Open Access Article
Open Access ArticleExploring the styrene metabolism by aerobic bacterial isolates for the effective management of leachates in an aqueous system
Ebciba C.,
Pavithra N.,
Chris Felshia S. and
Gnanamani A. *
*
Microbiology Division, CSIR-CLRI, Adyar, Chennai 600 020, Tamil Nadu, India. E-mail: gnanamani3@gmail.com
First published on 15th July 2020
Abstract
In the present study, the styrene metabolic profile of three aerobic bacterial isolates explored in a batch mode study. The isolates found application in the management of elachates in the waste dump yard. These three bacterial species have different origins and were studied as a single and mixed consortia. The Lysinibacillus strain M01 (from marine sources), Lysinibacillus strain WD03 (from a waste dump yard), and Pseudomonas strain BG07 (from bovine gut) were used in the present study. The styrene concentration was fixed in the range between 0.5 and 1.5 mL L−1. The metabolites obtained upon microbial degradation were assessed using high-performance liquid chromatography (HPLC), UV-visible spectroscopy, and FTIR spectroscopy (Fourier transform infrared spectroscopy). Furthermore, the genes (Sty A, B, C, D, and E) responsible for the degradation of styrene by the three abovementioned isolates were identified using PCR with respective designed primers. Instrumental analyses revealed the presence of phenylacetic acid (PAA) at significant levels in the growth medium after the scheduled experimental period and confirmed the metabolism of styrene by the chosen isolates. Compared to the case of individual cultures, the results of the mixed consortia support the metabolism of styrene at appreciable levels. The present study provides a suitable biological solution for the management of leachates containing styrene and a way to achieve industrially important chemicals (PAA) through a microbially mediated process.
Introduction
Styrene is a starting material in the production of synthetic polymers such as polystyrene and styrene–butadiene rubber.1 The petrochemical, resin, rubber, and polymer industries generate significant amounts of polystyrene wastes, which accumulate in the soil upon disposal. Leaching of styrene molecules contaminates the aqueous system and exerts toxic effects on living organisms.2 Studies have been reported on the presence of styrene monomers, trimers, and oligomers in the leachates of waste dump yards.3–5 In addition, the presence of styrene in water bodies, industrial effluents, and soil has been reported in the literature.6,7 Detailed studies on the harmful effects of styrene, toluene, phenol, and formaldehyde on the environment have also been reported in the literature.8–11 Since long-time exposure to the leachates of polystyrene has adverse effects on human health, the removal of polystyrene from the environment12 is indispensable. Over the past few decades, several thermal, chemical, and biological techniques have been implemented for the removal of polystyrene wastes.13 Moreover, techniques including biofiltration,12 UV-based photodegradation,14 thermo-oxidative degradation,15 and hydrolytic degradation16 have been adopted to reduce polystyrene contamination. However, the high cost, low efficiency, and long duration of the degradation process and the toxic nature of the degradation products are the major drawbacks. In the past decade, an attempt was made towards the microbial degradation of styrene in the leachate.17 Although the degradation of styrene by single cultures, viz., Pseudomonas sps., Corynebacterium strains, Xanthobacter sps., and Rhodococcus sps. are in reports,1,18 however, meagre reports are on the mixed aerobic mixed culture system. Some methanogenic microbial consortia (Clostridium and Enterobacter spp.) isolated from anaerobic sewage sludge transform styrene under strict anaerobic conditions.19 However, research initiatives are required for the microbial degradation of the styrene molecules in the leachates. Hence, herein, an attempt was made to explore the efficacy of aerobic bacterial isolates of three different origins as a single culture and a mixed consortium for the effective metabolism of styrene, and then, these bacterial isolates were applied for the management of leachates containing styrene.In brief, in the present study, the efficacy of three potential bacterial isolates of three different origins was screened for styrene metabolism. Subsequently, the degradation of styrene was assessed based on the characterization of the metabolized products. Since the styrene metabolism of the two of the identified species of the present study has not been in reports till date, exploring the genes responsible for the degradation of styrene was accordingly executed. Based on the observations, the representative styrene metabolic pathways of the isolates were elucidated.
Materials and methods
Isolation of styrene-degrading organisms
Initial studies on the selection of site and the screening of potential bacterial isolates were carried out during the years 2013–2017, and three potential sites, viz., municipal solid waste dump yard located at Perungudi, Chennai, Tamil Nadu (latitude and longitude coordinates are 12.965365 and 80.246109, respectively), marine sediment sludge samples, Ennore, Tamil Nadu, Chennai; (latitude and longitude coordinates are 13.206524 and 80.327225, respectively), and the bovine gut (slaughter house, Kodungaiyur slaughter house, Tamil Nadu, Chennai) as the cattle in the developing countries ingest plastic wastes along with food, were examined for the possibility of having styrene-degrading bacterial species.20Bacterial isolation was carried out by a serial dilution method. The isolates exhibiting growth in a mineral salt medium in the presence of styrene at 0.5 mL L−1 concentration were screened, isolated, and identified (as described hereinafter) based on their morphology, biochemical analysis, and 16S rDNA sequence. The identified isolates were then stored in a nutrient agar slant at 4 °C until use. The composition of the basal mineral medium used in the present study is as follows (per liter of dis. H2O): 0.80 g NH4Cl, 0.70 g K2HPO4, 0.1 g MgSO4·7H2O, 9.20 mg disodium EDTA, 7.0 mg FeSO4·7H2O, 2.0 mg CaSO4, 0.10 mg boric acid, 0.10 mg ZnSO4·7H2O, 0.02 mg MnSO4·4H2O, 0.01 mg cobalt nitrate, 0.01 mg sodium molybdate dihydrate, and 0.50 μg CuSO4·5H2O.
Identification of the isolates
Styrene metabolism
Styrene metabolism studies were conducted with the single isolate as well as with the mixed consortia containing all the three cultures. The styrene concentration utilized herein varied from 0.5 to 1.5 mL L−1. Styrene at the respective concentration was added to 250 mL of a sterile mineral salt medium in a 500 mL capacity Erlenmeyer flask and inoculated with an 18 hour-old culture at 2.0% (OD600 1.0) concentration for both the single and the mixed consortia. After the inoculation, the samples were incubated at 37 °C and 150 rpm for 72 hours. The assessment of the growth profile (at 4 hours intervals), biomass production (wet and dry), and nature of the released metabolites was performed according to the protocols summarized hereinafter. In brief, the growth profile was assessed by measuring the optical density (600 nm) at 4 hour intervals, where the plain medium served as a blank. For biomass determination, the samples were subjected to centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and 4 °C, and the measured weight of the cell mass free from supernatant was taken as the wet weight. The wet mass was then exposed to drying at 70 °C for two hours, and the mass of the dried cells was accordingly measured. The nature of the released metabolites was evaluated as described hereinafter.
000 rpm and 4 °C, and the measured weight of the cell mass free from supernatant was taken as the wet weight. The wet mass was then exposed to drying at 70 °C for two hours, and the mass of the dried cells was accordingly measured. The nature of the released metabolites was evaluated as described hereinafter.
Extraction and analysis of the metabolized products
After 72 hours of incubation, the styrene-amended cultures of both the single and the mixed consortia were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 min at 4 °C, and the bacterial cells were discarded. The supernatant was taken, extracted with equal volumes of ethyl acetate, and further evaporated with nitrogen gas. The concentrated samples were dissolved in acetonitrile and subjected to the following analyses.25,26
000 rpm for 20 min at 4 °C, and the bacterial cells were discarded. The supernatant was taken, extracted with equal volumes of ethyl acetate, and further evaporated with nitrogen gas. The concentrated samples were dissolved in acetonitrile and subjected to the following analyses.25,26
The extracts of the metabolized products were primarily analyzed using UV-visible spectroscopy (Shimadzu 2450) followed by FTIR spectroscopy (Fourier transform infrared spectroscopy-Jasco 4700) and HPLC analysis.27 For high-performance liquid chromatography (HPLC) analysis, the samples were filtered with 0.22 μm pore-sized PVDF syringe filters (Merck Millipore) and analyzed using a C18 reverse phase column (Waters, India, Spherisorb (100 × 4.6 mm) column), with 5 μm particles as the stationary phase, connected to a photodiode detector and fraction collector. The mobile phase used herein was 70% (v/v) methanol with 0.1% (w/v) phosphoric acid in an isocratic mode at the flow rate of 0.8 mL min−1.
Molecular analysis
In order to identify the genes responsible for the metabolism of styrene by the chosen isolates, PCR amplification studies were carried out for the genes Sty A, Sty B, Sty C, Sty D, and Sty E. The Sty A and Sty B genes are the styrene monooxygenase genes, which are accountable for the transformation of styrene to epoxy styrene. On the other hand, Sty C is responsible for the production of an enzyme, epoxy styrene isomerase, which in turn helps in the conversion of epoxy styrene to phenyl acetaldehyde; moreover, Sty D encodes an enzyme, phenyl acetaldehyde dehydrogenase, which is responsible for oxidizing phenyl acetaldehyde to phenylacetic acid.28 The Sty E gene has been found to be responsible for the production of phenylacetyl CoA, which in turn helps in the transportation of styrene across membranes.29 The DNA extracted from E. coli, MTCC 433 strain, acts as a negative control. Table 1 summarizes the details of the primers designed for the respective genes selected for this study.| Genes | Forward primer (5′ to 3′) | Start–stop | Reverse primer (5′ to 3′) | Start–stop | Tm | GC content |
|---|---|---|---|---|---|---|
| Sty A | TCCGTTTGCGACGAAGGTA | 6361–6380 | CGAGCGTATCGGTGATTCCA | 7019–7000 | 59.97 | 55 |
| Sty B | ATAATGCTGCCCAGCAAGC | 3216–3235 | CTCCATCAGTCTGGATCCGC | 3931–3912 | 59.97 | 55–60 |
| Sty C | GATAGCGGTGGCATTCGTT | 8203–8222 | CCGCGATTGGGAGAGAAGTT | 8797–8816 | 59.97 | 55 |
| Sty D | CGCTACTGCGGAATGACAA | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 626–11 626–11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 645 645 |
CAAAGTATCGCCCGTAGCC | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 943–1962 943–1962 |
59.97 | 55 |
| Sty E | CGCTGATCGATGGAAGCTCT | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 043–16 043–16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 422 422 |
GCTCTCGGCATAGAAGGGAC | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 023–17 023–17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 004 004 |
59.97 | 55 |
PCR amplification was carried out in duplicate using 0.25 U Taq polymerase, 10× Taq buffer, 0.25 mM dNTP mix, 1.5 μM of each forward and reverse primers, and 2.5 μl (10–100 ng) of template DNA in a total volume of 20 μl. The PCR amplification was performed for 40 cycles under the following conditions: initial denaturation at 94 °C for 3 min, at 95 °C for 45 s (DNA denaturation), at various annealing temperatures for each gene for 45 s (primer annealing), at 72 °C for 45 s (DNA extension), at 59 °C for 30 s (extra annealing), and a final extension at 72 °C for 10 minutes in a thermal cycler (Eppendorf, Hamburg, Germany). Herein, ten μl of each reaction product was mixed with 10 μL of 2× loading buffer, fractionated in a 2% agarose gel containing ethidium bromide (1 μg mL−1) using a 100-bp DNA ladder (Promega) as a size marker, and visualized under UV light according to the protocol followed elsewhere.
Statistical analysis
Statistical analysis was performed for the experiments conducted on the growth of the isolates and metabolic studies. One-way ANOVA with multiple comparisons was performed using GraphPad prism (Version 5.0) to compare the means between the experimental groups. A p value of <0.05 was considered significant.Results and discussion
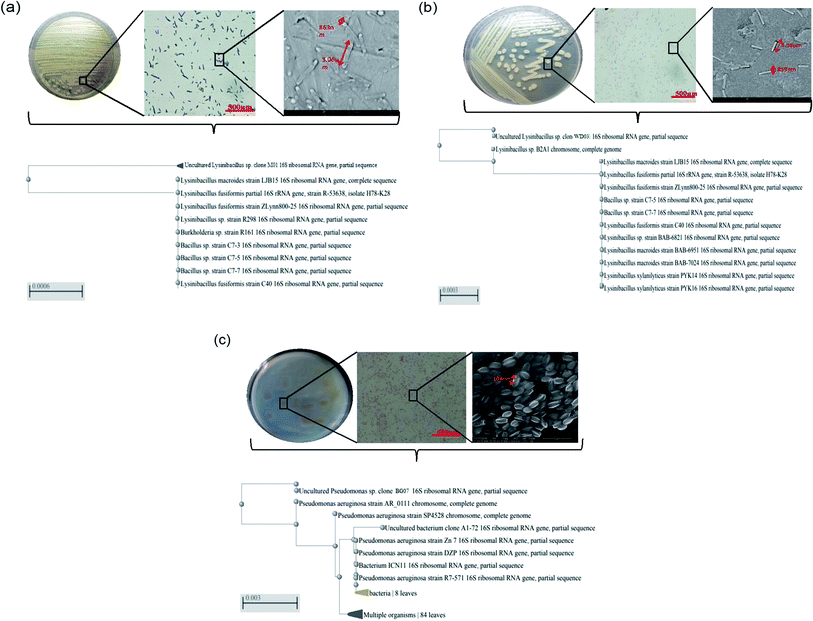
Recently, for the effective management of pollutants, researchers have suggested numerous methods; however, only few of these methods have been extended to the field scale with promising results. Since most of the pollutants are of petrochemical origin30 and polymeric in nature, their removal is highly challenging. Studies have indicated the presence of polymeric substances in leachates; this is an alarming signal as these substances can have adverse effects on the biological life in the ecosystem. The biological methods are considered eco-friendly and viable; nevertheless, the identification of a potential bacterial isolate for the management of pollutants of polymeric nature is a challenging mission.In the present study, a batch-mode approach was designed to achieve potential bacterial isolates for the management of styrene molecules present in the leachates of a dump yard. The sample site and the methodology followed in the present study provided an appreciable number of organisms belonging to various families and genera. However, based on styrene utilization, only three potential isolates (each from one site) were selected: M01 (isolate of marine origin), WD03 (isolate of waste dump yard origin), and BG07 (isolate of bovine gut origin). The isolates WD03 and M01 were Gram positive, spore forming, and motile strains, whereas BG07 was a Gram negative, non-spore forming, and motile strain. Table 2 presents the biochemical profile of all the three isolates chosen for the present study. The biochemical analysis revealed that the two isolates M01 and WD03 belong to Bacillus genera and the isolate BG07 belongs to Pseudomonas genera.31–33 Fig. 1a–c illustrate the plate morphology, microscopic image, scanning electron microscopy image, and the phylogenetic profile of all the three isolates M01, WD03 and BG07, respectively. From the gene sequence and blast analysis results, the isolates were accordingly identified as Lysinibacillus sp., strain WD03; Lysinibacillus sp., strain M01; and Pseudomonas aeruginosa strain BG07, the sequences were deposited in the GenBank, and the accession numbers were received as MN747168, MF755294.1, and MN737147.1 respectively.
| Biochemical and morphological characteristics | Lysinibacillus sp. WD03 | Lysinibacillus sp. M01 | Pseudomonas sp. BG07 |
|---|---|---|---|
| Gram staining | + | + | − |
| Shape | Irregular rod | Irregular rod | Short rod |
| Motility | Motile | Motile | Motile |
| Spore staining | + | + | − |
| Indole | − | − | − |
| Methyl red | − | − | − |
| Voges–Proskauer | − | − | − |
| Citrate | − | − | + |
| Urease | + | + | − |
| Glucose | − | − | − |
| Arabinose | − | − | − |
| Mannitol | − | − | + |
| Starch | − | − | − |
| Gelatin | + | + | + |
| NaCl 5% | + | + | + |
| Oxidase | + | + | + |
| Catalase | + | + | + |
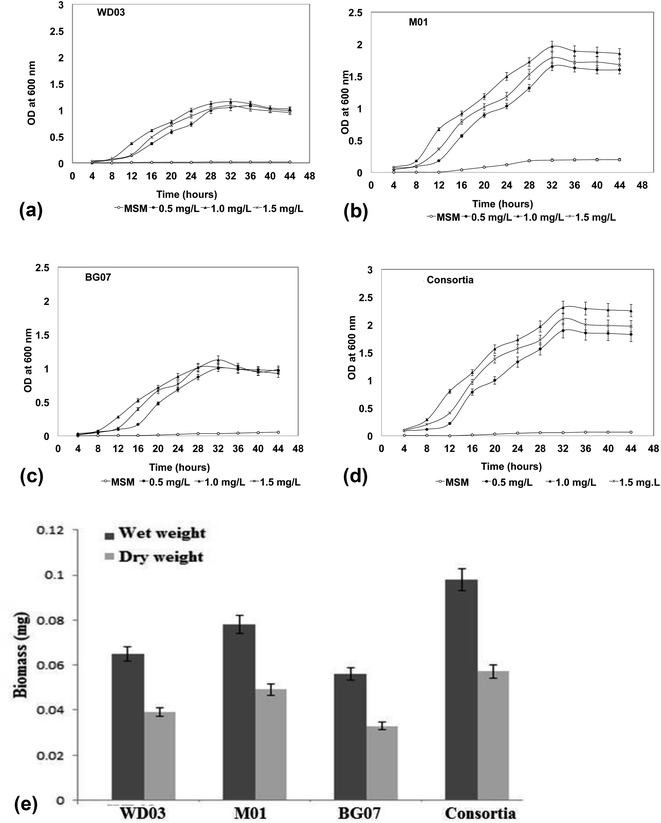
Fig. 2a–d depict the growth profile of the three isolates and the mixed consortium with respect to time and the increasing concentration of styrene. The optical density measured at 600 nm at 4 hour time intervals showed significant variations in the growth profile of the chosen isolates with respect to the styrene concentration. The growth profile of all the organisms displayed an extended lag phase. Compared to the case of the minimal salt medium, the growth of the isolates in the presence of styrene at lower concentrations ranging from 0.5 to 1.5 mL L−1 was appreciable at a significant level (P < 0.05). When the optical densities were compared, the M01 strain (marine origin) showed higher optical density as compared to the other isolates, which has been well-reflected by the biomass measurement results (Fig. 2e). Higher values of both the wet and the dry mass were obtained for the M01 strain as compared to those for WD03 and BG01. These observations suggested that all the three isolates were able to utilize styrene as a carbon source for their growth and energy and showed growth at 1.5 mL L−1 styrene concentration. The noticeable growth of the organisms at 1.5 mL L−1 is a significant and promising finding for the treatment of aqueous leachates. A study conducted on 63 industrial effluents showed that the level of styrene released to the environment ranges from 6.3 to a maximum of 970 pg L−1.34 The results of the studies reported by Aalam et al. on styrene degradation using Pseudomonas aeruginosa35 and León et al. (1999) on the degradation of toluene and xylene with an extended lag phase corroborate well with the present findings.36 Most of the studies on styrene degradation have been carried out using the gaseous form of styrene,37–41 and only few studies have discussed the degradation of styrene in the aqueous phase.42–45
With respect to the mixed consortium, its growth profile was similar to that of the single culture; however, a significant variation was observed in the optical density (Fig. 2d) and the produced biomass (Fig. 2e). The enhanced growth in the consortia indicated a possible synergism between the cultures; moreover, the competition for the substrate may be responsible for the increased growth and metabolism of styrene.46 According to Aalam et al.,47 the consortia containing four different microbial strains for the degradation of styrene resulted in the survival of only one bacterial strain, Pseudomonas aeruginosa, which exhibited the utilization of styrene as the sole carbon source. Other than the abovementioned study, no study has been reported on the role of Lysinibacillus sp. in styrene degradation in the literature; therefore, in the present study, we investigated the ability of Lysinibacillus sp. and Pseudomonas sp. in the form of a single and as a mixed system to metabolize the styrene molecule.
The metabolism of styrene in aerobic bacteria occurs via two types of pathways: lower and upper pathways; the former pathway involves vinyl side chain oxidation that converts styrene to phenylacetic acid, and the latter pathway involves the degradation of phenylacetic acid.14,32 The enzymes in the organism play a crucial role in the metabolism of styrene. As described above, five different genes are responsible for the metabolism of styrene.
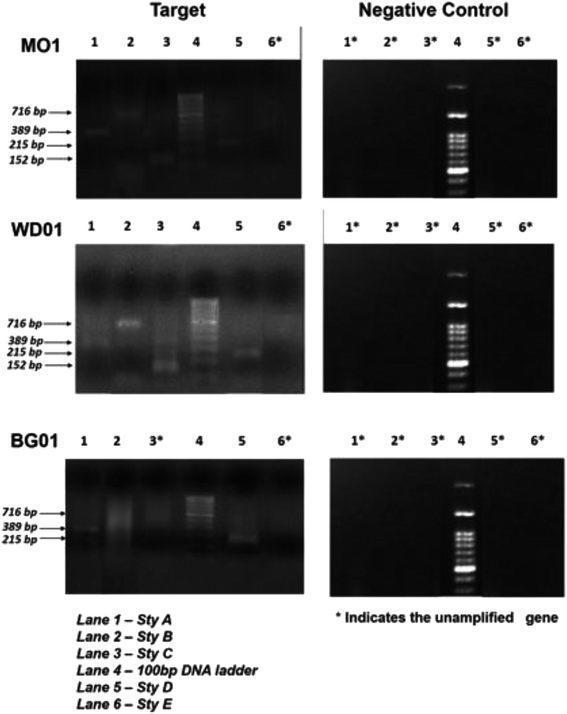
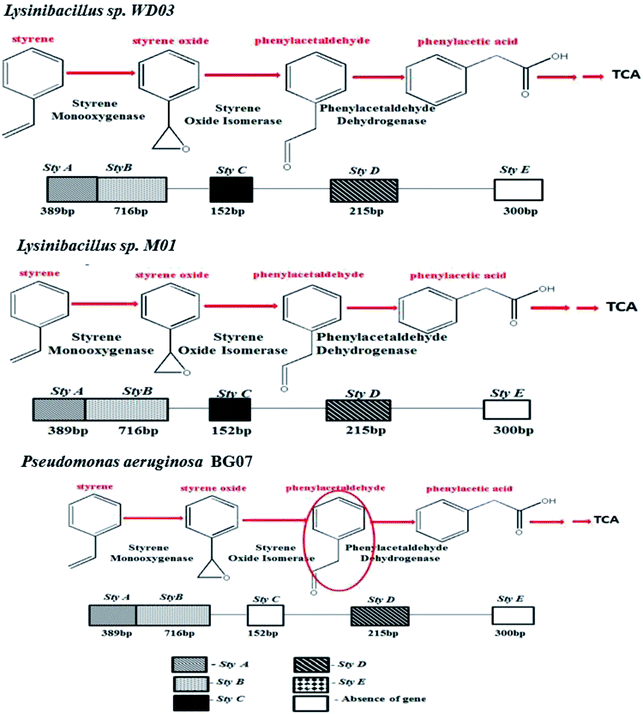
Fig. 3 depicts an image of the PCR-amplified target genes, corresponding to styrene degradation, for the isolates Lysinibacillus sp., strain M01 and WD03, and Pseudomonas aeruginosa, strain BG07, with respect to those of the negative control (E. coli). The Lysinibacillus strain M01 showed amplification of the Sty A, B, C, and D genes expressed at 389, 716, 152, and 215 base pairs, respectively, and no amplification of the Sty E gene. In the case of WD03, clear bands were observed, corresponding to 152, 716, 215, and 389 bp. Some of the lanes displayed primer dimers, which could be due to either increased volume of the primers or lack of amplification. In the case of the Pseudomonas aeruginosa strain, the amplification of Sty A (389 bp), Sty B (716 bp), and Sty D (215 bp) was observed, and nil amplification was noticed for Sty C and E. The abovementioned observations match with those of the studies on the Pseudomonas sp. strain Y2, Pseudomonas sp. VLB120, P. putida SN1, and P. putida CA-3.17,18 In addition, the roles of Actinobacteria, α-, β-, and γ-Proteobacteria, Sphingobacteria, and Bacillus species, Bacillus megaterium and B. cereus43 support the findings of the present study. The expressions of styrene-degrading genes were not observed with the negative control.
Fig. 4a and b depict the UV-visible spectra of standard styrene and the cell-free medium after 72 hours experimental period for all the experimental samples. The substrate styrene showed a plateau from 200 to 250 nm and a single maximum absorption peak at 281 nm (Fig. 4b (inset)). However, the cell-free medium of the single culture showed absorption peaks in the range of 252–274 nm, and the mixed cultures showed peaks at 268 and 274 nm (Fig. 4a), indicating that the substrate styrene was metabolized by the isolates. These observations imply that as compared to a single isolate, the consortia played a significant role. The absorption peak observed at 254 may be attributed to the conjugation of the benzene ring with a less extended secondary band. Furthermore, the peak observed at 274 nm matches with the absorption peak of aromatic acetic acid.33
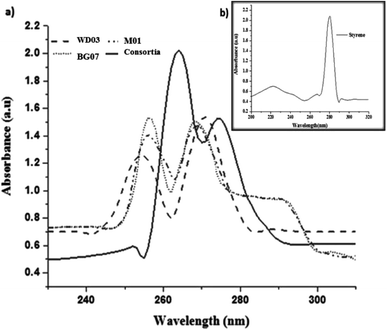 | ||
| Fig. 4 (a) UV-vis spectra of the metabolites obtained from each bacterial isolate; (b) (inset image) UV-vis spectra of standard styrene. | ||
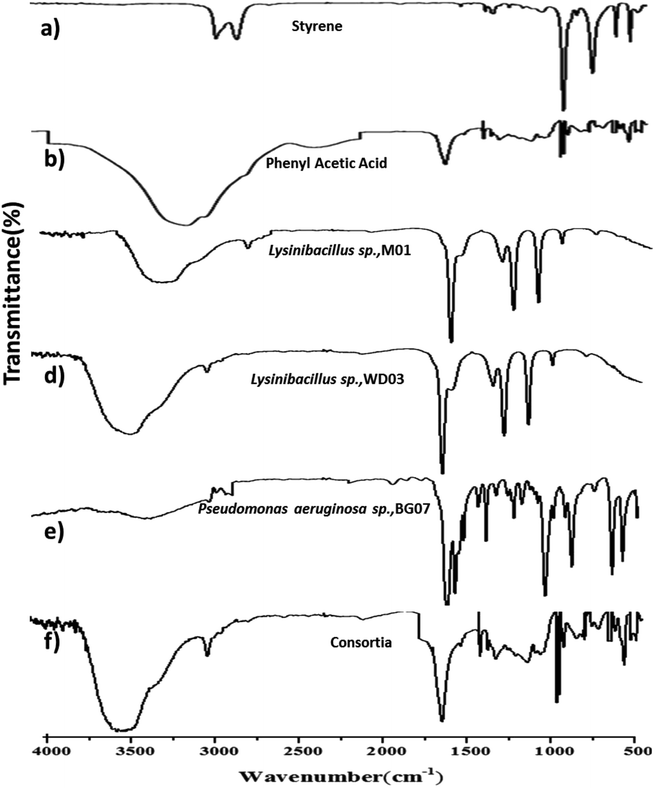
Fig. 5a–f depict the FTIR spectra of the concentrated cell-free extract obtained from three different isolates and the consortia samples after 72 hours of growth (Fig. 5a–d) along with those of the standards (Fig. 5a and b). Infrared peaks were observed at 690–750 cm−1 irrespective of the samples and accordingly indicated the presence of mono-substituted aromatic compounds. Moreover, the peaks observed at 1650–1400 cm−1 support the observations implying the presence of an aromatic compound. The medium-to-very broad band observed between 3400 and 2700 cm−1 indicated the presence of –OH stretch.48–50 The peaks observed for the cell-free medium match with those of standard PAA (Fig. 5b). The samples of BG07 displayed additional peaks, which also indicated the presence of an aromatic compound. The prominent peaks observed at 1100, 1270, and 1300 cm−1 for all the single isolates may be attributed to the incomplete metabolism of the styrene monomer or the presence of respective aldehydes or ketones.
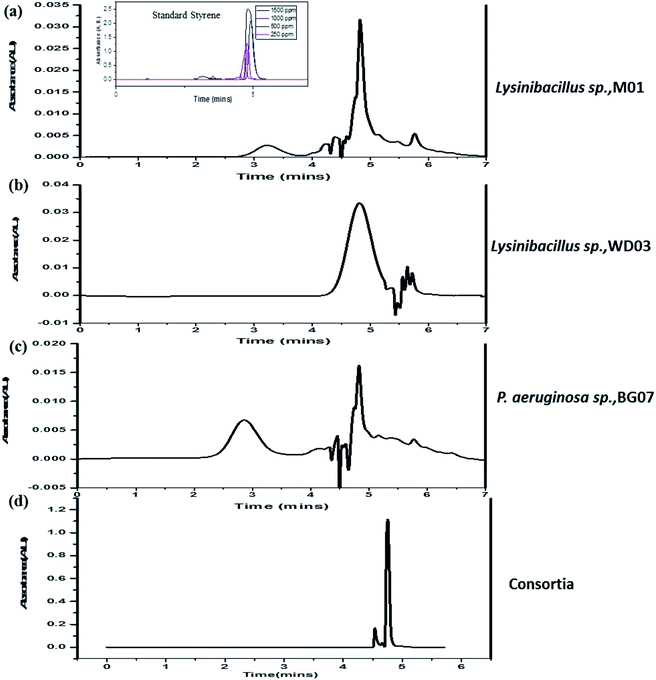
Fig. 6a–d illustrate the HPLC chromatograms of the cell-free experimental samples respective to those of the single isolates (M01, WD03, and BG07) and the consortia; in addition, Fig. 6a has an inset that depicts the chromatograms of phenylacetic acid at different concentrations. All the experimental samples invariably displayed a well-defined and high intensity peak at the retention time of 4.7–5.0 minutes and also showed less intense peaks between 3.0–4.5 and 5.5–6.0 minutes. Interestingly, the consortia sample showed a single high intensity peak at 4.9 minutes. These observations suggested that the intended styrene molecule was effectively metabolized by the chosen isolates into an industrially important chemical, namely phenylacetic acid. Previously, studies51 have been reported on the degradation of styrene and the formation of phenylacetic acid using bacterial strains such as Rhodococcus opacus 1CP, Pseudomonas fluorescens ST, Sphingopyxis sp. Kp5.2, and Gordonia sp. CWB2, which corroborate well with the present study. The less intense peaks observed at different retention times along with the high intensity peak could be due to the presence of remnants of the incomplete metabolism. This conclusion has been drawn based on the observations obtained for the consortia samples, which showed only a single high intensity peak, which more or less matches with the observations for phenylacetic acid. Based on the obtained analytical and PCR results, it has been hypothesized that the applied styrene may be metabolized to phenylacetic acid. No further degradation of phenylacetic acid was observed throughout the experimental period.
Table 3 depicts the list of studies reported on the degradation of styrene in an aqueous medium. In all these studies, the bacterial strains employed were Xanthobacter sp., Pseudomonas putida, Streptomyces halstedii, Bacillus megaterium, Sphingobacterium spiritivorum, and Bacillus cereus. Although 50% of the abovementioned studies revealed phenylacetic acid as the product of the degradation of styrene, no discussions were reported on the molecular level for the genes responsible for styrene degradation. Based on the observations made in the present study and the available literature, the styrene metabolism pathway for all the three isolates chosen for the present study was elucidated and is shown in Fig. 7. In brief, Lysinibacillus spp. metabolizes styrene to phenylacetic acid through its monooxygenase, oxide isomerase, and dehydrogenase genes. Moreover, Pseudomonas sp. metabolizes the applied styrene to phenylacetic acid despite the absence of oxide isomerase, and the observed ambiguity necessitates further research.
| S. no. | Name of the organisms | Concentration of styrene | By-product formed | Gene study | Reference |
|---|---|---|---|---|---|
| 1 | Xanthobacter sp. strain 124X | 0.01% to 0.1% (w/v) | Phenylacetic acid | Not done | 42 |
| 2 | 14 strains of aerobic bacteria and 2 strains of fungi | 10–500 μM | Phenylacetic acid | Not done | 40 |
| 3 | Pseudomonas putida S12 | 2 μL to 0.25 mL | No by-product formed | Not done | 40 |
| 4 | Streptomyces halstedii, Bacillus megaterium, Sphingobacterium spiritivorum, Bacillus cereus | 0.5–2.0 mL L−1 | No by-product formed | Not done | 43 |
| 5 | Lysinibacillus sp., M01, Lysinibacillus sp., WD03, Pseudomonas aeruginosa BG07, consortium of all three | 0.5 to 1.5 mL L−1 | Phenylacetic acid | Done | Present study |
Conclusion
In the present study, an attempt was made to achieve potential aerobic bacterial isolates to metabolize styrene in a batch mode study. Herein, three potential isolates from three different origins showed growth at 0.5 mL L−1 styrene and were subjected to styrene metabolism using a mineral medium as a single culture and as a mixed consortium. The growth profile studies suggested that the isolates were able to effectively utilize styrene at 0.5–1.5 mL L−1 concentration, and at 2.0 mL L−1 concentration, the growth was almost nil (not shown). Among the three isolates, the marine isolate showed an appreciable growth and biomass, as demonstrated. Analyses on the nature of the metabolized product suggested the presence of phenylacetic acid. The relative gene expression studies for styrene metabolism by the chosen isolates demonstrated the presence of Sty A, Sty B, Sty C, and Sty D except for Sty E in Lysinibacillus spp. and absence of Sty C and Sty E in Pseudomonas sp. On comparing the metabolic profiles of the single culture and the consortia, it was concluded that the consortia should be preferred for the complete metabolism of styrene. The findings of the present study suggested the possibility of aerobic management of styrene present in the leachates of the waste dump yard and provide a microbially mediated process to obtain the industrially important chemical phenylacetic acid.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the Department of Biotechnology for providing financial support in the form of a sponsored project. The first author acknowledges the Department of Science and Technology-Innovation in Science Pursuit for Inspired Research (INSPIRE) for providing fellowship in the form of JRF and SRF and also acknowledges University of Madras for the PhD program. The second author acknowledges CSIR, New Delhi, for providing fellowship in the form of CSIR-SRF. The authors hereby acknowledge the financial support given by DBT, New Delhi.References
- N. Itoh, K. Yoshida and K. Okada, Biosci., Biotechnol., Biochem., 1996, 60, 1826–1830 CrossRef CAS PubMed.
- C. Thaysen, K. Stevack, R. Ruffolo, D. Poirier, H. De Frond, J. DeVera, G. Sheng and C. M. Rochman, Front. Mar. Sci., 2018, 5, 71 CrossRef.
- S. Hirano, M. Tanaka, K. Date, K. Ohno, K. Kobayashi, T. Sakurai, Y. Nagao, Y. Nobuhara and T. Yamada, J. Agric. Food Chem., 2001, 49, 4127–4131 CrossRef CAS PubMed.
- M. Ahmad and A. S. Bajahlan, J. Environ. Sci., 2007, 19, 421–426 CrossRef CAS.
- B. G. Kwon, K. Amamiya, H. Sato, S. Y. Chung, Y. Kodera, S. K. Kim, E. J. Lee and K. Saido, Chemosphere, 2017, 180, 500–505 CrossRef CAS.
- K. O'Connor, C. M. Buckley, S. Hartmans and A. Dobson, Appl. Environ. Microbiol., 1995, 61, 544–548 CrossRef.
- S. Hartmans, Biotransformations: microbiological degradation of health risk compounds, Elsevier Science, 1995, vol. 32, pp. 227–238 Search PubMed.
- H. A. Kolstad, K. Juel, J. Olsen and E. Lynge, Occup. Environ. Med., 1995, 52, 320–327 CrossRef CAS.
- V. Benignus, Neurotoxicology, 1981, 2, 567–588 CAS.
- I. C. McCall, A. Betanzos, D. A. Weber, P. Nava, G. W. Miller and C. A. Parkos, Toxicol. Appl. Pharmacol., 2009, 241, 61–70 CrossRef CAS PubMed.
- A. Songur, O. A. Ozen and M. Sarsilmaz, in Reviews of environmental contamination and toxicology, Springer, 2010, pp. 105–118 Search PubMed.
- K. Okamoto, M. Izawa and H. Yanase, J. Biosci. Bioeng., 2003, 95, 633–636 CrossRef CAS PubMed.
- M. Suzuki and C. A. Wilkie, Polym. Degrad. Stab., 1995, 47, 217–221 CrossRef CAS.
- W. R. Waldman and M. A. De Paoli, Polym. Degrad. Stab., 2008, 93, 273–280 CrossRef CAS.
- J. D. Peterson, S. Vyazovkin and C. A. Wight, Macromol. Chem. Phys., 2001, 202, 775–784 CrossRef CAS.
- M. Grigore, Recycling, 2017, 2, 24 CrossRef.
- N. D. O'Leary, K. E. O'Connor and A. D. Dobson, FEMS Microbiol. Rev., 2002, 26, 403–417 CrossRef PubMed.
- M. S. Park, J. H. Han, S. S. Yoo, E. Y. Lee, S. G. Lee and S. Park, Korean J. Chem. Eng., 2005, 22, 418–424 CrossRef CAS.
- D. Grbić Galić, N. Churchman Eisel and I. Mraković, J. Appl. Bacteriol., 1990, 69, 247–260 CrossRef PubMed.
- M. Priyanka and S. Dey, Vet. World, 2018, 11, 1307 CrossRef CAS PubMed.
- C. S. Lee, Y. T. Jung, S. Park, T. K. Oh and J. H. Yoon, Int. J. Syst. Evol. Microbiol., 2010, 60, 281–286 CrossRef CAS PubMed.
- J. B. Gita, A. Aishwarya, N. Pavithra, S. Chandrasekaran, A. V. George and A. Gnanamani, Anaerobe, 2018, 49, 1–4 CrossRef PubMed.
- M. H. Abdelhai, H. A. Hassanin and X. Sun, Am. J. Biosci. Bioeng., 2016, 4, 1–8 CAS.
- B. G. Hall, Mol. Biol. Evol., 2013, 30, 1229–1235 CrossRef CAS PubMed.
- M. E. Migaud, J. C. Chee Sanford, J. M. Tiedje and J. Frost, Appl. Environ. Microbiol., 1996, 62, 974–978 CrossRef CAS.
- L. Zhang, C. Zhang, Z. Cheng, Y. Yao and J. Chen, Chemosphere, 2013, 90, 1340–1347 CrossRef CAS PubMed.
- S. C. Felshia, N. A. Karthick, R. Thilagam, A. Chandralekha, K. Raghavarao and A. Gnanamani, J. Environ. Manage., 2017, 197, 373–383 CrossRef.
- F. Beltrametti, A. M. Marconi, G. Bestetti, C. Colombo, E. Galli, M. Ruzzi and E. Zennaro, Appl. Environ. Microbiol., 1997, 63, 2232–2239 CrossRef CAS.
- A. Mooney, N. D. O'Leary and A. D. Dobson, Appl. Environ. Microbiol., 2006, 72, 1302–1309 CrossRef CAS PubMed.
- R. H. Shie and C. C. Chan, J. Hazard. Mater., 2013, 261, 72–82 CrossRef CAS PubMed.
- A. Cabuk and S. Ilhan, Eur. J. Miner. Process. Environ. Prot., 2003, 3, 67–74 Search PubMed.
- B. Kalmath, A. Prabhuraj, P. Dhakephalkar, H. Supreet and R. Giraddi, J. Biol. Control, 2014, 28, 24–30 Search PubMed.
- K. Z. Lae, S. S. Su, N. N. Win, N. N. Than and H. Ngwe, SciMedicine Journal, 2019, 1(4), 199–216 CrossRef.
- Z. A. Rosemond, Toxicological profile for styrene, Agency for Toxic Substances and Disease Registry, 2010 Search PubMed.
- S. E. Aalam, A. Pauss and J. M. Lebeault, Appl. Microbiol. Biotechnol., 1993, 39, 696–699 CrossRef.
- E. León, N. Adler and P. Peringer, Biodegradation, 1999, 10, 245–250 CrossRef PubMed.
- A. Gąszczak, G. Bartelmus and I. Greń, Appl. Microbiol. Biotechnol., 2012, 93, 565–573 CrossRef PubMed.
- R. Babaee, B. Bonakdarpour, B. Nasernejad and N. Fallah, J. Hazard. Mater., 2010, 184, 111–117 CrossRef CAS PubMed.
- H. Jorio, L. Bibeau and M. Heitz, Environ. Sci. Technol., 2000, 34, 1764–1771 CrossRef CAS.
- H. Cox, R. Moerman, S. Van Baalen, W. Van Heiningen, H. Doddema and W. Harder, Biotechnol. Bioeng., 1997, 53, 259–266 CrossRef CAS PubMed.
- I. G. Jung and C. H. Park, Chemosphere, 2005, 61, 451–456 CrossRef CAS PubMed.
- S. Hartmans, M. Van der Werf and J. de Bont, Appl. Environ. Microbiol., 1990, 56, 1347–1351 CrossRef CAS PubMed.
- F. J. Weber, L. P. Ooijkaas, R. Schemen, S. Hartmans and J. De Bont, Appl. Environ. Microbiol., 1993, 59, 3502–3504 CrossRef CAS PubMed.
- S. Hartmans, J. Smits, M. Van der Werf, F. Volkering and J. De Bont, Appl. Environ. Microbiol., 1989, 55, 2850–2855 CrossRef CAS PubMed.
- K. Przybulewska, A. Wieczorek and A. Nowak, Pol. J. Environ. Stud., 2006, 15(5), 669–676 Search PubMed.
- Y. J. Deng and S. Y. Wang, J. Microbiol., 2016, 54, 23–30 CrossRef.
- S. E. Aalam, A. Pauss and J. Lebeault, Mededelingen-Faculteit Landbouwkundige en Toegepaste Biologische Wetenschappen, Universiteit Gent, Belgium, 1993 Search PubMed.
- T. F. Herpin and G. C. Morton, in Methods in enzymology, Elsevier, 2003, vol. 369, pp. 75–99 Search PubMed.
- V. Herman, H. Takacs, F. Duclairoir, O. Renault, J. Tortai and B. Viala, RSC Adv., 2015, 5, 51371–51381 RSC.
- J. Khanjani, S. Pazokifard and M. J. Zohuriaan-Mehr, Prog. Org. Coat., 2017, 102, 151–166 CrossRef CAS.
- M. Oelschlägel, J. A. Gröning, D. Tischler, S. R. Kaschabek and M. Schlömann, Appl. Environ. Microbiol., 2012, 78(12), 4330–4337 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2020 |