Open Access Article
Open Access ArticleEnhanced reductive removal of ciprofloxacin in pharmaceutical wastewater using biogenic palladium nanoparticles by bubbling H2†
Peipei Hea,
Tianyu Maoa,
Anming Wang *a,
Youcheng Yinb,
Jinying Shena,
Haoming Chena and
Pengfei Zhanga
*a,
Youcheng Yinb,
Jinying Shena,
Haoming Chena and
Pengfei Zhanga
aCollege of Materials, Chemistry and Chemical Engineering, Hangzhou Normal University, Hangzhou 310014, P. R. China. E-mail: waming@hznu.edu.cn
bHolistic Integrative Pharmacy Institutes, College of Medicine, Hangzhou Normal University, Hangzhou, Zhejiang, China
First published on 10th July 2020
Abstract
To treat waste with waste and efficiently remove the organic pollutant, waste palladiums(II) were adsorbed and reduced on microorganism surface to catalyze the reductive removal of ciprofloxacin in pharmaceutical wastewater. By optimizing conditions such as pH and temperature, the amount of biogenic palladium adsorbed and reduced on E. coli reached 139.48 mg g−1 (Pd/microorganisms). Moreover, most of the Pd(II) was reduced to nanometer-sized Pd(0) as characterized by TEM and SEM with EDXA. Using the obtained biogenic palladium, the reductive removal of ciprofloxacin is up to 87.70% at 25 °C, 3.03 folds of that achieved in the absence of H2. The results show that waste E. coli microorganisms can efficiently adsorb and remove waste Pd(II) and produce Bio-Pd nanoparticle catalysts in the presence of H2. This biogenic palladium presents high catalytic activity and great advantages in the reductive degradation of ciprofloxacin. Our method can also be applied to other waste metal ions to prepare the biogenic metals, facilitate their recovery and reuse in degrading organic pollutants in wastewater to achieve “treating waste using waste”.
1. Introduction
Palladium (Pd) is a ‘‘precious metal” and is one of the most important elements in chemical catalysis,1 electronic circuits, jewelry, semiconductors, ornaments, corrosion-resistant equipment and thermocouples.2 With the increasing use of palladium in the past 30 years, a substantial amount of waste Pd is discharged into the environment, which results in the presence of higher levels of Pd in road dust, airborne particulates, groundwater tables and soil. Moreover, the chemical catalysis and electroplating industries generate a large amount of wastewater containing palladium. In epidemiological studies, exposure to Pd has been verified to cause not only acute toxicity, hypersensitivity with respiratory symptoms, and urticaria, but also some immune system diseases, indicating it is a significant hazard to human health because Pd ions are one of the most significant metal sensitizers. Thus, the recovery and reuse of this metal, based on the notion of recycling waste,3 are necessary to recover this valuable material and to treat wastewater4 to promote green chemistry, economic efficiency and sustainability.1Currently, a variety of methods have been developed to recycle Pd, such as solvent extraction, ion exchange chromatography, and pyrometallurgical processes.5 Among these methods, ion exchange is a simple way to separate Pd(II), but the process is expensive due to the use of resin.6 Compared with traditional physical or chemical methods, absorption based on biomaterials (biosorption) is considered one of the most promising options for recycling Pd from wastewater. Biosorption is defined as the removal of compounds, metals or metalloid species particles from solutions by living or dead biological materials, and it is a cost-effective and useful tool to for recovering precious metals.7,8 It is independent of metabolism and often occurs in the cell wall9 when bacteria, fungi, algae, plants and chitosan, etc. are usually used as bio-sorbents. Previous reports have shown that the biological palladium (Bio-Pd) obtained by biological adsorption is always a nanoscale catalyst, and its catalytic reaction rates and efficiency are better than those of Pd/C and colloidal Pd nanoparticles. Additionally, the obtained bio-palladium can be efficiently recycled and easily separated from reaction mixture with a slight decrease in its catalytic activity over sequential examinations and tests.10 When adsorbed on microorganism bio-sorbents, Pd(II) was verified to be reduced to Pd(0) by [NiFe] hydrogenases.11 Currently, increasing attention has been paid to this new, effective biological method; not only can a variety of biomaterials bind precious metals, but some cell-supported metals show good potential in the reductive degradation and removal of several persistent and bio-refractory environmental contaminants, such as Cr(VI), halogenated materials, and azo dyes.12–15
Ciprofloxacin (CIP) is an important fluoroquinolone antibiotic that is widely used in the treatment of diseases. It can inhibit DNA replication by interacting with topoisomerases II (DNA gyrase) and IV in microorganisms.16 Because it is too chemically stable to be fully metabolized by humans, CIP has become the most frequently detected fluoroquinolone in European wastewater treatment plant (WWTP) effluents because of its release to the environment and excretion into wastewater.17,18 The increased exposure to fluoroquinolone antibiotics during the last fifty years has also increased bacterial resistance to these compounds, which typically are found at low concentrations.19,20 Ciprofloxacin-resistant P. aeruginosa strains have been found in clinical and municipal wastewaters.21 In recent years, advanced oxidation technologies and processes using strong oxidant, such as UV/H2O2, UV/O3, and UV/TiO2, have been developed for CIP degradation.22,23 Advanced oxidation processes (AOPs) includes a series of technologies, ultraviolet (UV) radiation, hydrogen peroxide (H2O2), ozone (O3), Fenton (Fe(II)/H2O2).24 Among them, ozonation is very efficient in removing contaminants from wastewater effluents, but it is sure to be responsible for multiple occupational health hazards like headaches, sore throats, irritation in eyes and nose. In addition, the residual ozone reacts with water containing bromite and produces bromate which is a potential genotoxic human carcinogen.22 Fenton process is also used for the removal of antibiotics.24 But, this process takes longer time and Fe2+ salt is utilized as a catalyst, which makes the process economically non-feasible or a costly affair and imperfect for the remained iron salt.
In this work, we use waste E. coli to adsorb the metal ions to achieve the recovery of Pd(II) from wastewater, which not only means treating both microorganism-containing biological waste and palladium-containing wastewater preliminary, but also economically feasible to produce efficient catalyst. The Pd(II) adsorbed on the surface of waste E. coli microorganisms were reduced in the presence of H2 to prepare biological palladium (Bio-Pd). The obtained biogenic palladium was characterized using TEM, SEM and XRD and then used to catalyse ciprofloxacin (CIP) degradation in simulated wastewater. Bio-Pd-catalysed reductive degradation was expected to occur to CIP in the presence of H2. The effects of some factors, such as the pH, the amount of palladium on the microorganism and the degradation time, on the degradation efficiency were examined and evaluated carefully.
2. Materials and methods
2.1. Materials
The waste Escherichia coli BL21(DE3) strain was the host harbouring aldehyde ketone reductase and ethanol dehydrogenase used in these experiments. Ciprofloxacin was purchased from Energy Chemical (Hangzhou), and palladium chloride was purchased from Adamas-beta. All other biological reagents not mentioned above were purchased from Sangon Biotech (Shanghai), while all other chemical reagents were purchased from Sinopharm Chemical Reagent Ltd. (Hangzhou).2.2. Biosorption of Pd2+ on the waste Escherichia coli cells
The obtained waste E. coli cells were harvested by centrifugation (8000 rpm, 8 min), and the wet cells were washed with ultrapure water three times. One gram of wet cells was dispersed in 200 mL of PdCl2 solution (200 mg L−1), and the pH of the solutions was adjusted to 2.0. The mixture was shaken (170 rpm 30 °C) for 6 h to allow for metal ion biosorption. 3 mL of the reaction mixture solution was removed per hour via syringe and passed through a syringe filter with a pore size of 0.22 μm, and the amount of Pd2+ in the supernatant was detected by UV-Vis spectrophotometry. After confirmation that the Pd2+ was almost completely precipitated, the biogenic palladium nanoparticles were recovered by centrifugation (8000 rpm, 8 min) and washed three times with ultrapure water. The amount adsorbed by E. coli was calculated by the formula
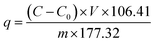 | (1) |
2.3. Synthesis of the biogenic palladium nanoparticles under anaerobic conditions
One gram of wet cells was added to 200 mL of PdCl2 solution (200 mg L−1), the pH of the solution was adjusted to 2.0, and the container was then sealed. The solutions were incubated under anaerobic conditions by flushing with N2 for five minutes. After incubation at 30 °C for 1 h to allow for Pd2+ biosorption, the biogenic palladium was harvested and collected by centrifugation (8000 rpm, 8 min) and resuspended in distilled water. The Bio-Pd(II) was transferred to a 100 mL three-necked flask. Then, the three-necked flask was flushed with H2 and left at 25 °C with stirring (150 rpm) in a H2 atmosphere. Five hours later, the biogenic palladium (metal) used in the next experiment was collected by centrifugation (8000 rpm, 8 min) and washed twice with distilled water. 50 mL of PdCl2 solution (pH 2.0) added to a 100 mL round-bottomed flask, flask was flushed with H2 and left at 25 °C with stirring (150 rpm) in a H2 atmosphere, collected by centrifugation (8000 rpm, 8 min) and washed twice with distilled water after 5 h to preparation Pd(0) without E. coli.2.4. Characterization of the biogenic palladium nanoparticles
The obtained biogenic palladium was collected and harvested by centrifugation (8000 rpm, 5 min), washed twice with distilled water, and resuspended in ethanol and dripped onto a copper grid. Vacuum drying then occurred to the Bio-Pd(0) for 24 h at room temperature. The dry samples on the copper grid were viewed and examined by TEM (HT-7700) under an acceleration voltage of 100 kV. Using TEM, we can identify the size and structure of the biogenic palladium nanoparticles.The morphology of the biogenic palladium nanoparticles was determined by scanning electron microscopy (SEM, Zeiss Sigma 500). The elements on the surface of the biogenic palladium nanoparticles were analysed using energy dispersive X-ray analysis (EDXA). In this work, the biological palladium nanoparticles on E. coli were harvested and collected by centrifugation (8000 rpm, 5 min), resuspended in ethanol and dripped onto a thin film. The samples of the biological palladium nanoparticles on E. coli were dried overnight in a vacuum drying oven at 30 °C and analysed by SEM and EDXA.
We investigated and analyzed the valence state of the biogenic palladium nanoparticles using XPS (ESCALAB 250Xi, England). XPS used an Al Kα source and surface chemical composition and reduction state analyses was done, with the core levels recorded using a pass energy of 30 eV (resolution approx. 0.10 eV). The peak fitting of the individual core-levels was done using XPS-peak 41 software, achieving better fitting and component identification. All binding energies were calibrated to the C 1s peak originating from C–H or C–C groups at 284.6 eV.
2.5. Ciprofloxacin removal using biological palladium nanoparticles on E. coli cells
A certain amount of ciprofloxacin was dissolved in ultrapure water acidified with glacial acetic acid (5 mL L−1), and the pH of the solution was adjusted to 3.2 with hydrochloric acid (1 mol L−1). Thirty milligrams of biological palladium was resuspended in 20 mL of this ciprofloxacin solution. Catalytic removal of ciprofloxacin using this biological palladium was carried out in the dark at 25 °C with stirring at 150 rpm. 3 mL of the reaction mixture was removed every 5 h with a syringe and passed through a syringe filter with a pore size of 0.22 μm to remove biological palladium(II) or biological palladium(0). An ultraviolet spectrophotometer was used to detect the residual content of ciprofloxacin in the supernatant at 280 nm, and the removal of ciprofloxacin was calculated by comparison with a standard curve. When the removal of ciprofloxacin was conducted under anaerobic conditions, 30 mg of biological palladium was transferred into the ciprofloxacin solution, and then H2 was bubbled into the suspension for at least 10 min from the bag, and a hose was used to connect a balloon of H2 and the reactor vessel.2.6. Effects of pH and the amount of catalyst on ciprofloxacin removal
The general procedure for reductive degradation and removal of ciprofloxacin removal was as follows: a 50 mL two-neck round bottom flask equipped with a condenser and magnetic stirrer, was charged with 20 mL of this ciprofloxacin solution at different pH values (2.0, 2.4, 2.8, 3.2 and 3.6) and 30 mg of biological palladium, pH was controlled by the addition of hydrochloric acid aqueous solution (1 mol L−1). After resuspending certain amount of biological palladium on E. coli (10 mg, 20 mg, 30 mg and 60 mg) in 20 mL of ciprofloxacin solution at pH 3.2, the mixture was stirred (150 rpm, 25 °C) for up to 25 h in the presence of H2. The ciprofloxacin remaining in the reaction mixture was determined using high-performance liquid chromatography (HPLC, Agilent 1260). The remained ciprofloxacin was separated by liquid chromatography on a Daicel IC column (250 mm by 4.6 mm, 5 μm particle size). Data were collected at 226 nm and the corresponding standard was used to verify retention times of ciprofloxacin.3. Results and discussion
3.1. Synthesis of biological palladium nanoparticles
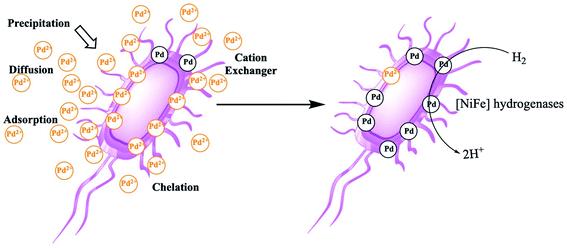
Platinum group metals (PGM), all precious metals, are usually used as catalysts and functioned as a significant part of modern chemistry.25 The high price of PGMs varies with fluctuating supply which is caused by the limited and uneven geographical distribution of this metal mines. Additionally, when released to environment after its usage, it is also a contamination to water and soil. Therefore, the recovery of waste PGMs such as Pd is of prime importance. Microorganisms such as E. coli can reduce the soluble Pd(II) in stock solutions or acid extracts from the spent catalysts, producing metallic nanocrystals of Pd(0),26 which can be used as catalysts in organic synthesis and organic pollutant removal. This simple process involves biosorption of Pd(II) cations on the surface of E. coli and the reduction of Pd(II) to Pd(0) crystals by an electron donor. The formed Pd(0) nanoparticles are supported either in the periplasmic space or on the outer membrane of the E. coli and thus remain attached to the cells.27,28Owing to the chelation of metal ions with the functional groups (carboxyl, phosphate, hydroxyl, amino, thiol, etc.) and biological structures involved, the mechanisms of biosorption and reduction of Pd(II) cations on the E. coli cell surface are fair complicated.29,30 The binding of the sorbate and sorbent depends on numerous factors, such as the number of binding sites on the E. coli cell surface, the density and type of the functional groups on microorganism cell surface, and the availability and accessibility of the sites. The mechanisms of biosorption can be divided into various criteria, which include cell metabolism and the location of the biosorption sites (Fig. 1).31,32 At first, Pd(II) cations in the bulk solution approach the microorganism by diffusion due to the concentration gradient. Then, physical adsorption of the cations can occur through van der Waals and electrostatic forces. Even dead biomasses have been shown to adsorb heavy metals ions such as Pd(II), Cu(II), Cd(II) and Zn(II) through electrostatic interactions.33 The polysaccharides in the microbial cell walls can function as counter ions to facilitate the exchange of bivalent metal ions. Thus, these cation exchangers can adsorb heavy metals such as Pd(II), Cu(II), Cd(II) and Zn(II) by counter ion exchange.34 Additionally, the complexation between Pd(II) cations and active groups on the polymers and polysaccharides of the cell surface can mediate their removal from bulk solutions by forming the complexes on the E. coli cell surface. Once exposed to toxic metals, as a stress reaction, microbes tend to produce compounds favouring the precipitation as a defense mechanism, and metal cations such as Pd(II) can often be reduced through microbiological metabolism and enzymatic catalysis in stained cells.35
In addition, functional groups such as –SH, –OH, and –COOH on E. coli cell surfaces and deprotonated ligands, e.g., –RCOO−, can also function as Lewis bases. The competitive complex formation may attributed to the adsorption of the above metal cations. By exploiting these synergistic effects, Pd(II) can be rapidly adsorbed onto the surface of E. coli. Then, several types of hydrogenases in E. coli36 can reduce Pd(II) on the surface of the cells. E. coli produce H2 during fermentation and can reduce Pd(II) to Pd(0); this fermentatively produced H2 (referred to as ‘biohydrogen’) serves as the reductant for Pd(II) reduction in the process of biosorption.37,38 In this work, no culture medium except the model wastewater was supplied to the waste E. coli, and the production of H2 during fermentation is impossible. However, when extrinsic H2 is bubbled into the mixture, Pd(II) reduction does occur and results in Pd(0) formation.
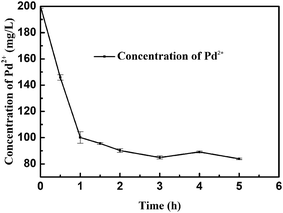
Fig. 1 indicates the effect of exposure time on the adsorption of Pd(II) onto E. coli cell. As shown in the resulting kinetic plots, Pd(II) adsorption on E. coli increased rapidly at first and then slowed down gradually, finally reaching the maximum adsorption capacity at approximately 2 h. Langmuir equation can also be used to describe the isotherms for the adsorption of Pd(II) on this microorganism cell surface and the adsorption kinetics investigations gave that the adsorption rate of Pd(II) on the bacteria can be well interpreted as a pseudo-second-order process, and the adsorption capacities calculated with a pseudo-second-order rate model were close to those determined experimentally.25 With the decrease in the number of free sorption sites and the concentration of palladium(II), the surfaces of the cells were saturated with Pd(II), and a dynamic equilibrium may be established between Pd(II) adsorption and desorption. What's more, we explored the influence of other common metal ions on the adsorption of Pd(II), including Na(I), Ca(II), Mg(II), Zn(II), Cu(II) in the process of adsorption respectively (Fig. S1 and Table S1†). The affinity of cations for the functional groups presented on the cellular surface is dependent on the pH of the solution. The carboxyl groups may play the most important role in the pH range between 2 and 5, because of their pKa value, and the pH 5 to 7 is optimal for the adsorption for Zn(II), Ca(II), Mg(II), Cu(II).39–41
3.2. Characterization of biological palladium (metal) nanoparticles by TEM and SEM with EDXA
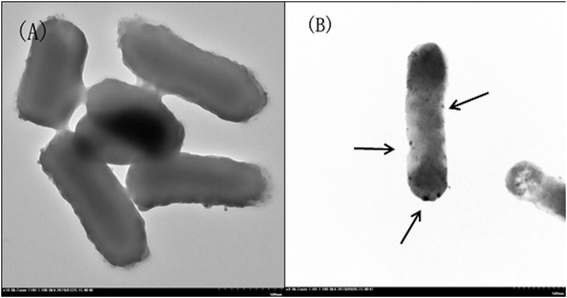
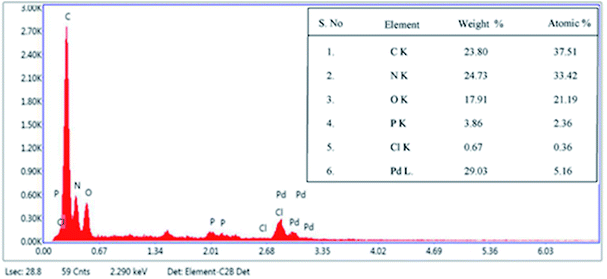
Fig. 2 shows TEM images of E. coli (A) and palladium (metal)-loaded E. coli (B) cells. This can be interpreted as the adsorbed Pd(II) being reduced and Pd(0) nanoparticles being successfully formed in the periplasmic space and on the surface of the cells. Furthermore, most of the Pd(0) particles are on the nanoscale, which is consistent with previous studies that reported dense Pd(0) precipitates on the periphery of E. coli.38 Metal precipitates were observed at the bacterial surface, and the cell surfaces provided excellent sites for Pd(II) nucleation. Early studies have shown that E. coli produces at least three hydrogenases,36 which play significant roles in the reduction of Pd(II). The Pd(0) in the periplasmic space may be formed because the catalytic subunits of the hydrogenases were exposed to the Pd(II) in this periplasmic inside of the cell membrane. After reductive conversion within the periplasm, a core could formulate and the metallic precipitates grew and erupted through the outer membrane to accumulate on the cell surface (Fig. 2).To produce high-resolution images of the cell surface and determine the content of different elements with high precision, we applied SEM with EDXA as a reference method to evaluate the adsorption and reduction of Pd(II) on the E. coli surface. The samples were made conductive by gold sputtering before morphological analysis by SEM. Fig. 3 shows that these precipitated nanoparticles ranged in size from 10 to 30 nm. Smaller Pd-NPs could exhibit higher catalytic activities when they are used to initiate the chemical reaction. EDXA indicated the relevant areas of E. coli, and its components were recognized through the analysis of the wave, confirming that elemental palladium is the main component of biological palladium (Fig. 4).
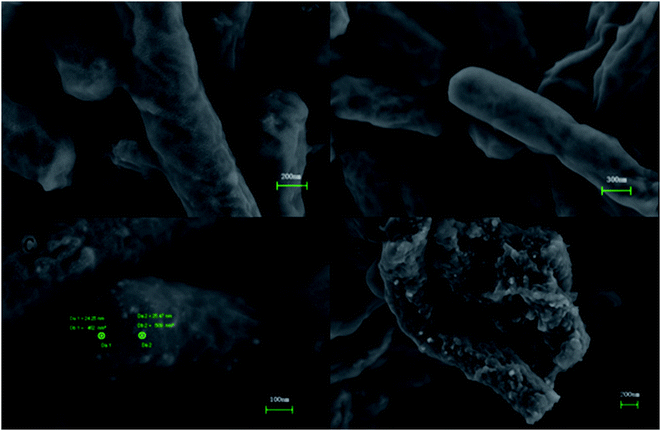 | ||
| Fig. 3 SEM images of untreated pure E. coli (A and B) and palladium nanoparticles-loaded E. coli cells surface (C and D). | ||
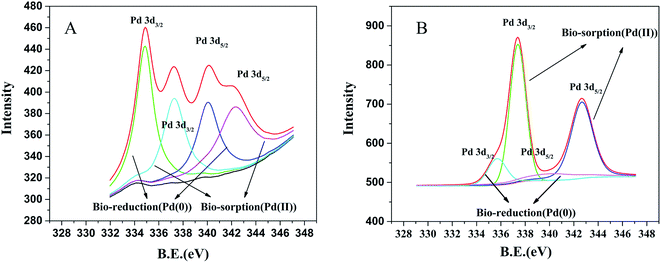
According to the XPS spectra of the prepared biogenic palladium nanoparticles, we further confirmed the deposition of Pd(0) on the surface of E. coli, which was shown in Fig. 5. The result indicates that in the presence of H2, most of palladium substrate exist as metal even if both Pd(II) and Pd(0) were found on the E. coli. The peaks at 340.05 eV and 334.87 eV are attributed to Pd(0) 3d3/2 and Pd(0) 3d5/2, respectively. The peaks at 342.28 eV and 337.28 eV are attributed to Pd(II) 3d3/2 and Pd(II) 3d5/2, respectively, which are consistent with the previous reports.42,43
3.3. Effect of pH on the reductive removal of ciprofloxacin
The initial pH in CIP solution played a significant role in its reductive removal (Fig. 6). The functional groups on the cell walls of E. coli microorganism are apt to be protonated under acidic conditions, which means that protons occupied the major binding sites resulting in the increase of the mass transfer resistance.44 Biological palladium can uniquely be adsorbed by microorganisms, while palladium can catalyse the reduction and removal of ciprofloxacin. In the process of CIP removal, biological palladium was first formed complexes with contaminants through different mechanisms, such as adsorption and surface complexation.45 The reductive remove of CIP were carried out at different pH values (2.0, 2.4, 2.8, 3.2 and 3.6). The solution pH significantly affects the presence of CIP.46 The fastest degradation of ciprofloxacin was observed at pH 3.6 due to the effect of the substrate activity. At pH 3.6, both the N1 atom of the piperazinyl substituent (pKa ∼ 5.05) and the N4 atom of the piperazinyl substituent are protonated (pKa ∼ 8.24–8.95), and the N atom of the quinolone moiety (pKa ∼ 3.64) is just partially protonated.47,48 As the pH decreased, the removal of ciprofloxacin also decreased. Because the solubility of ciprofloxacin in water is poor and further decreases with increasing pH, it is difficult to prepare a 5 mg L−1 ciprofloxacin solution with a pH higher than 3.6. Based on our results, we speculate that the degradation of ciprofloxacin is a complex reaction that may be triggered by the protonation of the piperazine N atom.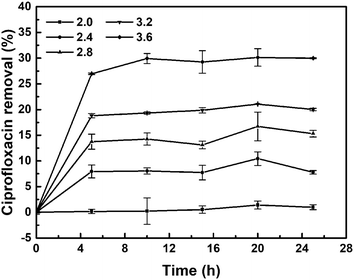 | ||
| Fig. 6 Removal of ciprofloxacin (5 mg L−1, 20 mL) by Bio-Pd at different pH values, 30 mg catalyst, 25 °C without the presence of H2. | ||
3.4. Effect of the amount of catalysts on the reductive removal of ciprofloxacin
In this work, we evaluated the influence of the amount of catalyst on the removal of CIP. The removal of CIP is slightly dependent on the catalyst loading and follows a trend similar to that of the high dependence on pH value in solution. The removal of ciprofloxacin increased as the catalyst loading increased (Fig. 7). It was speculated that when sufficient catalyst is present, the removal may start with the adsorption of ciprofloxacin by E. coli, reducing the mass transfer resistance, and allowing the ciprofloxacin to fully interact with the Pd metal and undergo catalysis. In this work, the result is promising for the further technological development. Remediation processes based on the use of biological metals are receiving much attention owing to their many advantages over traditional methods, such as their cost effectiveness, selectivity and efficient removal of contaminants. On an industrial scale, it is possible to increase the amount of Bio-Pd to achieve a high removal of CIP and reuse of the Bio-Pd as an immobilized, recoverable catalyst.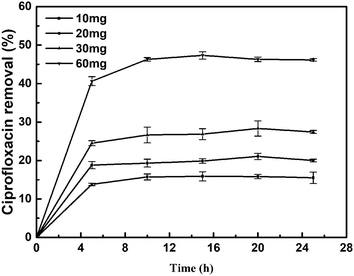 | ||
| Fig. 7 Removal of ciprofloxacin (5 mg L−1, 20 mL) with different amounts of Bio-Pd, pH 3.2, 25 °C, without the presence of H2. | ||
3.5. Reductive removal of ciprofloxacin compounds by biological palladium
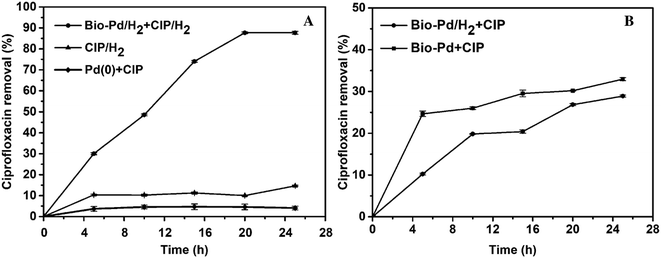
Due to the production of various mutagenic and toxic compounds by advanced oxidation technologies and methods, such as UV/H2O2, UV/O3, and UV/TiO2, an increasing number of studies have focused on reductive catalysis. Focus is also drifting away from traditional Pd/C or colloidal Pd catalysis towards palladium nanoparticles (NPs) since NPs are more active and thus less catalyst is needed.49 Martins and his co-workers have reported that biological palladium (metal) was effective in the catalytic reductive removal of diatrizoate depending on the H2 gas produced using microbial electrolysis cell (MEC).50 The catalytic property and performance of Pd nanoparticles is often influenced by the particle size, crystallinity and dispersion.51 Especially, this can be enhanced and improved by creating and forming Pd(0) clusters in the presence of H2.52 E. coli has the ciprofloxacin-acetylating variant gene aac(6′)-Ib-cr, which can transform ciprofloxacin by N-acetylation to produce N-acetyl-ciprofloxacin, which is a major metabolite of ciprofloxacin.53 The removal may occur to the CIP solution as the following four mechanisms: bio-adsorption to microorganism cell surface, extracellular reactive removal by the produced metabolites during microorganism cell growth, biodegradation by co-metabolism with other substrate, or biodegradation by CIP contamination sole consumption in which microorganism just uses CIP contamination as the sole carbon and electron source.54In this work, using waste E. coli microorganism we synthesized and obtained biological palladium (Bio-Pd) nanoparticles in the presence of H2 gas and also examined and evaluated their property and performance in the reductive catalytic removal of ciprofloxacin for the first time. Fig. 8A shows the catalytic reductive removal of CIP contamination by Bio-Pd in the presence of H2. However, no H2 was bubbled into the reaction mixture when CIP was degraded and removed using Bio-Pd catalyst, even though this hybrid catalyst was obtained by adsorption and reduction in the presence of H2 on the waste E. coli cells (Fig. 8B). When no H2 was bubbled in the reaction mixture, just approximately 28.90% and 32.96% of ciprofloxacin was removed, which may also be partially attributed to the adsorption of CIP to the surface of Bio-Pd(II) and Bio-Pd(0), respectively. A portion of the free Pd(II) ions on the E. coli cells could complex with the CIP molecules in solution, and then attributed to the some removal of them.55,56
However, when H2 gas was bubbled into the reaction solution, after 25 h, approximately 87.70% of the ciprofloxacin was reductively degraded and removed by Bio-Pd(0), approximately 2-fold of that reported by Martins,57 44.4% (0.2 mg mL−1 Bio-Pd at an initial CIP of 1 mg L−1) after 24 h. This removal of ciprofloxacin is also 3.03 folds of that achieved in the absence of H2. Although in each case the exact mechanism running still keeps unknown, based on the hydrogenation power of palladium, we can speculate that H2 may react with the Pd nanoparticles and clusters to form hydrogen hydride which is a strong reductant to catalyse the conversion of organic molecules. General concept of hydrogen spillover could be defined as dissociative chemical adsorptions of hydrogen on metal nanoparticles and subsequent migration of dissociated hydrogen atoms onto adjacent receptor surfaces via diffusion.58 Palladium has been intensively researched for hydrogen storage and hydrogen-related catalytic reactions as hydrogen easily dissociates on the surface of Pd, and the hydrogen atoms can permeate into the metal lattice.59 This highly active hydride may react with CIP and catalyse its reductive degradation when the electron donor H2 could supply constantly.57 As heterogeneous catalysts, both biological palladium(0) and biological palladium(II) on waste E. coli cells have the advantage that they can be separated, recovered from the solution and reused. However, to achieve continuous reductive degradation of CIP, Pd(0) hydrogen hydride must be constantly generated and the H2 need to be bubbled in the solution and transferred to the substrate (Scheme 1).52
3.6. Possible mechanism for CIP degradation by Bio-Pd in the presence of H2
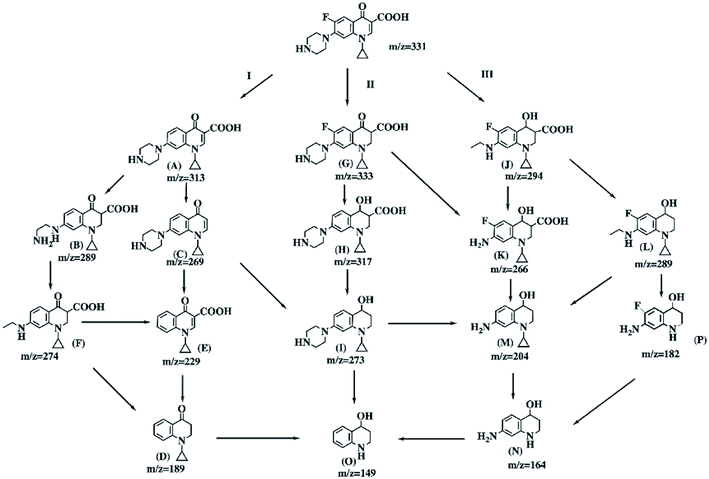
Scheme 2 illustrates the three possible major degradation pathways of CIP with the Bio-Pd catalyst in the presence of H2. In addition, we propose the possible reductive catalytic degradation pathways for CIP contamination combing the examination and analysis results with the previous reports. In pathway I, dehalogenation is the first step of the degradation process, and this result is in line with a previous study.60 Three intermediate products, A (m/z 313), B (m/z 289) and C (m/z 269), were all detected and observed (Fig. S2 and S3†). Afterwards, intermediate C was converted to E (m/z 229) through the loss of an amino group. Finally, all the above three intermediate products could be degraded to D (m/z 189). Pd(0) is an efficient catalyst not only for the chemo-selective hydrogenation of olefinic bonds but also for the hydrogenation of carbonyl compounds at room temperature under benign reaction conditions.61 Pathway II was initiated by the selective reduction of the olefinic bond and resulted in intermediate product G (m/z 333). Then, the carbonyl groups of intermediate G could be converted to alcohols, resulting in the further transformation to the intermediate H, and the losses of the carboxylic groups could produce intermediate I. Pathway III involves the piperazine side chain reduction of the CIP molecule while preserving the fluorine atom. In this pathway, the intermediate J (m/z 294) was detected, which would produce intermediate K (m/z 266) or L (m/z 289) through elimination of the amino group or carboxyl group in the presence of H2, respectively. Both could be further degraded into intermediate M (m/z 204), which could go on to produce compounds N (m/z 164) and O (m/z 149) through the successive loss of the amino group or hydroxyl group.4. Conclusions
In summary, Pd(II) was successfully adsorbed and reduced on the E. coli cells surface to form the Bio-Pd to catalyse the efficient and reductive degradation of ciprofloxacin. Although this degradation mechanism is still unclear, the possibility that organic pollutants can be reduced and degraded using a catalyst derived from waste is sufficient to attract our attention. On the route to “treating waste using waste”, ciprofloxacin was removed and degraded, and palladium was recovered and utilized by adsorption on waste E. coli cells at a low cost, which contributes greatly to environmental protection and ecological improvement.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by the National Natural Science Foundation of China (21576062), the Natural Science Foundation of Zhejiang Province (LY18B060009, LY15B060011), the Technology Research and Development Program of Hangzhou (20191203B09), the Key projects of National Natural Science Foundation of China (81730108), the Research Plan for Sprout Talents in University in Zhejiang Province (2019R426081), the National Innovation and Entrepreneurship Training Program for Undergraduate (201910346044), the “Star and light” Project for Talent Students in Hangzhou Normal University (2019).References
- C. Xu, M. Nasrollahzadeh, M. Selva, Z. Issaabadi and R. Luque, Waste-to-wealth: biowaste valorization into valuable bio(nano)materials, Chem. Soc. Rev., 2019, 48, 4791–4822 RSC.
- K. Folens, S. Van Hulle, F. Vanhaecke and G. Du Laing, Chemical fractionation and speciation modelling for optimization of ion-exchange processes to recover palladium from industrial wastewater, Water Sci. Technol., 2016, 73, 1738–1745 CrossRef CAS PubMed.
- V. Sattayarut, C. Chanthad, P. Khemthong, S. Kuboon, T. Wanchaem, M. Phonyiem, M. Obata, M. Fujishige, K. Takeuchi, W. Wongwiriyapan, P. Khanchaitit and M. Endo, Preparation and electrochemical performance of nitrogen-enriched activated carbon derived from silkworm pupae waste, RSC Adv., 2019, 9, 9878–9886 RSC.
- N. F. M. Noah, N. Othman and N. Jusoh, Highly selective transport of palladium from electroplating wastewater using emulsion liquid membrane process, J. Taiwan Inst. Chem. Eng., 2016, 64, 134–141 CrossRef CAS.
- D. Huang, B. Li, J. Ou, W. Xue, J. Li, Z. Li, T. Li, S. Chen, R. Deng and X. Guo, Megamerger of biosorbents and catalytic technologies for the removal of heavy metals from wastewater: preparation, final disposal, mechanism and influencing factors, J. Environ. Manage., 2020, 261, 109879 CrossRef CAS PubMed.
- W. Pronk, A. Ding, E. Morgenroth, N. Derlon, P. Desmond, M. Burkhardt, B. Wu and A. G. Fane, Gravity-driven membrane filtration for water and wastewater treatment: a review, Water Res., 2019, 149, 553–565 CrossRef CAS PubMed.
- X. Gong, D. Huang, Y. Liu, Z. Peng, G. Zeng, P. Xu, M. Cheng, R. Wang and J. Wan, Remediation of contaminated soils by biotechnology with nanomaterials: bio-behavior, applications, and perspectives, Crit. Rev. Biotechnol., 2018, 38, 455–468 CrossRef CAS PubMed.
- Y. Shen, H. Li, W. Z. Zhu, H. Ho, W. Q. Yuan, J. F. Chen and Y. P. Xie, Microalgal-biochar immobilized complex: a novel efficient biosorbent for cadmium removal from aqueous solution, Bioresour. Technol., 2017, 244, 1031–1038 CrossRef CAS PubMed.
- M. W. Ullah, Z. J. Shi, X. D. Shi, D. Zeng, S. X. Li and G. Yang, Microbes as Structural Templates in Biofabrication: Study of Surface Chemistry and Applications, ACS Sustainable Chem. Eng., 2017, 5, 11163–11175 CrossRef CAS.
- T. Parandhaman, M. D. Dey and S. K. Das, Biofabrication of supported metal nanoparticles: exploring the bioinspiration strategy to mitigate the environmental challenges, Green Chem., 2019, 21, 5469–5500 RSC.
- T. Hennebel, S. De Corte, W. Verstraete and N. Boon, Microbial production and environmental applications of Pd nanoparticles for treatment of halogenated compounds, Curr. Opin. Biotechnol., 2012, 23, 555–561 CrossRef CAS PubMed.
- H. Zhang and X. Hu, Biosynthesis of Pd and Au as nanoparticles by a marine bacterium Bacillus sp GP and their enhanced catalytic performance using metal oxides for 4-nitrophenol reduction, Enzyme Microb. Technol., 2018, 113, 59–66 CrossRef CAS PubMed.
- G. Kratosova, V. Holisova, Z. Konvickova, A. P. Ingle, S. Gaikwad, K. Skrlova, A. Prokop, M. Rai and D. Placha, From biotechnology principles to functional and low-cost metallic bionanocatalysts, Biotechnol. Adv., 2019, 37, 154–176 CrossRef CAS PubMed.
- H. Chitam, N. Zhu, R. Shang, C. Shi, J. Cui, I. Sohoo, P. Wu and Y. Cao, Biorecovery of palladium as nanoparticles by Enterococcus faecalis and its catalysis for chromate reduction, Chem. Eng. J., 2016, 288, 246–254 CrossRef.
- Y. N. Hou, B. Zhang, H. Yun, Z. N. Yang, J. L. Han, J. Z. Zhou, A. J. Wang and H. Y. Cheng, Palladized cells as suspension catalyst and electrochemical catalyst for reductively degrading aromatics contaminants: roles of Pd size and distribution, Water Res., 2017, 125, 288–297 CrossRef CAS PubMed.
- D. C. Hooper, Mode of action of fluoroquinolones, Drugs, 1999, 58(suppl. 2), 6–10 CrossRef CAS PubMed.
- B. Tiwari, B. Sellamuthu, Y. Ouarda, P. Drogui, R. D. Tyagi and G. Buelna, Review on fate and mechanism of removal of pharmaceutical pollutants from wastewater using biological approach, Bioresour. Technol., 2017, 224, 1–12 CrossRef CAS PubMed.
- M. Rusch, A. Spielmeyer, H. Zorn and G. Hamscher, Degradation and transformation of fluoroquinolones by microorganisms with special emphasis on ciprofloxacin, Appl. Microbiol. Biotechnol., 2019, 103, 6933–6948 CrossRef CAS PubMed.
- M. Gavrilescu, K. Demnerova, J. Aamand, S. Agathoss and F. Fava, Emerging pollutants in the environment: present and future challenges in biomonitoring, ecological risks and bioremediation, New Biotechnol., 2015, 32, 147–156 CrossRef CAS PubMed.
- J. Schulz, N. Kemper, J. Hartung, F. Janusch, S. A. I. Mohring and G. Hamscher, Analysis of fluoroquinolones in dusts from intensive livestock farming and the co-occurrence of fluoroquinolone-resistant Escherichia coli, Sci. Rep., 2019, 9, 5117 CrossRef PubMed.
- D. Hocquet, A. Muller and X. Bertrand, What happens in hospitals does not stay in hospitals: antibiotic-resistant bacteria in hospital wastewater systems, J. Hosp. Infect., 2016, 93, 395–402 CrossRef CAS PubMed.
- S. K. Mondal, A. K. Saha and A. Sinha, Removal of ciprofloxacin using modified advanced oxidation processes: kinetics, pathways and process optimization, J. Cleaner Prod., 2018, 171, 1203–1214 CrossRef CAS.
- R. Anjali and S. Shanthakumar, Insights on the current status of occurrence and removal of antibiotics in wastewater by advanced oxidation processes, J. Environ. Manage., 2019, 246, 51–62 CrossRef CAS PubMed.
- A. R. Ribeiro, O. C. Nunes, M. F. R. Pereira and A. M. T. Silva, An overview on the advanced oxidation processes applied for the treatment of water pollutants defined in the recently launched Directive 2013/39/EU, Environ. Int., 2015, 75, 33–51 CrossRef CAS PubMed.
- B. Godlewska-Zylkiewicz, S. Sawicka and J. Karpinska, Removal of Platinum and Palladium from Wastewater by Means of Biosorption on Fungi Aspergillus sp. and Yeast Saccharomyces sp, Water, 2019, 11(7), 1522 CrossRef CAS.
- A. J. Kora and L. Rastogi, Catalytic degradation of anthropogenic dye pollutants using palladium nanoparticles synthesized by gum olibanum, a glucuronoarabinogalactan biopolymer, Ind. Crops Prod., 2016, 81, 1–10 CrossRef CAS.
- W. De Windt, P. Aelterman and W. Verstraete, Bioreductive deposition of palladium (0) nanoparticles on Shewanella oneidensis with catalytic activity towards reductive dechlorination of polychlorinated biphenyls, Environ. Microbiol., 2005, 7, 314–325 CrossRef CAS PubMed.
- I. P. Mikheenko, M. Rousset, S. Dementin and L. E. Macaskie, Bioaccumulation of palladium by Desulfovibrio fructosivorans wild-type and hydrogenase-deficient strains, Appl. Environ. Microbiol., 2008, 74, 6144–6146 CrossRef CAS PubMed.
- S. Kim, M.-H. Song, W. Wei and Y.-S. Yun, Selective biosorption behavior of Escherichia coli biomass toward Pd(II) in Pt(IV)-Pd(II) binary solution, J. Hazard. Mater., 2015, 283, 657–662 CrossRef CAS PubMed.
- K. Chojnacka and M. Mikulewicz, Green analytical methods of metals determination in biosorption studies, TrAC, Trends Anal. Chem., 2019, 116, 254–265 CrossRef CAS.
- L. Guzzella, D. Feretti and S. Monarca, Advanced oxidation and adsorption technologies for organic micropollutant removal from lake water used as drinking-water supply, Water Res., 2002, 36, 4307–4318 CrossRef CAS PubMed.
- N. Ahalya, R. D. Kanamadi and T. V. Ramachandra, Cr(VI) and Fe(III) removal using Cajanus cajan husk, J. Environ. Biol., 2007, 28, 765–769 CAS.
- G. Crini, E. Lichtfouse, L. D. Wilson and N. Morin-Crini, Conventional and non-conventional adsorbents for wastewater treatment, Environ. Chem. Lett., 2019, 17, 195–213 CrossRef CAS.
- S. K. Sharma, A. Mudhoo, G. Jain and J. Sharma, Corrosion inhibition and adsorption properties of Azadirachta indica mature leaves extract as green inhibitor for mild steel in HNO3, Green Chem. Lett. Rev., 2010, 3, 7–15 CrossRef CAS.
- N. Gaur, G. Flora, M. Yadav and A. Tiwari, A review with recent advancements on bioremediation-based abolition of heavy metals, Environ. Sci.: Processes Impacts, 2014, 16, 180–193 RSC.
- S. Abinandan, S. R. Subashchandrabose, K. Venkateswarlu and M. Megharaj, Nutrient removal and biomass production: advances in microalgal biotechnology for wastewater treatment, Crit. Rev. Biotechnol., 2018, 38, 1244–1260 CrossRef CAS PubMed.
- K. Deplanche, I. Caldelari, I. P. Mikheenko, F. Sargent and L. E. Macaskie, Involvement of hydrogenases in the formation of highly catalytic Pd(0) nanoparticles by bioreduction of Pd(II) using Escherichia coli mutant strains, Microbiology, 2010, 156, 2630–2640 CrossRef CAS PubMed.
- T. Hennebel, S. Van Nevel, S. Verschuere, S. De Corte, B. De Gusseme, C. Cuvelier, J. P. Fitts, D. Van der Lelie, N. Boon and W. Verstraete, Palladium nanoparticles produced by fermentatively cultivated bacteria as catalyst for diatrizoate removal with biogenic hydrogen, Appl. Microbiol. Biotechnol., 2011, 91, 1435–1445 CrossRef CAS PubMed.
- J. B. Omajali, J. Gomez-Bolivar, I. P. Mikheenko, S. Sharma, B. Kayode, B. Al-Duri, D. Banerjee, M. Walker, M. L. Merroun and L. E. Macaskie, Novel catalytically active Pd/Ru bimetallic nanoparticles synthesized by Bacillus benzeovorans, Sci. Rep., 2019, 9, 4715 CrossRef PubMed.
- F. Dusa, W. Chen, J. Witos and S. K. Wiedmer, Calcium Dependent Reversible Aggregation of Escherichia coli Biomimicking Vesicles Enables Formation of Supported Vesicle Layers on Silicon Dioxide, Front. Mater., 2019, 6, 23 CrossRef.
- D. P. Teran Valdez, O. Monge Amaya, M. T. Certucha Barragan, F. J. Almendariz Tapia, P. Zavala Rivera and R. Sierra Alvarez, Bioadsorption of copper and zinc with pre-treated and untreated dry biomass of Escherichia coli, Rev. Int. Contam. Ambiental, 2019, 35, 45–55 CrossRef.
- Z. Shen, D. Liu, G. Peng, Y. Ma, J. Li, J. Shi, J. Peng and L. Ding, Electrocatalytic reduction of nitrate in water using Cu/Pd modified Ni foam cathode: High nitrate removal efficiency and N-2-selectivity, Sep. Purif. Technol., 2020, 241, 116743 CrossRef CAS.
- Y. Soni, S. Pradhan, M. K. Bamnia, A. K. Yadav, S. N. Jha, D. Bhattacharyya, T. S. Khan, M. A. Haider and C. P. Vinod, Spectroscopic Evidences for the Size Dependent Generation of Pd Species Responsible for the Low Temperature CO Oxidation Activity on Pd-SBA-15 Nanocatalyst, Appl. Catal., B, 2020, 272, 118934 CrossRef CAS.
- M. M. Areco, S. Hanela, J. Duran and M. D. Afonso, Biosorption of Cu(II), Zn(II), Cd(II) and Pb(II) by dead biomasses of green alga Ulva lactuca and the development of a sustainable matrix for adsorption implementation, J. Hazard. Mater., 2012, 213, 123–132 CrossRef PubMed.
- G. M. Gadd, Biosorption: critical review of scientific rationale, environmental importance and significance for pollution treatment, J. Chem. Technol. Biotechnol., 2009, 84, 13–28 CrossRef CAS.
- Q. Mao, Y. Zhou, Y. Yang, J. Zhang, L. Liang, H. Wang, S. Luo, L. Luo, P. Jeyakumar, Y. S. Ok and M. Rizwan, Experimental and theoretical aspects of biochar-supported nanoscale zero-valent iron activating H2O2 for ciprofloxacin removal from aqueous solution, J. Hazard. Mater., 2019, 380, 120848 CrossRef CAS PubMed.
- M. C. Dodd, M. O. Buffle and U. Von Gunten, Oxidation of antibacterial molecules by aqueous ozone: Moiety-specific reaction kinetics and application to ozone-based wastewater treatment, Environ. Sci. Technol., 2006, 40, 1969–1977 CrossRef CAS PubMed.
- B. De Witte, J. Dewulf, K. Demeestere, M. De Ruyck and H. Van Langenhove, Critical points in the analysis of ciprofloxacin by high-performance liquid chromatography, J. Chromatogr. A, 2007, 1140, 126–130 CrossRef CAS PubMed.
- S. De Corte, T. Hennebel, B. De Gusseme, W. Verstraete and N. Boon, Bio-palladium: from metal recovery to catalytic applications, Microb. Biotechnol., 2012, 5, 5–17 CrossRef CAS PubMed.
- B. De Gusseme, T. Hennebel, L. Vanhaecke, M. Soetaert, J. Desloover, K. Wille, K. Verbeken, W. Verstraete and N. Boon, Biogenic Palladium Enhances Diatrizoate Removal from Hospital Wastewater in a Microbial Electrolysis Cell, Environ. Sci. Technol., 2011, 45, 5737–5745 CrossRef CAS PubMed.
- J. Cui, N. Zhu, N. Kang, C. Ha, C. Shi and P. Wu, Biorecovery mechanism of palladium as nanoparticles by Enterococcus faecalis: From biosorption to bioreduction, Chem. Eng. J., 2017, 328, 1051–1057 CrossRef CAS.
- L. Kuai, Z. Chen, S. Liu, E. Kan, N. Yu, Y. Ren, C. Fang, X. Li, Y. Li and B. Geng, Titania supported synergistic palladium single atoms and nanoparticles for room temperature ketone and aldehydes hydrogenation, Nat. Commun., 2020, 11, 48 CrossRef PubMed.
- C. M. Jung, T. M. Heinze, R. Strakosha, C. A. Elkins and J. B. Sutherland, Acetylation of fluoroquinolone antimicrobial agents by an Escherichia coli strain isolated from a municipal wastewater treatment plant, J. Appl. Microbiol., 2009, 106, 564–571 CrossRef CAS PubMed.
- M. Martins, S. Sanches and I. A. C. Pereira, Anaerobic biodegradation of pharmaceutical compounds: new insights into the pharmaceutical-degrading bacteria, J. Hazard. Mater., 2018, 357, 289–297 CrossRef CAS PubMed.
- G. A. A. Al-Hazmi and F. A. Saad, A Comparative Antimicrobial Study In Between a Quinoline Drug and Its Complexes: Spectral, Kinetic, and Molecular Modeling Investigations, Synth. React. Inorg., Met.-Org., Nano-Met. Chem., 2015, 45, 1743–1757 CrossRef CAS.
- M. F. Li, Y. G. Liu, S. B. Liu, D. Shu, G. M. Zeng, X. J. Hu, X. F. Tan, L. H. Jiang, Z. L. Yan and X. X. Cai, Cu(II)-influenced adsorption of ciprofloxacin from aqueous solutions by magnetic graphene oxide/nitrilotriacetic acid nanocomposite: competition and enhancement mechanisms, Chem. Eng. J., 2017, 319, 219–228 CrossRef CAS.
- M. Martins, C. Mourato, S. Sanches, J. P. Noronha, M. T. Barreto Crespo and I. A. C. Pereira, Biogenic platinum and palladium nanoparticles as new catalysts for the removal of pharmaceutical compounds, Water Res., 2017, 108, 160–168 CrossRef CAS PubMed.
- H. Chen, L. F. Wang, J. Yang and R. T. Yang, Investigation on Hydrogenation of Metal-Organic Frameworks HKUST-1, MIL-53, and ZIF-8 by Hydrogen Spillover, J. Phys. Chem. C, 2013, 117, 7565–7576 CrossRef CAS.
- G. Q. Li, H. Kobayashi, J. M. Taylor, R. Ikeda, Y. Kubota, K. Kato, M. Takata, T. Yamamoto, S. Toh, S. Matsumura and H. Kitagawa, Hydrogen storage in Pd nanocrystals covered with a metal-organic framework, Nat. Mater., 2014, 13, 802–806 CrossRef CAS PubMed.
- I. Ali, C. S. Peng, Z. M. Khan, I. Naz, M. Sultan, M. Ali, I. A. Abbasi, T. Islam and T. Ye, Overview of microbes based fabricated biogenic nanoparticles for water and wastewater treatment, J. Environ. Manage., 2019, 230, 128–150 CrossRef CAS PubMed.
- M. L. Kantam, T. Parsharamulu and S. V. Manorama, Layered double hydroxides supported nano palladium: an efficient catalyst for the chemoselective hydrogenation of olefinic bonds, J. Mol. Catal. A: Chem., 2012, 365, 115–119 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra03783d |
| This journal is © The Royal Society of Chemistry 2020 |