Open Access Article
Open Access ArticleOrigami-based “Book” shaped three-dimensional electrochemical paper microdevice for sample-to-answer detection of pathogens†
Tao Hea,
Jingwen Liab,
Lisheng Liucd,
Shenguang Ge a,
Mei Yanae,
Haiyun Liu
a,
Mei Yanae,
Haiyun Liu *af and
Jinghua Yu
*af and
Jinghua Yu ae
ae
aCollaborative Innovation Center of Technology and Equipment for Biological Diagnosis and Therapy in Universities of Shandong, Institute for Advanced Interdisciplinary Research (IAIR), University of Jinan, Jinan 250022, P. R. China. E-mail: chm_liuhy@ujn.edu.cn; Fax: +86-531-82765956; Tel: +86-531-82767161
bCollege of Biological Sciences and Technology, University of Jinan, Jinan 250022, P. R. China
cKey Laboratory of Animal Resistance Research, College of Life Science, Shandong Normal University, Jinan, 250014, P. R. China
dClinical Laboratory, Shandong Cancer Hospital Affiliated to Shandong University, Shandong Academy of Medicine Science, Jinan, 250117, P. R. China
eSchool of Chemistry and Chemical Engineering, University of Jinan, Jinan, 250022, P. R. China
fCollege of Chemistry, Chemical Engineering and Materials Science, Key Laboratory of Molecular and Nano Probes, Ministry of Education, Shandong Provincial Key Laboratory of Clean Production of Fine Chemicals, Shandong Normal University, Jinan, 250014, P. R. China
First published on 8th July 2020
Abstract
Herein, an ease-of-use and highly sensitive origami-based “book” shaped three-dimensional electrochemical paper microdevice based on nucleic acid testing (NAT) methodology was developed for sample-to-answer detection of pathogens from whole blood and food samples. The whole steps of NAT, including sample preparation, amplification and detection, were performed by alternately folding the panels of the microdevice, just like flipping a book. The screen-printing electrodes were combined with wax-printing technology to construct a paper-based electrochemical unit to monitor Loop-mediated isothermal amplification (LAMP) reaction with an electrochemical strategy. After nucleic acid extraction and purification with the glass fiber, the LAMP reaction was performed for 45 min to amplify the extracted nucleic acid sequence, followed by the execution of the electrochemical interrogation reaction based on methylene blue (MB) and double-stranded LAMP amplicons. Starting with whole blood and food samples spiked with Salmonella typhimurium, this microdevice was successfully applied to identify pathogens from biological samples with satisfactory sensitivity and specificity. Therefore, the proposed origami-based “book” shaped three-dimensional paper microdevice has great potential for disease diagnosis, food safety analysis applications in the future.
1 Introduction
The incidence and mortality of infectious diseases caused by pathogens remains extraordinarily high and constitutes a global health threat,1–3 especially in developing countries.4 With the increasing incidence of infectious diseases,5 nucleic acid testing (NAT) has played an essential role in controlling and monitoring the worldwide infectious disease due to superior sensitivity and specificity.6–10 For example, loop-mediated isothermal amplification (LAMP),11–13 rolling circle amplification (RCA),14,15 and recombinase polymerase amplification (RPA)16,17 have been reported to perform the detection of pathogens. However, these strategies require precise and specialized laboratory equipment, such as qPCR thermocyclers, turbidimeters, centrifuges, fluorescent microscopes, and so on, which impose restriction on their applications of rapid identification of pathogens.18–21Recently, paper-based microfluidic technologies integrated with NAT methodology have garnered much attention for sample-to-answer detection of pathogens because they are cost-effective, portable and disposable.22–27 The paper-based microfluidic technologies eliminate the requirement of pumps or other fluid flow instrumentation due to paper's natural ability to wick fluids through capillary action.28–30 For instance, Whitesides' group has reported a “paper machine” for fluorescent detection of E. coli cells by sliding the device.31 Integrated sample-to-answer paper-based microfluidic devices have been developed to incorporate FTA card-based extraction, amplification and lateral flow assay (LFA) to achieve colorimetric readout.32–34 Despite the attractiveness of these paper-based microfluidic NAT systems, there are multiple limitations render their applications. Most of these NAT systems involve gray value, colorimetric, fluorescence and LFA detection, and the sensitivity and specificity need to be further improved. Besides, complicated assay procedures were also an obstacle for researcher to complete sample-to-answer analysis. The electrochemical method with the distinct virtues of speed and sensitivity has shown good promise to achieve the qualitative and quantitative detection of the isothermal amplification products. There are currently fewer NAT devices based on electrochemical strategies for sample-to-answer detection of pathogens. Consequently, there is a strong demand to develop a convenient operating paper-based electrochemistry NAT device which can achieve higher sensitivity and specificity.
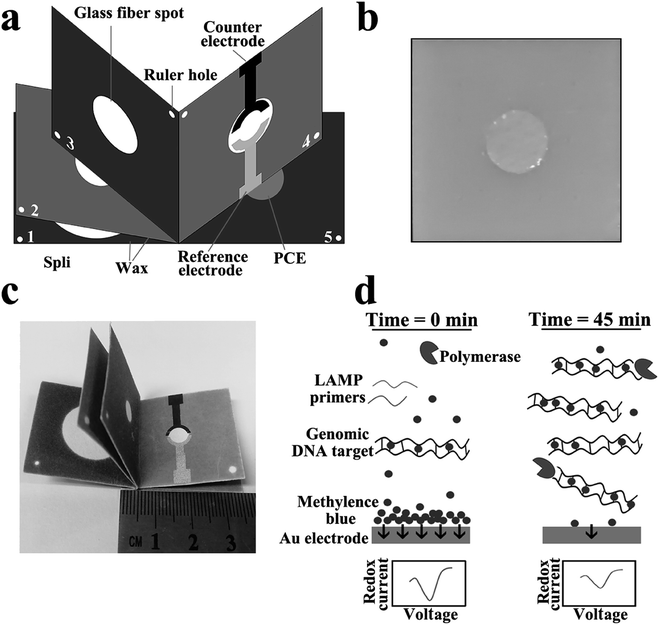
In this study, we develop an origami-based “book” shaped three-dimensional electrochemical paper microdevice with simple operation and high sensitivity, which realizes the detection of pathogens from sample-to-answer by using electrochemical strategy. This electrochemical paper microdevice was composed of wicking panels (Fig. 1a, panel 1 & 2), extraction panel (Fig. 1a, panel 3), amplification pad (Fig. 1b), and paper-based electrochemical unit panel (Fig. 1a, panel 4 & 5). As shown in Fig. 1, all panels were stacked and stapled into a “book”-like structure so that the sequential steps of sample preparation, LAMP reaction and electrochemical detection could be accomplished with simply flipping the panels. The filter paper was first printed with wax and heated to melt the printed wax to form hydrophobic pattern of channels and vias. The glass fiber was selected to perform sample preparation due to its properties of high flow rate, high nucleic acid loading and low protein adsorption. After nucleic acid extraction and purification, the LAMP reaction was conducted directly within the sealed amplification chamber via a commercial hotplate. At the end of LAMP reaction, the electrodes of paper-based electrochemical unit were connected to potentiostat and the redox current was measured. The electrochemical interrogation based on a redox active molecule methylene blue (MB) and double-stranded LAMP reaction products was executed to monitoring the LAMP reaction. The electrochemical strategy has distinct virtues of speed and sensitivity compare with colorimetric and fluorescence methods. Using screen printing electrodes combined with wax printing technology, we constructed a paper-based electrochemical device to perform electrochemical monitoring of the LAMP reaction. With Salmonella typhimurium as a template, our microdevice has achieved satisfactory sensitivity and specificity. Besides, the detection results of our microdevice consistent with the benchmark PCR, which demonstrated that our microdevice could be an attractive substituted tool for the determination of pathogens in the future.
2 Material and methods
2.1 Chemicals and materials
10× Isothermal Amplification Buffer, Bst 2.0 DNA polymerase and dNTPs were obtained from NEB (New England Biolabs). The electrophoresis regents, TE buffer, betaine, DNase-free and methylene blue (MB) were purchased from Sigma-Aldrich (Shanghai, China). Carbon ink (ED423ss) and Ag/AgCl ink (CNC-01) were purchased from Acheson. The glass fiber and fiber paper were purchased from Whanman (Beijing, China). The drinking water and milk were purchased from local supermarkets. The clinical serum samples used in here were obtained from Shandong Tumor Hospital under the prior approval from serum donors. All experiments were conducted following national laws and relative regulation and permitted by Shandong Tumor Hospital. The LAMP primers for the S. typhimurium were ordered from Takara Biotechnology Co., Ltd (Dalian, China) with HPLC purification, and the sequences of primers were shown in ESI Table S1.†2.2 Bacteria culture
S. typhimurium was cultured on a Luria-Bertani (LB) plate at 37 °C overnight. A single colony was pricked and grown at 37 °C overnight in 10 mL of LB medium, with agitation at 110 rpm. The turbidity of bacteria suspension at a wavelength of 600 nm (OD 600) and the colony counting method was combined to determine the concentrations of bacteria.2.3 Design and fabrication of the “book” shaped three-dimensional electrochemical paper microdevice
The shape for wax-printing of this microdevice was designed using Adobe Illustrator CS4. The fabrication process was performed without specialised facilities or a clean room by simply using a wax printer and a hotplate (the latter can also be used to carry out the LAMP reaction). In order to assemble the microdevice, the filter paper was first printed with wax using an office printer, then heated at 120 °C for 1 min on the hotplate to melt the printed wax, which diffused through the paper to form the same hydrophobic pattern of channels and vias on both sides. The paper-based electrochemical unit was manufactured using screen printing electrodes combined with wax-printing technology. The circle hydrophilic zone of panel 4 was designed for screen-printing carbon counter electrode and Ag/AgCl reference electrode, and the circle hydrophilic zone of panel 5 was designed for fabricating gold nanocomposite modified paper cathode electrode (Au-PCE). The SEM images of bare paper and Au-PCE was showed in Fig. S1.† The detailed manufacturing process was shown in ESI.†35 All panels were stacked and stapled into a “book” as shown in Fig. 1c. Subsequently the glass fiber spots were manually positioned to glass fiber spot (panel 3) and amplification chamber (Fig. 1b) so that appropriate folding manipulations allowed the easy transfer of reagents and sample. Finally the device was then sealed using two acetate films or cling film, preventing liquid evaporation during the amplification.2.4 General operation of the electrochemical paper microdevice
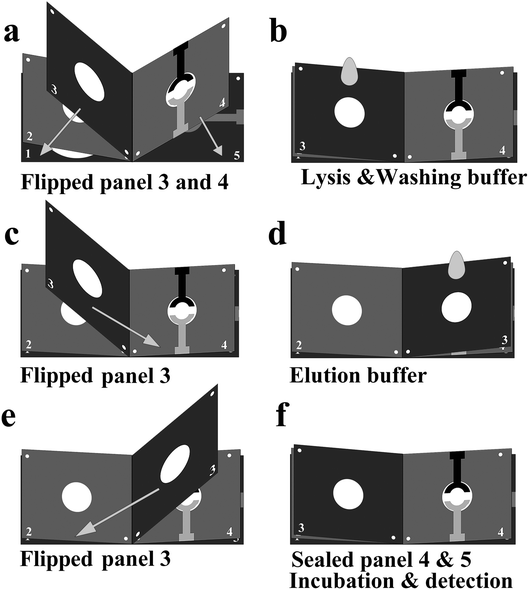
The whole procedure for sample-to-answer assays was illustrated in Fig. 2. After flipped the panel 3 and 4 (Fig. 2a), the samples and washing buffer were continuously added onto the glass fiber spot (panel 3) enable the first steps of the assay, involving cell lysis and DNA extraction, to yield purified DNA on the glass fiber spots (Fig. 2b). The panel 1 and 2 were used to absorb cell lysates through capillary action. To transfer the extracted DNA from the glass fiber spot to amplification chamber, the panel 3 was flipped onto the panel 4 (Fig. 2c) and elution buffer was added into the glass fiber spot (Fig. 2d). After eluting DNA, the panel 3 was flipped on the opposite side (Fig. 2e) and the panel 4, plastic plate and panel 5 were sealed by an acetate film to prevent evaporation during amplification (Fig. 2f). Before starting the LAMP reaction, the electrodes of paper-based electrochemical unit (panel 4 & 5) were first connected to a potentiostat for three per-LAMP electrochemical measurements at room temperature. At the end of LAMP reaction, the microdevice was removed from the hotplate, and the currents were measured three times as the microdevice cooled down to room temperature (Fig. 2f).2.5 Paper-based extraction procedure
Paper-based nucleic acid extraction eliminated the need of centrifuges or other sample treatment equipment. 10 μL of blood or food samples was mixed with 70 μL lysis buffer (L6 – 1.20 g mL−1 of GuSCN, 0.1 M Tris–hydrochloride, 0.04 M EDTA, adjusted with NaOH to pH 8.0, 26 mg mL−1 Triton X-100), before heating at 95 °C for 5 minutes. The mixtures were dispensed onto the glass fiber spot (Fig. 1, panel 3) and absorbed by capillary action into the panel 1 & 2 (Fig. 1). The DNA was drawn in the flow of liquid from panel 3 to panel 1 and captured by the glass fiber. Subsequently, 50 μL washing buffer containing 70% ethanol and 30 mM NaOAc was introduced to wash cell residues and other inhibitors away. After washing, 20 μL elution buffer (10 mM TE buffer, pH 8.0) was used to eluted the nucleic acid from the glass fiber onto the amplification chamber where the LAMP primers and reagents were previous deposited (Fig. 1b).We compared the DNA recovery rate of paper extraction with traditional centrifuge extraction using serially diluted genomic DNA from 0.04 to 5 ng μL−1. The diluted DNA was put through the extraction process on the paper device and the traditional centrifugation extraction methods, respectively. After elution, 1 μL of liquid of paper extractions and traditional centrifugation extractions was analysed by Quantitative Real-Time PCR (qRT-PCR) to determine the extraction efficiency, respectively, compared to the original dilutions.
2.6 On-chip LAMP amplification and detection
We based our LAMP system on previously published primer sequences for Salmonella typhimurium (S. typhimurium).22 The LAMP primers were ordered from Takara Biotechnology Co., Ltd (Dalian) with HPLC purification, and the sequences of primers were shown in ESI Table S1.† The 20 μL LAMP reaction mixture containing 1× isothermal amplification buffer, 4 mM MB, 6 mM MgSO4, 1.4 mM dNTPs, 1.6 μM of each inner primer (FIP and BIP), 0.2 μM of each outer primer (F3 and B3), 0.8 μM of each loop primer (LF and LB), 0.8 U of Bst 2.0 DNA polymerase was preloaded in the LAMP chamber. After nucleic acid extraction, the paper electrodes were first connected to a potentiostat for three per-LAMP electrochemical measurements at room temperature. After that, the LAMP amplification was conducted at 65 °C for 45 min in a hotplate. Differential pulse voltammetry (DPV) was selected for monitoring the electrochemical interrogation based on MB and double-stranded LAMP reaction products. The DPV measurement was taken: the potential ranges from −0.3 to −0.1 V, modulation amplitude was 0.05 V, pulse width was 0.05 s, and sample width was 0.0167 s.2.7 Determination of analytical sensitivity with purified genomic DNA
The analytical sensitivity of “book” shaped three-dimensional electrochemical paper microdevice was determined using purified genomic DNA of S. typhimurium. The concentration of purified genomic DNA was confirmed by spectrometry using the absorbance at 260 nm. After being serially diluted from 0.1 to 104 pg μL−1, the purified genomic DNA were denatured at 95 °C for 5 minutes and annealed at room temperature. 10 μL diluted genomic DNA was applied to dry glass fiber spot, which had been pretreated with 10 μL of human blood, 70 μL lysis buffer and 50 μL washing buffer. The glass fiber spots were then dried completely, and the general operations outlined above were followed starting from the application of 20 μL elution buffer.2.8 Integrated assay with biological samples
For a demonstration of the sample-to-answer capability of the “book” shaped three-dimensional electrochemical paper microdevice, the platform was investigated with S. typhimurium-spiked human whole blood samples over a range concentration of bacteria from 0 to 105 CFU mL−1. All the steps are as described in the operation process of paper microdevice. To further show the potential of the microdevice to be applied in food safety analysis, the S. typhimurium-spiked drinking water and milk were also tested. The blood samples were obtained from anonymous healthy donors, drinking water and milk were purchased from local supermarkets.3 Results and discussion
3.1 “Book” shaped three-dimensional electrochemical paper microdevice
This electrochemical paper microdevice was designed to perform DNA preparation, isothermal amplification and electrochemical detection with minimal user steps. An overview of the “book” shaped three-dimensional electrochemical paper microdevice is schematically illustrated in Fig. 1a. Each microdevice contained four components, namely: a filter paper based fluidic device where the liquid was constrained by printed hydrophobic wax (Fig. 1a); a plastic plate with a glass fibre spots (diameter 6 mm), which form amplification chamber for LAMP reaction (Fig. 1b); one glass fiber spot (panel 3, Fig. 1a) (GFF, Whatman) with diameter 6 mm for absorbing nucleic acids from samples in the presence of high concentration of the chaotropic agent, guanidine thiocyanate (GuSCN); a paper-based electrochemical unit (panel 4 and 5, Fig. 1a) for monitoring LAMP reaction.As shown in Fig. 1c, this microdevice is small, only 3.5 cm long and 3 cm wide. By alternatively flipping the panel (Fig. 2), the steps of DNA release, isolation and purification was performed with this microdevice. After sample preparation, the LAMP reaction was conducted at 65 °C in the sealed amplification chamber via a hotplate. Temperature calibration curve for the hotplate and the temperature of the amplification chamber during amplification reaction was investigated in the support material (Fig. S2†). The electrochemical interrogation based on MB and double-stranded LAMP reaction products was executed through the paper-based electrochemical unit to monitoring the LAMP reaction. Initially, the MB molecules are free to encounter the gold working electrode and transfer electrons, producing a high peak current (Fig. 1d, left). As the LAMP reaction progresses, the MB intercalated into double-stranded LAMP reaction products and thus reduced the free MB concentration, decreasing the peak current (Fig. 1d, right). By combining the origami setting like a flipping book and the electrochemical strategies, we constructed a sample-to-answer detection device with direct electrochemical readout, high sensitivity, and low detection limit.
3.2 DNA recovery rates of paper extraction
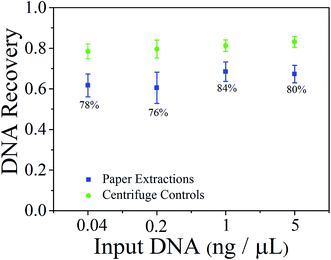
The DNA recovery rates of the paper extraction were compared with that of traditional centrifuge (Fig. 3). The DNA recovery rates of paper extraction and traditional centrifugation extraction (1 denotes a perfect recovery without loss) obtained by real-time PCR with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 serial diluted S. typhimurium genomic DNA form 25 ng μL−1. Results from triplicate paper extraction experiments show the DNA recovery rates for different concentrations were between 60–70%. The DNA recovery rates through the glass fiber ranged from 75% to 85% compared to centrifugation control. These results demonstrate that paper extraction method is slightly lower than that of traditional centrifuge, but satisfactory DNA recovery rates have also been obtained.
5 serial diluted S. typhimurium genomic DNA form 25 ng μL−1. Results from triplicate paper extraction experiments show the DNA recovery rates for different concentrations were between 60–70%. The DNA recovery rates through the glass fiber ranged from 75% to 85% compared to centrifugation control. These results demonstrate that paper extraction method is slightly lower than that of traditional centrifuge, but satisfactory DNA recovery rates have also been obtained.
3.3 Optimization of experimental parameters
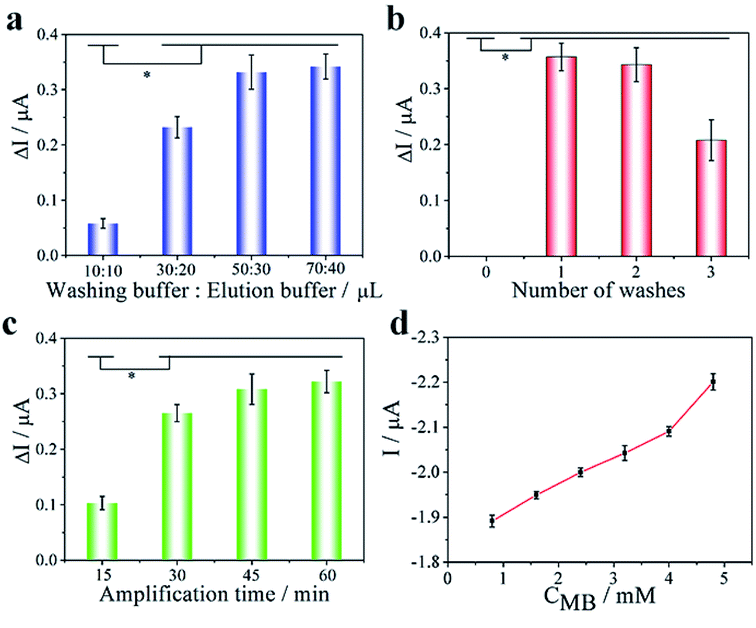
To achieve the optimum analytical performance of the platform, the experimental parameters were investigated with the changes of current intensity between different peak current and background (ΔI) as an evaluation criterion (Fig. 4). Due to superior extraction efficiency and simple processing steps, the glass fiber was selected to complete cell lysis, nucleic acid extraction and purification from biological samples. The washing buffer and elution buffer were tested with volumes of 10 and 5 μL, 30 and 10 μL, 50 and 20 μL, 70 and 30 μL, respectively. As depicted in (Fig. 4a), 50 μL washing buffer and 20 μL elution buffer produced a higher change of current intensity (ΔI) as compared to others. Therefore, 50 μL purification reagent and 20 μL TE buffer were employed as the optimum volume of purification and washing buffer. Furthermore, the effect of number of washes was also compared under the optimum volume of two buffers (Fig. 4b). There was no significant difference in the changes of current intensity between one and two washes. However, due to the excessive purification and washing buffer causes the loss of DNA, the change of current intensity (ΔI) of the biological samples washed for three times was almost half of the sample that is washed for one times and two times.Amplification time and temperature were crucial for the amplification efficiency of the LAMP reaction. LAMP amplification was performed on glass fiber with a range of incubation time (15, 30, 45 and 60 min) at 65 °C to optimize the amplification time. As depicted in Fig. 4c, the maximum ΔI reached at the incubation time of 45 min, which indicated that the optimum amplification time was 45 min. Additionally, a range of amplification temperature (58, 60, 63, 65 and 68 °C) were also evaluated at the optimum amplification time. With SYBR Green I fluorescence staining, agarose gel electrophoresis was used to verify the amplification efficiency of LAMP amplification at different temperatures. We found that the bands of amplification at 65 °C produce the most clearly visible light among all temperatures tested (Fig. S4†), which indicated that 65 °C was the highest efficiency of amplification temperature.
The concentration of MB was crucial for the sensitivity of biosensor. We investigated the concentration of MB in a range from 0.8 to 4.8 mM to optimize the sensitivity of biosensor (Fig. S3†). With increasing of MB concentration from 0.8 to 4.0 mM, the current response changed slowly, while suddenly increased at the concentration of 4.8 mM (Fig. 4d). This could be attributed to MB concentration was supersaturated when MB concentration reached 4.8 mM. Therefore, 4.0 mM was employed as the optimum concentration of MB.
3.4 Determination of analytical sensitivity with purified genomic DNA
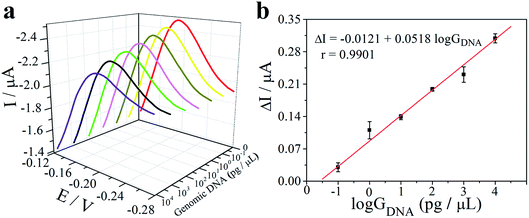
The analytical sensitivity of the electrochemical paper microdevice for pathogens detection was tested using purified genomic DNA of S. typhimurium. The S. typhimurium genomic DNA was isolated and purified from bacterial culture. By drying S. typhimurium genomic DNA onto the glass fiber spot (panel 3, Fig. 1a), which had been treated with whole blood, lysis buffer and washing buffer. 10 μL serially diluted purified genomic DNA (0.1–104 pg μL−1) was introduced through the glass fiber spot into amplification chamber, where the specific LAMP primers for the target pathogens were preloaded. After incubation at 65 °C for 45 minutes, the electrodes of paper-based electrochemical unit were connected to potentiostat, and the redox current was measured.As shown in Fig. 5a (red curve), the relatively large peak current was measured due to the high concentration of free MB in reaction mixture when S. typhimurium genomic DNA was not added. When the target sequence was added in reaction mixture and triggered the LAMP amplification, the LAMP amplification products could combined with MB and reduced the concentration of free MB in reaction mixture and thereby diminished the peak current (Fig. 5a; purple curve). With the increase of genomic DNA concentration, the amplicon produced by the LAMP reaction also increases, more MB molecules are embedded in the amplicon, and the intensity of the current also decreases (Fig. 5a). Moreover, a linear correlation between the changes of current intensity between different peak current and background (ΔI) and the logarithm of the concentration of genomic DNA (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GDNA) was obtained. The linear equation was ΔI = −0.0121 + 0.0518
GDNA) was obtained. The linear equation was ΔI = −0.0121 + 0.0518![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GDNA with a correlation coefficient of 0.9901 and the detection limit (LOD) was calculated to be 42 fg μL−1 (3σ, n = 11).
GDNA with a correlation coefficient of 0.9901 and the detection limit (LOD) was calculated to be 42 fg μL−1 (3σ, n = 11).
3.5 Integrated assay with spiked whole blood and food samples
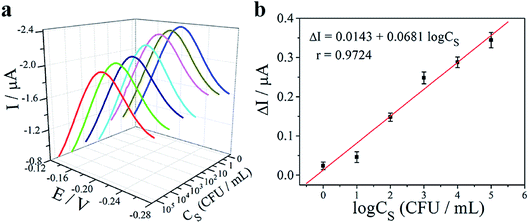
To demonstrate the potential use of the “book” shaped three-dimensional electrochemical paper microdevice for sample-to-answer diagnostics of pathogens, the assays were challenged with S. typhimurium-spiked human whole blood. The whole blood samples were subjected to the whole procedure, in the order as described in General operation of the electrochemical paper microdevice, by alternately flipping the panels for application of lysis buffer, washing buffer and elution buffer. As depicted in Fig. 6a, with the increasing concentration of S. typhimurium (CS) from 102 to 105 CFU mL−1, the peak current was synchronously declined. However, the current peaks at 100 and 101 CFU mL−1 were not significantly lower than the background due to the extracted genomic DNA concentration was too low to trigger the LAMP reaction. The linear correlation between ΔI and the logarithm of the concentration of S. typhimurium (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CS) was also obtained and the LOD was calculated to be 44.2 CFU mL−1 (Fig. 6b).
CS) was also obtained and the LOD was calculated to be 44.2 CFU mL−1 (Fig. 6b).
For further to verify the versatility of our electrochemical paper microdevice, S. typhimurium were spiked into water and food samples, e.g. drinking water and milk, with a serial concentration from 100 to 105 CFU mL−1. As illustrated in Fig. S5,† it is found that the detection limit of S. typhimurium was 1.57, 5.51 CFU mL−1 in drinking water and milk, respectively. The detection limit in drinking water and milk were lower than that in human blood. This could be attributed to the whole blood contains more polymerase inhibitors may potentially blocking subsequent amplification process. These results indicated that our electrochemical paper microdevice is capable of target analysis in whole blood, drinking water and milk, which offering great potential for clinical diagnosis, food and water safety control.
3.6 Analytical sensitivity and specificity in electrochemical paper microdevice
To demonstrate the specificity and sensitivity of our electrochemical paper microdevice for sample-to-answer detection of pathogens, sixty biological samples were tested using our method and benchmark PCR. Our method showed satisfactory specificity and sensitivity for identifying S. typhimurium in blood, drinking water and milk samples (Table 1), when compared against the benchmark PCR. Three biological samples all have a sensitivity of over 80% and exhibit high specificity (>90%). In addition, this work was compared with the existing proposals reported in previous literatures (Table 2). The ability of electrochemical strategies to detect 44.2, 1.57, 5.51 CFU mL−1 bacterial in whole blood, drinking water and milk was comparable or even more sensitive than other methods (fluorescence, LFA, grayscale).| Benchmark PCR | Paper microdevice | Sensitivity | Specificity | ||
|---|---|---|---|---|---|
| Positive | Negative | ||||
| Whole blood | Positive | 14 | 3 | 82% | 91% |
| Negative | 4 | 39 | |||
| Drinking water | Positive | 17 | 2 | 89% | 98% |
| Negative | 1 | 40 | |||
| Milk | Positive | 22 | 4 | 85% | 94% |
| Negative | 2 | 32 | |||
| Pathogen | Biological samples | Methods | LOD | Ref. |
|---|---|---|---|---|
| Salmonella spp | Milk | Fluorescence | 170 CFU mL−1 | 25 |
| S. typhimurium | Artificial urine | Fluorescence | 200 cells per μL | 22 |
| E. coli | Human plasma | Grayscale | 0.5 cells per μL | 31 |
| P. falciparum | Whole blood | Fluorescence | 0.5 parasites per μL | 6 |
| P. falciparum | Whole blood | Fluorescence | 5 parasites per mL | 29 |
| Escherichia coli | Whole blood | LFA | 102 CFU mL−1 | 36 |
| S. typhimurium | Whole blood | Electrochemistry | 14.2 CFU mL−1 | This work |
4 Conclusion
In summary, a single-use format and highly sensitive electrochemical paper microdevice was developed by integrating electrochemical strategy, paper-based nucleic acid extraction and LAMP reaction. We combined wax-printing technology and screen printing electrodes to construct a paper electrochemical unit, and successfully applied the electrochemical strategy to the sample-to-answer detection of pathogens. This electrochemical paper microdevice was successfully applied to identify pathogens from whole blood, milk and water samples with satisfactory sensitivity and specificity.Moreover, the microdevice could be easily modified to analyze multiple samples or analyze multiple indicators of the same sample at the same time. Our next work will improve the multiple detection capability of the microdevice. The origami-based “book” shaped three-dimensional electrochemical paper microdevice exhibited great potential for various sample-to-answer diagnostics in the future.
Ethical statement
The clinical serum samples used in here were obtained from Shandong Tumor Hospital under the prior approval from serum donors. All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of University of Jinan, approved by the Animal Ethics Committee of University of Jinan. All experiments were conducted following national laws and relative regulation and permitted by Shandong Tumor Hospital.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the program for Key Research and Development Program of Shandong Province (2018GSF118023), A Project of Shandong Province Higher Educational Science and Technology Program (J18KA068), Shandong Provincial Key Laboratory of Clean Production of Fine Chemicals (2019FCCEKL03), Taishan Scholar of Shandong Province (ts201712048), the National Natural Science Foundation of China (21874055), the 111 Project of International Corporation on Advanced Cement-based Materials (No. D17001), Major Program of Shandong Province Natural Science Foundation (ZR2017ZC0124) and the project of “20 items of University” of Jinan (No. 2018GXRC001).References
- M. Amiri, A. Bezaatpour, H. Jafari, R. Boukherroub and S. Szunerits, ACS Sens., 2018, 3, 1069–1086 CrossRef CAS PubMed.
- A. H. Havelaar, M. D. Kirk, P. R. Torgerson, H. J. Gibb, T. Hald, R. J. Lake, N. Praet, D. C. Bellinger, N. R. de Silva, N. Gargouri, N. Speybroeck, A. Cawthorne, C. Mathers, C. Stein, F. J. Angulo, B. Devleesschauwer and G. World Health, Organization Foodborne Disease Burden Epidemiology Reference, PLoS Med., 2015, 12, e1001923 CrossRef PubMed.
- M. De Rycker, B. Baragana, S. L. Duce and I. H. Gilbert, Nature, 2018, 559, 498–506 CrossRef CAS PubMed.
- I. Pereiro, A. Bendali, S. Tabnaoui, L. Alexandre, J. Srbova, Z. Bilkova, S. Deegan, L. Joshi, J. L. Viovy, L. Malaquin, B. Dupuy and S. Descroix, Chem. Sci., 2017, 8, 1329–1336 RSC.
- Y. Tepeli and A. Ülkü, Sens. Actuators, B, 2018, 254, 377–384 CrossRef CAS.
- G. Choi, T. Prince, J. Miao, L. Cui and W. Guan, Biosens. Bioelectron., 2018, 115, 83–90 CrossRef CAS PubMed.
- X. Xu, X. Wang, J. Hu, Y. Gong, L. Wang, W. Zhou, X. Li and F. Xu, Electrophoresis, 2019, 40, 914–921 CrossRef CAS PubMed.
- M. G. Krebs, R. L. Metcalf, L. Carter, G. Brady, F. H. Blackhall and C. Dive, Nat. Rev. Clin. Oncol., 2014, 11, 129–144 CrossRef CAS PubMed.
- A. Akhtar, E. Fuchs, T. Mitchison, R. J. Shaw, D. St Johnston, A. Strasser, S. Taylor, C. Walczak and M. Zerial, Nat. Rev. Mol. Cell Biol., 2011, 12, 669–674 CrossRef CAS PubMed.
- J. Zhao, L. Chang and L. Wang, Eur. J. Clin. Microbiol. Infect. Dis., 2019, 38, 829–842 CrossRef CAS PubMed.
- Y. Chen, N. Cheng, Y. Xu, K. Huang, Y. Luo and W. Xu, Biosens. Bioelectron., 2016, 81, 317–323 CrossRef CAS PubMed.
- J. Chi, B. Ma, X. Dong, B. Gao, A. Elbaz, H. Liu and Z. Gu, Analyst, 2018, 143, 4559–4565 RSC.
- H. Yuan, Y. Chao, S. Li, M. Y. H. Tang, Y. Huang, Y. Che, A. S. T. Wong, T. Zhang and H. C. Shum, Anal. Chem., 2018, 90, 13173–13177 CrossRef CAS PubMed.
- J. Wen, W. Li, J. Li, B. Tao, Y. Xu, H. Li, A. Lu and S. Sun, Sens. Actuators, B, 2016, 227, 655–659 CrossRef CAS.
- L. He, Z. Shen, Y. Cao, T. Li, D. Wu, Y. Dong and N. Gan, Analyst, 2019, 144, 2755–2764 RSC.
- H. Ahn, B. S. Batule, Y. Seok and M. G. Kim, Anal. Chem., 2018, 90, 10211–10216 CrossRef CAS PubMed.
- J. Chen, Y. Xu, H. Yan, Y. Zhu, L. Wang, Y. Zhang, Y. Lu and W. Xing, Lab Chip, 2018, 18, 2441–2452 RSC.
- M. Dou, D. C. Dominguez, X. Li, J. Sanchez and G. Scott, Anal. Chem., 2014, 86, 7978–7986 CrossRef CAS PubMed.
- X. Fang, H. Chen, S. Yu, X. Jiang and J. Kong, Anal. Chem., 2011, 83, 690–695 CrossRef CAS PubMed.
- X. Jiang, J. C. Loeb, C. Manzanas, J. A. Lednicky and Z. H. Fan, Angew. Chem., Int. Ed., 2018, 57, 17211–17214 CrossRef CAS PubMed.
- N. A. Feasey, G. Dougan, R. A. Kingsley, R. S. Heyderman and M. A. Gordon, Lancet, 2012, 379, 2489–2499 CrossRef.
- L. Zhang, F. Tian, C. Liu, Q. Feng, T. Ma, Z. Zhao, T. Li, X. Jiang and J. Sun, Lab Chip, 2018, 18, 610–619 RSC.
- H. Deng, X. Zhou, Q. Liu, B. Li, H. Liu, R. Huang and D. Xing, ACS Appl. Mater. Interfaces, 2017, 9, 41151–41158 CrossRef CAS PubMed.
- P. Chen, C. Chen, Y. Liu, W. Du, X. Feng and B.-F. Liu, Sens. Actuators, B, 2019, 283, 472–477 CrossRef CAS.
- T. N. D. Trinh and N. Y. Lee, Lab Chip, 2018, 18, 2369–2377 RSC.
- Y. Fan, S. Liu, K. Gao and Y. Zhang, Microsyst. Technol., 2017, 24, 1783–1787 CrossRef.
- B. Pang, K. Fu, Y. Liu, X. Ding, J. Hu, W. Wu, K. Xu, X. Song, J. Wang, Y. Mu, C. Zhao and J. Li, Anal. Chim. Acta, 2018, 1040, 81–89 CrossRef CAS PubMed.
- J. Hiltunen, C. Liedert, M. Hiltunen, O. H. Huttunen, J. Hiitola-Keinanen, S. Aikio, M. Harjanne, M. Kurkinen, L. Hakalahti and L. P. Lee, Lab Chip, 2018, 18, 1552–1559 RSC.
- G. Xu, D. Nolder, J. Reboud, M. C. Oguike, D. A. van Schalkwyk, C. J. Sutherland and J. M. Cooper, Angew. Chem., Int. Ed., 2016, 55, 15250–15253 CrossRef CAS PubMed.
- J. H. Shin and J. K. Park, Anal. Chem., 2016, 88, 10374–10378 CrossRef CAS PubMed.
- J. T. Connelly, J. P. Rolland and G. M. Whitesides, Anal. Chem., 2015, 87, 7595–7601 CrossRef CAS PubMed.
- R. Tang, H. Yang, Y. Gong, M. You, Z. Liu, J. R. Choi, T. Wen, Z. Qu, Q. Mei and F. Xu, Lab Chip, 2017, 17, 1270–1279 RSC.
- J. R. Choi, J. Hu, Y. Gong, S. Feng, W. A. Wan Abas, B. Pingguan-Murphy and F. Xu, Analyst, 2016, 141, 2930–2939 RSC.
- N. M. Rodriguez, J. C. Linnes, A. Fan, C. K. Ellenson, N. R. Pollock and C. M. Klapperich, Anal. Chem., 2015, 87, 7872–7879 CrossRef CAS PubMed.
- L. Ge, P. Wang, S. Ge, N. Li, J. Yu, M. Yan and J. Huang, Anal. Chem., 2013, 85, 3961–3970 CrossRef CAS PubMed.
- J. R. Choi, J. Hu, R. Tang, Y. Gong, S. Feng, H. Ren, T. Wen, X. Li, W. A. Wan Abas, B. Pingguan-Murphy and F. Xu, Lab Chip, 2016, 16, 611–621 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra03833d |
| This journal is © The Royal Society of Chemistry 2020 |