Open Access Article
Open Access ArticleProperties of differentiated SH-SY5Y grown on carbon-based materials†
Sae-Bom Yoon‡
a,
Geonhee Lee‡b,
Sung Bum Parka,
Heeyeong Choa,
Jeong-O. Lee *b and
Byumseok Koh
*b and
Byumseok Koh *a
*a
aBiotechnology and Therapeutics Division, Korea Research Institute of Chemical Technology, 141 Gajeong-ro, Yuseong-gu, Daejeon 34114, Republic of Korea. E-mail: bkoh@krict.re.kr
bAdvanced Materials Division, Korea Research Institute of Chemical Technology, 141 Gajeong-ro, Yuseong-gu, Daejeon 34114, Republic of Korea. E-mail: jolee@krict.re.kr
First published on 20th May 2020
Abstract
Neural cell differentiation has been extensively studied in two-dimensional (2D) cell culture plates. However, the cellular microenvironment and extracellular matrix (ECM) are much more complex and flat 2D surfaces are hard to mimic in ECM. Carbon nanotubes (CNTs) and graphenes are multidimensional carbon-based nanomaterials and may be able to provide extra dimensions on cell growth and differentiation. To determine the effect of CNTs and graphene surfaces on the growth, gene expression, differentiation and functionality of neuroblastoma to a neural cell, SH-SY5Y cells were grown on a 2D (control) surface, a CNT network and a graphene film. The data suggest that SH-SY5Y cells grown on CNT surfaces show an average 20.2% increase in cell viability; 5.7% decrease in the ratio of cells undergoing apoptosis; 78.3, 43.4 and 38.1% increases in SOX2, GFAP and NeuN expression, respectively; and a 29.7% increase in mean firing rate on a multi-electrode array. SH-SY5Y cells grown on graphene film show little or no changes in cell properties compared to cells grown in 2D. The data indicate that the three-dimensional (3D) surface of CNTs provides a favorable environment for SH-SY5Y cells to proliferate and differentiate to neurons.
Introduction
Studies on available experimental models in neurobiology, such as animal models, neurons differentiated from embryonic cells, induced pluripotent stem cells and neuroblastoma cell lines, are important to understand neurodevelopmental and neurodegenerative diseases.1,2 Among the in vitro models used to study human neurological diseases, the SH-SY5Y neuroblastoma cell line is widely documented and used due to its advantages of low cost, ease of handling, reproducibility and possibility for neuronal differentiation.3–11Cells including neurons inside living specimens intensively interact with their surrounding microenvironment, and the communication between cells and their extracellular matrices is the basic, most critical element of all cellular processes.12–16 Recent advances in nanotechnology have provided synthetic bio-friendly materials that can closely mimic controllable microenvironments for cell growth, proliferation and directed differentiation.16–19 CNTs and graphene are an allotrope of carbon in the form of a single layer of atoms in 3D and 2D hexagonal lattices.20,21 Studies have shown that both CNTs and graphene are relatively nontoxic to cells and can provide extra surface dimensions that can mimic actual cellular microenvironments.22–24 It has been shown that CNTs and graphene can promote proliferation and differentiation of human mesenchymal stem cells.25,26 Both CNTs and graphene have also been used in other biomedical applications, such as cell tracking and labeling, nano sensors and the controlled delivery of drugs and bioactive agents.27–30 However, CNTs and graphene each have different properties of flat or irregular surfaces as well as mechanical, chemical and electrical characterizations.21,22 Due to the different characteristics, specific study on the interaction between cells and each carbon-based material is urgently needed. Currently, the majority of SH-SY5Y cells differentiation studies were conducted on 2D polystyrene and flat 2D surfaces.1–11 In addition, studies on the surface effect on cell viability and proliferation are rare. To bridge this knowledge gap and explore the effect of surface modification on SH-SY5Y cells proliferation and differentiation to neurons, we conducted systemic studies of SH-SY5Y cells culture on a control, CNT network and graphene films.
Results and discussion
Characteristics of CNT network and graphene films on cover glass
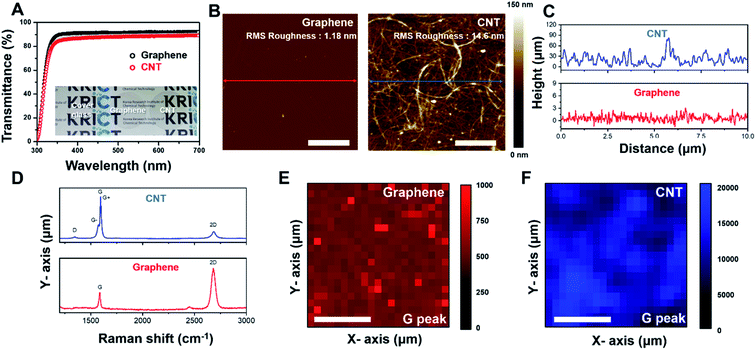
Fig. 1A shows the light transmittance data and camera image of the CNT network and graphene film on cover glass. The cover glass and graphene film show transmittance rates of 91.6% and 88.7%, respectively. Even after CNTs coating on the cover glass, ∼87.66% transmittance occurred in the 300–700 nm visible range (Fig. 1B and S1†). The AFM image of the CNT network and graphene film on cover glass is represented in Fig. 1B. Graphene film shows flat surface properties (roughness: 1.18 nm). A CNT network film formed a fibrous mesh network with a roughness of 14.6 nm (Fig. 1C), exhibiting a uniform wire network characterization of the CNT network film. To further confirm the characterization of the CNT network and graphene film, Raman spectroscopy was performed on each sample. As shown in Fig. 1D (bottom), the Raman spectrum of graphene mostly consists of a G band and a 2D band with few D peaks. The CNT network film consists of single-walled nanotubes, which show the G band divided into G+ and G− peaks (Fig. 1D (top)). Additionally, uniformity of the CNT network and graphene film was demonstrated by Raman mapping analysis of each G peak intensity (Fig. 1E and F), indicating that the film was well fabricated in cover glass. These results suggested that CNT network and graphene film on cover glass have high transparency properties, and the CNT network film forms a mesh structure to provide mechanical cues for cells like in an in vivo extracellular matrix (ECM) environment.31,32Effect of surface variation on SH-SY5Y cells differentiation and viability
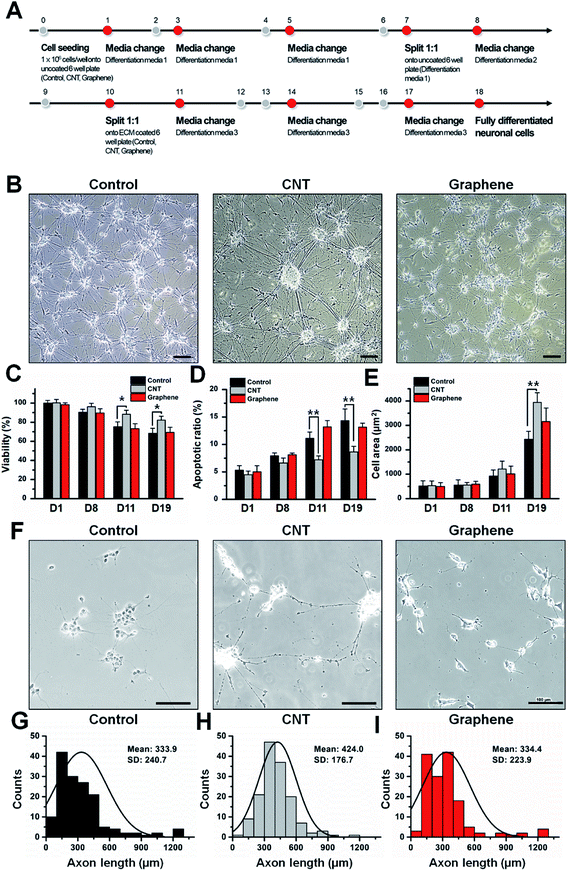
To determine the effect of the CNT network and graphene film on viability and differentiation, SH-SY5Y cells were differentiated following a previously reported protocol4 (Fig. 2A). The morphologies and viabilities of differentiated SH-SY5Y cells on different surfaces were monitored (Fig. 2B–I). During the adopted differentiation process, SH-SY5Y cells undergo a two subculture process. After each subculture process on day 7 and day 10, viability of SH-SY5Y cells decreased compared to that of day 1. The data suggest that the viability of differentiated SH-SY5Y cells on the control, CNT network and graphene film do not differ by >10% until day 8 (Fig. 2C); however, differentiated SH-SY5Y cells on the CNT network film showed an increase of 17.4 and 20.2% on day 11 and day 19 of differentiation, respectively. Differentiated SH-SY5Y cells on graphene film do not show >5% differences in viability compared to differentiated cells on the control surface. Differentiated SH-SY5Y cells on CNT network film showed a 3.9 and 5.7% decrease in cells undergoing apoptosis compared to differentiated cells on the control surface, while differentiated SH-SY5Y cells on graphene film showed >5% variation compared to the control (Fig. 2D). The size of the neuronal cell bodies of differentiated SH-SY5Y cells on the control, CNT network and graphene film showed gradual increases of 2423.5, 3942.5 and 3152.4 μm2, respectively, after 19 days of differentiation (Fig. 2E). Differentiated SH-SY5Y cells on the CNT network film showed longer axonal length with an average axon length of 424.0 μm compared to 333.9 and 334.4 μm of SH-SY5Y cells axon length differentiated on the control and graphene coated matrix, respectively (Fig. 2F–I). Overall, the data suggest that SH-SY5Y cells are most prolific on the CNT network film, and little or no differences were observed between the control and graphene film surfaces. No distinguishable differences on cellular morphologies, viabilities, ratio of cells undergoing apoptosis and size of cellular bodies were observed on undifferentiated SH-SY5Y cells grown on control, CNT network and graphene films (Fig. S2†).Neuronal marker expression of differentiated SH-SY5Y cells on control, CNTs and graphene-coated matrices
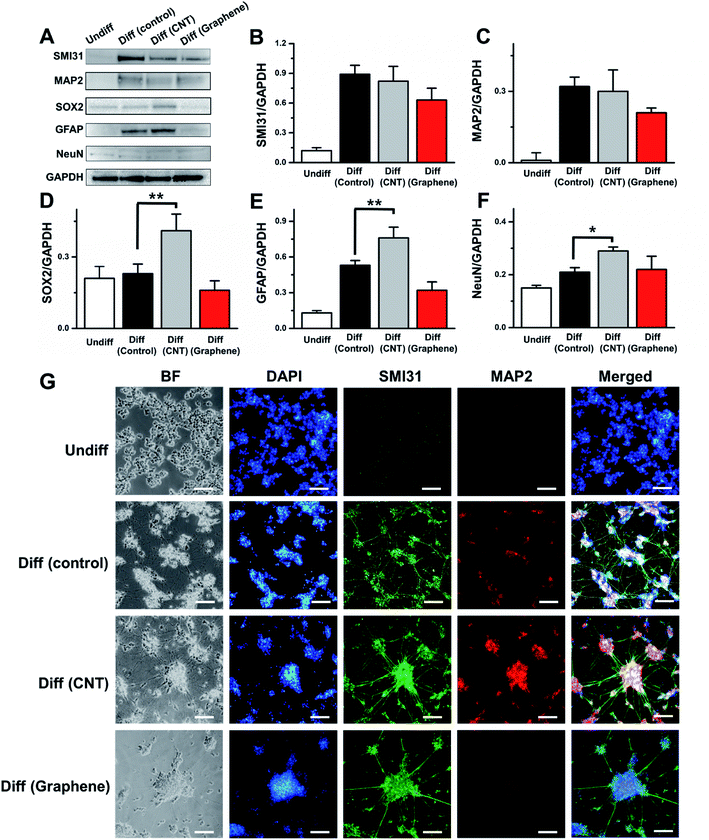
Next, we examined how the cell growing surface could influence the neural marker expression during the differentiation process (Fig. 3). SH-SY5Y cells underwent a full 18 day differentiation process on the control, CNT network and graphene film and were collected for western blot and immunofluorescence imaging. Neurofilament marker H (SMI31), microtubule-associated protein (MAP2), neural stem cell marker (SOX2), glial fibrillar acidic protein (GFAP) and neuronal marker (NeuN) all showed increases in expression during SH-SY5Y differentiation to neurons. Compared to non-differentiated SH-SY5Y cells, differentiated SH-SY5Y cells on the control surface showed a 7.4-, 32.0-, 1.1-, 4.1-, and 1.4-fold increase in SMI31, MAP2, SOX2, GFAP and NeuN expression, respectively. The expression level of SMI31 and MAP2 does not differ >10% in differentiated SH-SY5Y cells on the control and CNT network film, while differentiated SH-SY5Y cells on the graphene film showed a 29.2 and 34.4% decrease in the expression of these two genes compared to differentiated SH-SY5Y cells in the control matrix (Fig. 3B and C). Differentiated SH-SY5Y cells on the CNT network film showed 78.3, 43.4 and 38.1% increases in SOX2, GFAP and NeuN expression compared to that of control (Fig. 3D–F), respectively. Differentiated SH-SY5Y cells on the graphene surface showed a 30.4 and 39.6% decrease in SOX2 and GFAP expression compared to control, respectively, and >5% differences in NeuN expression compared to control (Fig. 3D–F). It has been reported that although GFAP often regarded as glial cell marker, differentiated neuron also expresses GFAP.33 The immunofluorescence data correlated with western blotting data and indicated that undifferentiated SH-SY5Y cells showed minimal expression levels of SMI31 and MAP2, while differentiated SH-SY5Y cells on the control, CNT network and graphene film express fluorescence from SMI31 and MAP2 (Fig. 3G). The increase in axon length (Fig. 2F–I) and the largest neuronal cell bodies area (Fig. 2E) of differentiated SH-SY5Y cells on the CNT network film compared to that from the control and graphene film can be associated with the neural marker expression. This occurred because SMI31 is vastly expressed in axons and MAP2 is expressed in neuronal cell bodies. Therefore, compared to non-differentiated SH-SY5Y cells, differentiated SH-SY5Y cells on the three different surfaces showed an increase in all 5 tested neural markers.Neural signal from differentiated SH-SY5Y cells on control, CNTs and graphene-coated matrices
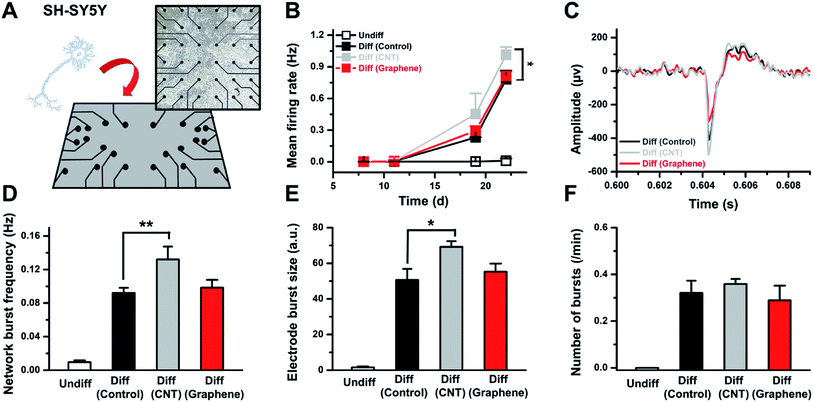
Next, differentiated SH-SY5Y cells for a designated amount of time on the control, CNT network and graphene film were placed on a microelectrode array, and the neural signals from the cells were monitored (Fig. 4A). The data show that SH-SY5Y cells started to show neural activity after 17 days of differentiation, and undifferentiated SH-SY5Y cells do not express activities (Fig. 4B and C). The mean firing rate for differentiated SH-SY5Y cells on the CNT network film after 22 days of differentiation showed a 29.7% increase compared to that of the control (Fig. 4B and C). The network burst frequency and electrode burst size both increased 43.4 and 36.7%, respectively, on differentiated SH-SY5Y cells on the CNT network film compared to the control (Fig. 4D and E). The number of bursts per minute does not differ by >10% in differentiated SH-SY5Y cells on the control, CNT network and graphene film (Fig. 4F). Differentiated SH-SY5Y cells on graphene film do not show >10% variation in the network burst frequency and electrode burst size compared to the control.The MEA results correlate with the neural marker expression level, and differentiated SH-SY5Y cells on the CNT network film showed increased activity. Differentiated SH-SY5Y cells on the control and graphene film revealed similar activities. We speculate that the interaction between SH-SY5Y cells and the CNT network film influenced the differentiation process of SH-SY5Y cells. Although one cannot completely rule out the existence of specific interaction between cells and unique carbon structures in CNT networks, it is likely that marked changes in SH-SY5Y differentiation originate from the nanoscale topography of CNT networks that is absent in both graphene and control substrate. We are currently investigating the effect of surface roughness on SH-SY5Y differentiation using non-carbon substrate.
Conclusions
In summary, we have conducted comparative and systemic studies of SH-SY5Y differentiation on CNT network film and graphene nano-carbon surfaces with polystyrene as a control surface. While both CNTs and graphene showed similar optical transparency and sp2 carbon behavior as evidenced from Raman spectroscopy, SH-SY5Y cells on CNT network film were more prolific and showed higher neural marker expression compared to the differentiated SH-SY5Y cells on polystyrene and graphene film. As expected from increased neural marker expressions, SH-SY5Y cells on CNT network film exhibited enhanced neural activity in MEA recording experiment as well. Our data suggest that activation of neural differentiation may be originated from the nanoscale morphology of CNTs, rather than from its carbonaceous nature.Experimental section
Fabrication of CNT network film on cover glass
CNT (SWCNT 0.4% in H2O) (a diameter of 1.6 (±0.4) nm and a length of 5 μm) of was purchased from OCSiAl Corporation (Luxembourg) and diluted 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in deionized-water (Di-water). Diluted CNT solution was centrifuged at 4 °C (13
1 in deionized-water (Di-water). Diluted CNT solution was centrifuged at 4 °C (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 30 min) to remove the surfactant. A supernatant solution was discarded, and remaining washed CNT was diluted in ethanol. Finally CNT solution was coated on a cover glass using spray coating system (SPINCUBE-100, Jaesung Engineering, Anyang, Korea) through injection of the CNT solution from a syringe pump (KDS 100, KD Scientific, Holliston, MA, USA). The cover glass was cleaned in acetone, isopropyl alcohol (IPA), and Di-water for 30 min each using ultra sonication and sprayed with the CNT solution. The sample stage was rotated at 100 rpm at 120 °C. The syringe pump speed was approximately 5.56 mL h−1. After spray coating, the CNT network film on cover glass was heated to 200 °C for 1 h to remove residual water on the surface.
000 rpm, 30 min) to remove the surfactant. A supernatant solution was discarded, and remaining washed CNT was diluted in ethanol. Finally CNT solution was coated on a cover glass using spray coating system (SPINCUBE-100, Jaesung Engineering, Anyang, Korea) through injection of the CNT solution from a syringe pump (KDS 100, KD Scientific, Holliston, MA, USA). The cover glass was cleaned in acetone, isopropyl alcohol (IPA), and Di-water for 30 min each using ultra sonication and sprayed with the CNT solution. The sample stage was rotated at 100 rpm at 120 °C. The syringe pump speed was approximately 5.56 mL h−1. After spray coating, the CNT network film on cover glass was heated to 200 °C for 1 h to remove residual water on the surface.
Fabrication of graphene film on cover glass
Graphene on copper foil was purchased from MCK Tech Co., Ltd. (Daejeon, Korea). Prior to graphene transfer, poly(methyl methacrylate) (495 PMMA C4, Microchem, Chungbuk, Korea) was spin-coated onto the top of the graphene on copper foil at 2000 rpm for 30 s. The PMMA/graphene on copper foil was then baked at 80 °C for 5 min. The Cu foil was etched with a copper etchant solution (CE-100, Transene Inc., Danvers, MA, USA) and washed with Di-water three times. Next, the PMMA/graphene layer was placed on cover glass. Finally, the PMMA film was dissolved by acetone and washed with isopropyl alcohol (IPA, Sigma-Aldrich, St. Louis, MO, USA).CNT network and graphene film characterization
Optical transmittance spectrum analysis was conducted by UV-vis spectroscopy (UV-2550, Shimadzu, Kyoto, Japan). Surface information was determined by atomic force microscopy (AFM) (Digital Instruments Dimension 3000, Veeco, Plainview, NY, USA). The Raman spectrum and mapping analysis was acquired by using a Raman system (inVia™, Renishaw, Wotton-under-Edge, UK). Mapping images were obtained from 441 spectra (a 50 μm × 50 μm area in the CNT network film). The interval of each spectrum point was every 2.5 μm horizontally and vertically.SH-SY5Y cells differentiation on CNTs and graphene-coated matrices
SH-SY5Y cells (American Type Culture Collection, ATCC, Manassas, VA, USA) differentiation was performed based on a previous study (medium and supplements information on Table S1†)4. In brief, 0.5 × 106 SH-SY5Y cells were seeded on a 6-well culture plate (Corning Life Sciences, Corning, NY, USA), and the CNT network film was placed on top of a 6-well plate. Cells were incubated with Eagle's Minimum Essential Medium (EMEM, ATCC) supplemented with fetal bovine serum (FBS, Thermo Fisher Scientific, Inc., Waltham, MA, USA), penicillin–streptomycin (Thermo Fisher Scientific, Inc.) and retinoic acid (RA, Sigma-Aldrich). After 11 days of differentiation, SH-SY5Y cells were further incubated with a neurobasal medium (Life Technologies, Carlsbad, CA, USA) supplemented with B-27 (Thermo Fisher Scientific, Inc.), potassium chloride (KCl, Sigma-Aldrich), glutamax I (Life Technologies), brain-derived neurotrophic factor (BDNF, Sigma-Aldrich), RA and dibutyryl cyclic AMP (db-cAMP, Sigma-Aldrich). After 18 days of differentiation, cells were characterized with immunofluorescence, western blotting and signal transduction functionality in MEA.Viability and apoptotic ratio of SH-SY5Y cells
SH-SY5Y viabilities after 1, 8, 11 and 19 days of differentiation were determined with a cell counting kit (CCK-8) assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Briefly, 200 μL of WST-8 [2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt] solution was added to each well of the plate and incubated for 1 h, following which the absorbance at 450 nm was measured using a microplate reader (SpectraMax M5e, Molecular Devices, San Jose, CA, USA). The apoptotic rate of SH-SY5Y cells was measured with a Muse® cell analyzer (MilliporeSigma, Burlington, MA, USA) using a Muse® Caspase-3/7 Assay kit.Western blot
After designated differentiation, SH-SY5Y cells were lysed in an ice-cold whole-cell extract buffer (pH 7.4) containing protease inhibitors. The protein concentrations were determined by a BCA protein assay kit (Thermo Fisher Scientific, Inc.). Equal amounts of protein were subjected to electrophoresis using 10% sodium dodecyl sulfate-polyacrylamide gels (Bio-Rad Laboratories, Inc., Hercules, CA, USA). After transfer to polyvinylidene fluoride membranes (Bio-Rad Laboratories, Inc.), the membranes were blocked with 5% bovine serum albumin (Sigma-Aldrich) at room temperature for 1 h. Western blotting was performed with primary antibody overnight at 4 °C, followed by horseradish peroxidase (HRP)-conjugated secondary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Thermo Fisher Scientific, Inc.) at room temperature for 2 h. Protein bands were visualized by a Chemidoc™ imaging system (Bio-Rad Laboratories, Inc.) and an ImageQuant™ LAS 500 (GE Healthcare, Chicago, IL, USA). GAPDH (Sigma-Aldrich) was used as the protein loading control. The following primary antibodies were diluted 1
1000, Thermo Fisher Scientific, Inc.) at room temperature for 2 h. Protein bands were visualized by a Chemidoc™ imaging system (Bio-Rad Laboratories, Inc.) and an ImageQuant™ LAS 500 (GE Healthcare, Chicago, IL, USA). GAPDH (Sigma-Aldrich) was used as the protein loading control. The following primary antibodies were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; SMI31 (Biolegend, San Diego, CA, USA), MAP2 (Cell Signaling Technology, Danvers, MA, USA), SOX2, GFAP, and NeuN (Abcam, Cambridge, UK).
1000; SMI31 (Biolegend, San Diego, CA, USA), MAP2 (Cell Signaling Technology, Danvers, MA, USA), SOX2, GFAP, and NeuN (Abcam, Cambridge, UK).
Imaging
Brightfield (BF) images were obtained using an Eclipse Ti2 inverted microscope (Nikon, Tokyo, Japan). For immunofluorescence imaging, differentiated SH-SY5Y cells were fixed and permeabilized with 1 mL of 0.01% Triton™ X-100 containing formaldehyde solution (Sigma-Aldrich) mixed with 1 mL of neurobasal medium for 1 h at room temperature. After careful washing with Dulbecco's Phosphate-Buffered Saline (DPBS, Thermo Fisher Scientific, Inc.), fixed and permeabilized cells were incubated with a primary antibody (SMI31 and MAP2) in 1% bovine serum albumin solution (BSA, Sigma-Aldrich) overnight at 4 °C. After careful washing with DPBS, cells were further incubated with secondary antibodies conjugated to Alexa Fluor 488 and 555 (Thermo Fisher Scientific, Inc.) in 1% BSA for 1 h, followed by incubation with 4′,6-diamidino-2-phenylindole (DAPI, Thermo Fisher Scientific, Inc.) solution for 5 min. Fluorescence images were obtained with an Eclipse Ti2 microscope.MEA recording
Microelectrode array (60MEA200/30iR/T, Multi-Channel Systems MCS GmbH, Reutlingen, Germany) and CytoView MEA 24 plate with 16 electrodes (Axion Biosystems, Atlanta, GA, USA) were used to record neuronal activity. In brief, microelectrode arrays were coated with 1 mL of 1% ECM solution (100 μL MaxGel™ ECM in 900 μL DMEM) for 1 h at room temperature. After removing ECM solution, microelectrode arrays were air-dried for 1 h at room temperature. 1 mL of 0.5 × 106 cells per mL SH-SY5Y cells differentiated for designated amount of time in control, CNT network and graphene film were seeded on recording electrode and maintained for designated amount of period by replacing new differentiation media. Neuronal activity was monitored with in vitro MEA-system (Multi Channel Systems MCS GmbH) and the Maestro Edge MEA system (Axion Biosystems).Statistical analysis
Statistical analysis was performed using GraphPad Prism (version 6, GraphPad Software, Inc., San Diego, CA, USA) and Origin 8.5 (OriginLab Corporation, Northampton, MA, USA). Each experiment was performed in triplicate, and values are expressed as the mean ± standard deviation (SD). Statistical significance is denoted as * for p < 0.05 and ** for p < 0.01.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
We would like to acknowledge the financial support from the R&D Convergence Program of National Research Council of Science and Technology (grant No. CAP-15-10-KRICT) and the core project (grant No. SKO1930-20 and SI2031-50) of the Korea Research Institute of Chemical Technology.References
- A. Chiocchetti, D. Haslinger, J. Stein, L. De La Torre-ubieta, E. Cocchi, T. Rothämel, S. Lindlar, R. Waltes, S. Fulda and D. Geschwind, Transl. Psychiatry, 2016, 6, e864 CrossRef CAS PubMed.
- M. S. Yap, K. R. Nathan, Y. Yeo, L. W. Lim, C. L. Poh, M. Richards, W. L. Lim, I. Othman and B. C. Heng, Neural Differentiation of Human Pluripotent Stem Cells for Nontherapeutic Applications, Stem Cells Int., 2015, 105172 Search PubMed.
- J. Forster, S. Köglsberger, C. Trefois, O. Boyd, A. Baumuratov, L. Buck, R. Balling and P. Antony, J. Biomol. Screening, 2016, 21, 496–509 CrossRef CAS PubMed.
- M. M. Shipley, C. A. Mangold and M. L. Szpara, J. Visualized Exp., 2016, e53193 Search PubMed.
- S. Kang, X. Chen, S. Gong, P. Yu, S. Yau, Z. Su, L. Zhou, J. Yu, G. Pan and L. Shi, Sci. Rep., 2017, 7, 1–11 CrossRef PubMed.
- Y.-H. Rhee, L. Puspita, Y. A. Sulistio, S. W. Kim, V. Vidyawan, R. Elvira, M.-Y. Chang, J. Shim and S.-H. Lee, Mol. Ther., 2019, 27, 1299–1312 CrossRef CAS PubMed.
- L. Gong, L. Cao, Z. Shen, L. Shao, S. Gao, C. Zhang, J. Lu and W. Li, Adv. Mater., 2018, 30, 1705684 CrossRef PubMed.
- J.-H. Chuang, L.-C. Tung and Y. Lin, World J. Stem Cell., 2015, 7, 437–447 CrossRef PubMed.
- Y. J. Hong and J. T. Do, Front. Bioeng. Biotechnol., 2019, e.2019.00400 Search PubMed.
- Y.-Q. Liu, L.-B. Zhan, T.-T. Bi, L.-N. Liang, X.-X. Sun and H. Sui, RSC Adv., 2016, 6, 34959–34969 RSC.
- A. P. Liu, O. Chaudhuri and S. H. Parekh, Integr. Biol., 2017, 9(5), 383–405 CrossRef PubMed.
- S. Soleman, M. Filippov, A. Dityatev and J. Fawcett, Neuroscience, 2013, 253, 194–213 CrossRef CAS PubMed.
- D. Jain D, S. Mattiassi, E. L. Goh and E. K. Yim, Extracellular matrix and biomimetic engineering microenvironment for neuronal differentiation, Neural Regener. Res., 2020, 15, 573–585 CrossRef PubMed.
- F. Gattazzo, A. Urciuolo and P. Bonaldo, Biochim. Biophys. Acta, Gen. Subj., 2014, 1840, 2506–2519 CrossRef CAS PubMed.
- M. Akhmanova, E. Osidak, S. Domogatsky, S. Rodin and A. Domogatskaya, Physical, Spatial, and Molecular Aspects of Extracellular Matrix of In Vivo Niches and Artificial Scaffolds Relevant to Stem Cells Research, Stem Cells Int., 2015, 167025 Search PubMed.
- S. Schulze, G. Huang, M. Krause, D. Aubyn, V. A. B. Quiñones, C. K. Schmidt, Y. Mei and O. G. Schmidt, Adv. Eng. Mater., 2010, 12, B558–B564 CrossRef.
- S. R. Shin, S. M. Jung, M. Zalabany, K. Kim, P. Zorlutuna, S. b. Kim, M. Nikkhah, M. Khabiry, M. Azize and J. Kong, ACS Nano, 2013, 7, 2369–2380 CrossRef CAS PubMed.
- Y. Xia, S. Li, C. Nie, J. Zhang, S. Zhou, H. Yang, M. Li, W. Li, C. Cheng and R. Haag, Appl. Mater. Today, 2019, 16, 518–528 CrossRef.
- M. Sebaa, T. Y. Nguyen, R. K. Paul, A. Mulchandani and H. Liu, Mater. Lett., 2013, 92, 122–125 CrossRef CAS.
- S. Ahmad, H. Jiang, K. Mustonen, Q. Zhang, A. Hussain, A. T. Khan, N. Wei, M. Tavakkoli, Y. Liao and E.-X. Ding, 2019, arXiv:1904.08323.
- C. Lee, X. Wei, J. W. Kysar and J. Hone, science, 2008, 321, 385–388 CrossRef CAS PubMed.
- Kenry, W. C. Lee, K. P. Loh and C. T. Lim, Biomaterials, 2018, 155, 236–250 CrossRef CAS PubMed.
- N. W. Shi Kam, T. C. Jessop, P. A. Wender and H. Dai, J. Am. Chem. Soc., 2004, 126, 6850–6851 CrossRef PubMed.
- P. Wu, X. Chen, N. Hu, U. C. Tam, O. Blixt, A. Zettl and C. R. Bertozzi, Angew. Chem., Int. Ed., 2008, 47, 5022–5025 CrossRef CAS PubMed.
- J.-R. Lee, S. Ryu, S. Kim and B.-S. Kim, Biomater. Res., 2015, 19, 3 CrossRef PubMed.
- E. K. Luong-Van, T. T. Madanagopal and V. Rosa, Mater. Today Chem., 2020, 16, 100250 CrossRef CAS.
- S. Ania, J. Igor, B. Cyrill, F. Chiara, R. Tariana da, P. Elzbieta, B. Belen, P. Maurizio, N. Klaas and K. Kosras, Carbon, 2016, 97, 126–133 CrossRef.
- B. Meghali, G. P. K. Ajay, M. jianmin and S. T. Michael, NPG Asia Mater., 2017, 9(10), e440 CrossRef.
- W. Ziran, H. Zhuang, G. D. M. Carlos, J. H. Lee, Z. Xuezeng and L. Qiao, Adv. Funct. Mater., 2019, 29, 1905202 CrossRef.
- J. Kim, S. H. Song, Y. Jin, H.-J. Park, H. Yoon, S. Jeon and S.-W. Cho, Nanoscale, 2016, 8, 8512–8519 RSC.
- T. Fujie, S. Ahadian, H. Liu, H. Chang, S. Ostrovidov, H. Wu, H. Bae, K. Nakajima, H. Kaji and A. Khademhosseini, Nano Lett., 2013, 13, 3185–3192 CrossRef CAS PubMed.
- M. A. Correa-Duarte, N. Wagner, J. Rojas-Chapana, C. Morsczeck, M. Thie and M. Giersig, Nano Lett., 2004, 4, 2233–2236 CrossRef CAS.
- E. M. Hol, R. F. Roelofs, E. Moraal, M. a. F. Sonnemans, J. A. Sluijs, E. A. Proper, P. N. E. de Graan, D. F. Fischer and F. W. van Leeuwen, Mol. Psychiatry, 2003, 8, 786–796 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra03383a |
| ‡ The authors equally contributed to this work. |
| This journal is © The Royal Society of Chemistry 2020 |