with different chamber configurations, which would enlarge the functionalities of magnetic devices. Moreover, due to the three-dimensionally cycling magnetic field generated according to the mechanism of ec-2MagRotors, the 3D-mixing module holds great potential of increasing the height of the capture chamber on the same area to accommodate an extra large volume of sample, while still maintaining highly efficient mixing of MBs in the entire chamber. This potential enables highly efficient enrichment and purification of rare subpopulations of cells of interest (such as CTCs29 and CD4+ cells11 in blood sample) for subsequent characterization. This is totally different from previous single layer of rotating plate with eccentrically embedded magnets,30 which generates a magnetic field periodically changing in X and Y dimentions, not including vertical direction, and is thought to further improve the MBs mixing by decreasing the height of the mixing chamber. To perform MB-based cell enrichment, other studies employed a sample chamber with a larger area, bringing about extra complexity for chip design and control.29,31 Besides, the active magnetic 3D mixing would be hardly affected by gravity conditions and can then be applied to in-orbit staining and detection of radiation biomarkers in space36 under microgravity, thus improving the adaptability to experimental environments.
Characterization of the on-chip magnetic 3D mixing module
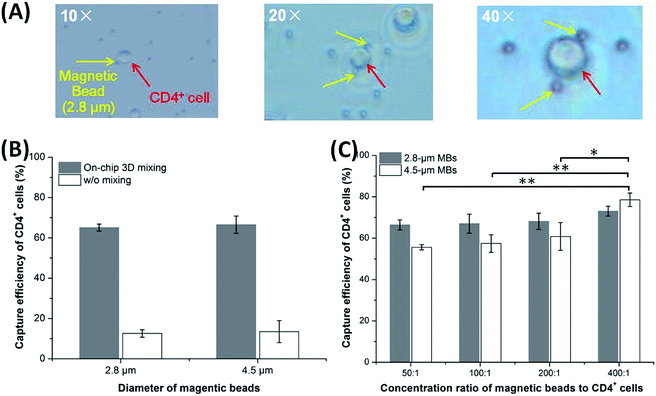
To characterize the magnetic 3D mixing module, the binding event and the capture efficiency of CD4+ cells using the functionalized MBs was tested. In Fig. 6(A), a CD4+ cell was bound on one (the 1st picture) or two (the 2nd and 3rd pictures) MBs (2.8 μm diameter), and the pictures were taken using a microscope at magnification factors of 10×, 20×, 40×, respectively. To demonstrate the effect of 3D mixing on the binding of MBs to cells, a small size (2.8 μm diameter) and a large size (4.5 μm diameter) of MBs were tested for capture efficiency of CD4+ cells at a bead-to-cell concentration ratio of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 under on-chip 3D mixing and without mixing (at 4 °C for 20 min), respectively. Result in Fig. 6(B) shows that the capture efficiency increases from less than 14% without mixing to more than 65% with on-chip 3D mixing for both of the two sizes of MBs, indicating a significant enhancement of capture efficiency for 3D-mixing module even at a very low bead-to-cell concentration ratio. Due to the aggregation of MBs in applied stationary magnetic field (bottom right in Fig. S4A†) and the relatively large size and low diffusion of CD4+ cells,31 reactions between MBs and CD4+ cells would be expected to benefit significantly from mixing30 provided by the 3D-mixing module.
1 under on-chip 3D mixing and without mixing (at 4 °C for 20 min), respectively. Result in Fig. 6(B) shows that the capture efficiency increases from less than 14% without mixing to more than 65% with on-chip 3D mixing for both of the two sizes of MBs, indicating a significant enhancement of capture efficiency for 3D-mixing module even at a very low bead-to-cell concentration ratio. Due to the aggregation of MBs in applied stationary magnetic field (bottom right in Fig. S4A†) and the relatively large size and low diffusion of CD4+ cells,31 reactions between MBs and CD4+ cells would be expected to benefit significantly from mixing30 provided by the 3D-mixing module.
The capture efficiency was also evaluated at room temperature over the MB sizes and bead-to-cell concentration ratios. In this study, the 2.8 μm and the 4.5 μm MBs were tested over bead-to-cell concentration ratios varying from 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 400
1 to 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 6C). For the small MBs, as the bead-to-cell concentration ratio increases, the capture efficiency increases slightly from 66.40 ± 2.45% to 73.03 ± 2.37%, but there is no statistically significant difference (p > 0.05, n = 3). This shows that the bead-to-cell concentration ratios in the studied range have no significant effect on the capture efficiency of CD4+ cells for the small MBs. For the large MBs, the capture efficiency increases from 55.60 ± 1.23% to 78.57 ± 3.24% as the bead-to-cell concentration ratio increases; the low bead-to-cell concentration ratio (50
1 (Fig. 6C). For the small MBs, as the bead-to-cell concentration ratio increases, the capture efficiency increases slightly from 66.40 ± 2.45% to 73.03 ± 2.37%, but there is no statistically significant difference (p > 0.05, n = 3). This shows that the bead-to-cell concentration ratios in the studied range have no significant effect on the capture efficiency of CD4+ cells for the small MBs. For the large MBs, the capture efficiency increases from 55.60 ± 1.23% to 78.57 ± 3.24% as the bead-to-cell concentration ratio increases; the low bead-to-cell concentration ratio (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–200
1–200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) has no significant effect on the capture efficiency, but the high bead-to-cell concentration ratio (400
1) has no significant effect on the capture efficiency, but the high bead-to-cell concentration ratio (400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) significantly increases the capture efficiency in contrast to that of low bead-to-cell concentration ratios (p < 0.05, n = 3). Comparing the capture efficiencies of cells using different sizes of MBs, it is obvious that the capture efficiency for small MBs is higher than that of the large MBs in the range of bead-to-cell concentration ratios from 50
1) significantly increases the capture efficiency in contrast to that of low bead-to-cell concentration ratios (p < 0.05, n = 3). Comparing the capture efficiencies of cells using different sizes of MBs, it is obvious that the capture efficiency for small MBs is higher than that of the large MBs in the range of bead-to-cell concentration ratios from 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 200
1 to 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, but lower at the bead-to-cell concentration ratio of 400
1, but lower at the bead-to-cell concentration ratio of 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, though none is statistically significant between the two sizes of MBs. As can be seen from Fig. 6(B) and (C), temperature significantly affects the binding of large MBs to CD4+ cells. It seems that the size and surface modification of MBs have an effect on the capture efficiency of CD4+ cells, but the specific reasons need to be further investigated. To enable traverse of oil barriers between reaction chambers with minimum loss of MBs and decrease the consumption of expensive functionalized MBs, the bead-to-cell concentration ratio of 100
1, though none is statistically significant between the two sizes of MBs. As can be seen from Fig. 6(B) and (C), temperature significantly affects the binding of large MBs to CD4+ cells. It seems that the size and surface modification of MBs have an effect on the capture efficiency of CD4+ cells, but the specific reasons need to be further investigated. To enable traverse of oil barriers between reaction chambers with minimum loss of MBs and decrease the consumption of expensive functionalized MBs, the bead-to-cell concentration ratio of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was selected in the following experiments.
1 was selected in the following experiments.
Validation of automatic actuation of MBs for the magnetic device
In order to realize on-chip automatic IF staining, it is required that there are enough cells carried by MBs to react successively with the reagents contained in reaction chambers on the fluidic chip. To verify the feasibility of the proposed magnetic device for automatic manipulation of MBs, the 2.8 μm and the 4.5 μm MBs were used to evaluate the recovery rate of CD4+ cells. A 90 μL mixture of CD4+ cells and MBs (with the bead-to-cell concentration ratio of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was loaded into the capture chamber. The reaction chambers and the isolation chambers were filled with 90 μL of PBS solution and 25 μL of mineral oil, respectively. The device was then powered to perform 3D mixing and programmed 1D moving of MBs. In this way, we focused on evaluating the functionality of magnetic actuation for the device and avoided influences of other conditions. After completion of the procedures, the MB–cell complexes were removed from chamber 6 to a centrifuge tube, and the bead dissociation solution was added, mixed and incubated. Subsequently, the supernatant with released cells was aspirated under the action of a permanent magnet. The recovered cells were washed, resuspended and counted using cell-count boards. Recovery rate of CD4+ cells was calculated according to the following formula,
1) was loaded into the capture chamber. The reaction chambers and the isolation chambers were filled with 90 μL of PBS solution and 25 μL of mineral oil, respectively. The device was then powered to perform 3D mixing and programmed 1D moving of MBs. In this way, we focused on evaluating the functionality of magnetic actuation for the device and avoided influences of other conditions. After completion of the procedures, the MB–cell complexes were removed from chamber 6 to a centrifuge tube, and the bead dissociation solution was added, mixed and incubated. Subsequently, the supernatant with released cells was aspirated under the action of a permanent magnet. The recovered cells were washed, resuspended and counted using cell-count boards. Recovery rate of CD4+ cells was calculated according to the following formula,
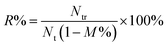 | (2) |
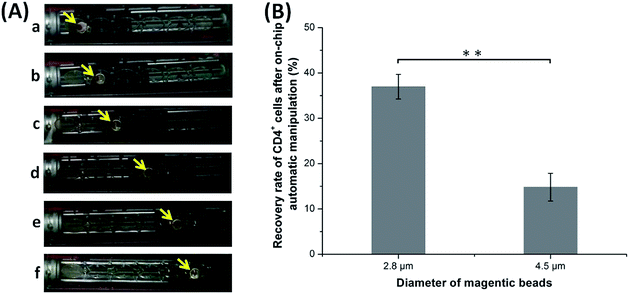
Fig. 7(A) shows images of the movement of the MB–CD4+ cell complexes passing through reaction chambers in sequence under the control of the magnetic device, indicating the feasibility of automatic magnetic manipulation. The result in Fig. 7(B) shows that the average recovery rate of CD4+ cells is 36.97 ± 2.72% for the small MBs, which demonstrates highly significant difference to that of the large MBs (with an average recovery rate of 14.80 ± 3.06%) (p < 0.01, n = 3). Therefore, the 2.8 μm MBs were selected as the carrier of CD4+ cells in the following experiments.
In this experiment, target cells were lost with every traverse of oil barrier,31 possibly due to nonspecific adsorption to chamber walls30 or dissociation of MB–cell complexes. Multiple traverses would obviously decrease the recovery rate of CD4+ cells. This issue could be addressed by further optimizations of the size and moving speed of the magnet in the 1D-moving module, as well as the structure and surface modification of the chambers.37
Automatic on-chip IF staining of γ-H2AX in UVC-irradiated CD4+ cells
The mortality rate of CD4+ cells was measured after UVC irradiation over the dose range of 0–64 J m−2 and then incubation for 30 minutes. It can be seen from Fig. S6† that as the radiation dose increases, the cell mortality rate increases accordingly, with a range of 14.93 ± 2.14%–62.27 ± 4.81%. The mortality rate (>60%) for the highest UVC radiation dose (64 J m−2) was too high to obtain accurate and reproducible experimental results. Therefore, we chose the UVC radiation dose range of 0–32 J m−2 in the subsequent experiment.Using the developed portable magnetic device and primarily optimized conditions, this proof-of-principle study achieved on-chip automatic IF staining of γ-H2AX in UVC-irradiated CD4+ cells. After measurement of fluorescence intensity using flow cytometry, the relative fluorescence intensity was calculated and a radiation dose–response relationship was established. In Fig. 8, it shows that the linear relationship of the obtained radiation dose–response curve is significant (R2 > 0.87) using the on-chip staining method, which is similar to that of the conventional in-tube staining protocol (R2 > 0.94), indicating the feasibility of on-chip automatic IF staining of γ-H2AX in CD4+ cells.
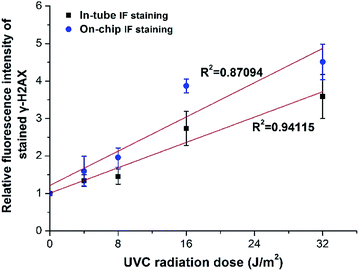 | ||
| Fig. 8 Relationship between UVC radiation dose and relative fluorescence intensity of γ-H2AX in CD4+ cells stained using both manual in-tube protocol and automatic on-chip protocol. | ||
On the other hand, the standard in-tube protocol for MB–CD4+ cell binding and γ-H2AX staining is time-consuming (entirely about 3 hours for at least 11 steps), half of which needed to be performed on ice (Fig. S7†), labor-intensive and operator-dependent (involving multiple manual operations, such as buffer exchange, cell centrifugation, etc.) according to the manufacturer's instructions. To speed up the MB-based IF staining process, a simplified protocol was presented for automatic on-chip IF staining of γ-H2AX in CD4+ cells.
The whole on-chip staining procedures could be completed within six chambers (Fig. 4A) by removing several steps of washing and combining the two steps of permeabilization and staining, as compared to in-tube protocol. This study shows that the automatic on-chip staining protocol is efficient (with 3D mixing of MBs and cells), automatic and rapid (approximately 40 min in total), and all reactions can be done at room temperature, which would substantially improve the practicability of this method. Further optimization of the on-chip procedures will be expected to reduce the time required for the automatic on-chip staining protocol.
Concluding remarks
This work presents a portable, cost-effective and integratable magnetic device, which contains a 3D-mixing module to perform on-chip efficient cell capture, and a 1D-moving module to conduct automatic intracellular IF staining. We propose a novel mechanism of on-chip magnetic 3D mixing, which is based on eccentrically-rotating upper and bottom circular plates embedded with staggeredly arranged magnets (ec-2MagRotors) to generate periodically changing 3D magnetic field, achieving thorough mixing of MBs and cells to obtain efficient cell capture at relatively low bead-to-cell concentration ratios. Automatic and rapid IF staining of γ-H2AX in UVC-irradiated CD4+ cells was conducted successfully, demonstrating the feasibility of the MB-based fully automatic IF staining protocol. Future studies would focus on fully characterizing properties of the portable magnetic device to obtain optimal conditions, and redesigning the capture chamber to enlarge its volume for efficient enrichment and staining of CD4+ cells from blood samples. Moreover, further integration of detection module like microfluidic cytometer14 or other approaches will be carried out for future development of μ-TAS for POC IF assays.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Fundamental Research Funds for the Central Universities (3132019335), Advance Research Project in Chinese Manned Spaceflight (040101), Aerospace Medicine Project in China Space Station Program (04002) and Scientific research project in key fields of Chinese manned space application system (2018 (28)).References
- R. H. Kim and N. K. Brinster, Am. J. Dermatopathol., 2020, 42, 75–85 CrossRef PubMed.
- A. Francisco-Cruz, E. R. Parra, M. T. Tetzlaff and I. I. Wistuba, Methods Mol. Biol., 2020, 2055, 467–495 CrossRef PubMed.
- K. Im, S. Mareninov, M. F. P. Diaz and W. H. Yong, Methods Mol. Biol., 2019, 1897, 299–311 CrossRef PubMed.
- A. Ghanadan, A. Saghazadeh, I. Jahanzad and N. Rezaei, Expert Rev. Clin. Immunol., 2015, 11, 597–616 CrossRef PubMed.
- M. Durdik, P. Kosik, J. Gursky, L. Vokalova, E. Markova and I. Belyaev, Cytometry, Part A, 2015, 87, 1070–1078 CrossRef PubMed.
- P. L. Meroni, N. Bizzaro, I. Cavazzana, M. O. Borghi and A. Tincani, BMC Med., 2014, 12, 38 CrossRef PubMed.
- K. Khan, S. Tewari, N. P. Awasthi, S. P. Mishra, G. R. Agarwal, M. Rastogi and N. Husain, Med. Hypotheses, 2018, 115, 22–28 CrossRef PubMed.
- L. Jezkova, M. Zadneprianetc, E. Kulikova, E. Smirnova, T. Bulanova, D. Depes, I. Falkova, A. Boreyko, E. Krasavin, M. Davidkova, S. Kozubek, O. Valentova and M. Falk, Nanoscale, 2018, 10, 1162–1179 RSC.
- P. Johansson, A. Fasth, T. Ek and O. Hammarsten, Cytometry, Part B, 2017, 92, 534–540 CrossRef PubMed.
- M. Wojewodzka, S. Sommer, M. Kruszewski, K. Sikorska, M. Lewicki, H. Lisowska, A. Wegierek-Ciuk, M. Kowalska and A. Lankoff, Radiat. Res., 2015, 184, 95–104 CrossRef PubMed.
- S. Wang, S. Tasoglu, P. Z. Chen, M. Chen, R. Akbas, S. Wach, C. I. Ozdemir, U. A. Gurkan, F. F. Giguel, D. R. Kuritzkes and U. Demirci, Sci. Rep., 2014, 4, 3796 CrossRef PubMed.
- H. Shafiee, S. Wang, F. Inci, M. Toy, T. J. Henrich, D. R. Kuritzkes and U. Demirci, Annu. Rev. Med., 2015, 66, 387–405 CrossRef.
- D. Wasserberg, X. Zhang, C. Breukers, B. J. Connell, E. Baeten, D. van den Blink, È. S. Benet, A. C. Bloem, M. Nijhuis, A. M. J. Wensing, L. Terstappen and M. Beck, Biosens. Bioelectron., 2018, 117, 659–668 CrossRef PubMed.
- J. Wang, Z. Fan, Y. Zhao, Y. Song, H. Chu, W. Song, Y. Song, X. Pan, Y. Sun and D. Li, Sci. Rep., 2016, 6, 23165 CrossRef.
- A. Manz, N. Graber and H. M. Widmer, Sens. Actuators, B, 1990, 1, 244–248 CrossRef.
- S. Haeberle and R. Zengerle, Lab Chip, 2007, 7, 1094–1110 RSC.
- A. C. Fernandes, K. V. Gernaey and U. Kruhne, Biotechnol. Adv., 2018, 36, 1341–1366 CrossRef.
- K. Ohno, K. Tachikawa and A. Manz, Electrophoresis, 2008, 29, 4443–4453 CrossRef PubMed.
- B. Nasseri, N. Soleimani, N. Rabiee, A. Kalbasi, M. Karimi and M. R. Hamblin, Biosens. Bioelectron., 2018, 117, 112–128 CrossRef.
- B. C. Dhar and N. Y. Lee, BioChip J., 2018, 12, 173–183 CrossRef.
- E. Iswardy, T. C. Tsai, I. F. Cheng, T. C. Ho, G. C. Perng and H. C. Chang, Biosens. Bioelectron., 2017, 95, 174–180 CrossRef PubMed.
- D. G. Dupouy, A. T. Ciftlik, M. Fiche, D. Heintze, B. Bisig, L. de Leval and M. A. Gijs, Sci. Rep., 2016, 6, 20277 CrossRef PubMed.
- J. Shen, Y. Zhou, T. Lu, J. Peng, Z. Lin, L. Huang, Y. Pang, L. Yu and Y. Huang, Lab Chip, 2012, 12, 317–324 RSC.
- S. Kwon, C. H. Cho, Y. Kwon, E. S. Lee and J. K. Park, Sci. Rep., 2017, 7, 45968 CrossRef PubMed.
- A. van Reenen, A. M. de Jong, J. M. den Toonder and M. W. Prins, Lab Chip, 2014, 14, 1966–1986 RSC.
- Q. Cao, X. Han and L. Li, Lab Chip, 2014, 14, 2762–2777 RSC.
- R. Zhong, K. Flack and W. Zhong, Analyst, 2012, 137, 5546–5552 RSC.
- H. M. Pezzi, D. J. Guckenberger, J. L. Schehr, J. Rothbauer, C. Stahlfeld, A. Singh, S. Horn, Z. D. Schultz, R. M. Bade, J. M. Sperger, S. M. Berry, J. M. Lang and D. J. Beebe, Lab Chip, 2018, 18, 3446–3458 RSC.
- B. P. Casavant, L. N. Strotman, J. J. Tokar, S. M. Thiede, A. M. Traynor, J. S. Ferguson, J. M. Lang and D. J. Beebe, Lab Chip, 2014, 14, 99–105 RSC.
- M. Berenguel-Alonso, X. Granados, J. Faraudo, J. Alonso-Chamarro and M. Puyol, Anal. Bioanal. Chem., 2014, 406, 6607–6616 CrossRef PubMed.
- B. P. Casavant, D. J. Guckenberger, S. M. Berry, J. T. Tokar, J. M. Lang and D. J. Beebe, Lab Chip, 2013, 13, 391–396 RSC.
- A. G. Gehring and S. I. Tu, Annu. Rev. Anal. Chem., 2011, 4, 151–172 CrossRef PubMed.
- Y. Gao, A. van Reenen, M. A. Hulsen, A. M. de Jong, M. W. Prins and J. M. den Toonder, Lab Chip, 2013, 13, 1394–1401 RSC.
- C. Murray, H. Miwa, M. Dhar, D. E. Park, E. Pao, J. Martinez, S. Kaanumale, E. Loghin, J. Graf, K. Raddassi, W. W. Kwok, D. Hafler, C. Puleo and D. Di Carlo, Lab Chip, 2018, 18, 2396–2409 RSC.
- Q. Xiong, C. Y. Lim, J. Ren, J. Zhou, K. Pu, M. B. Chan-Park, H. Mao, Y. C. Lam and H. Duan, Nat. Commun., 2018, 9, 1743 CrossRef PubMed.
- T. Lu, Y. Zhang, M. Wong, A. Feiveson, R. Gaza, N. Stoffle, H. Wang, B. Wilson, L. Rohde, L. Stodieck, F. Karouia and H. Wu, Life Sci. Space Res., 2017, 12, 24–31 CrossRef PubMed.
- K. Perez-Toralla, J. Champ, M. R. Mohamadi, O. Braun, L. Malaquin, J. L. Viovy and S. Descroix, Lab Chip, 2013, 13, 4409–4418 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available: (1) Pictures of the main components inside the device. (2) The circuit diagram and the driver board for stepper motors. (3) Schematic diagram of the UVC radiation box for CD4+ cell irradiation. (4) Schematic illustration and rotation images of the magnetic 3D-mixing module. (5) Computational modeling of magnetic field intensity and relative magnetic forces for the 3D mixing module. (6) Effect of UVC radiation dose on the mortality rate of CD4+ cells. (7) Standard in-tube protocol for magnetic bead–CD4+ cell binding and γ-H2AX immunofluorescence staining. See DOI: 10.1039/d0ra03925j |
| ‡ Present address: Department of Applied Biology and Chemical Technology, The Hong Kong Polytechnic University, Hung Hom, Kowloon, Hong Kong, China. |
| This journal is © The Royal Society of Chemistry 2020 |