Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Organic–inorganic nanocrystal reductase to promote green asymmetric synthesis†
Kotchakorn T.sriwong ,
Afifa Ayu Koesoema
,
Afifa Ayu Koesoema and
Tomoko Matsuda
and
Tomoko Matsuda *
*
Department of Life Science and Technology, School of Life Science and Technology, Tokyo Institute of Technology, 4259 Nagatsuta-cho, Midori-ku, Yokohama, 226-8501, Japan. E-mail: tmatsuda@bio.titech.ac.jp; Fax: +81-45-924-5757; Tel: +81-45-924-5757
First published on 20th August 2020
Abstract
An acetophenone reductase from Geotrichum candidum (GcAPRD) was immobilized by the organic–inorganic nanocrystal method. The GcAPRD nanocrystal presented improved stability and recyclability compared with those of the free GcAPRD. Moreover, the GcAPRD nanocrystal reduced broad kinds of ketones with excellent enantioselectivities to produce beneficial chiral alcohols such as (S)-1-(3′,4′-dichlorophenyl)ethanol with >99% yield and >99% ee. The robust and versatile properties of the GcAPRD nanocrystal demonstrated an approach to promote green asymmetric synthesis and sustainable chemistry.
1. Introduction
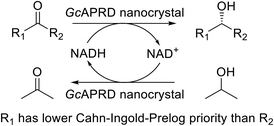
Biocatalysts have presented superior properties for green and sustainable synthesis with their high selectivity, turnover rate, and efficiency.1,2 For example, an alcohol dehydrogenase (ADH) from Rhodococcus ruber DSM 44541 expressed in Arxula adeninivorans is able to enantioselectively reduce p-chloroacetophenone with a remarkable turnover rate (Kcat) of 432 s−1.3,4 Thus, the biocatalysts have played an essential role in an extensive area of industries such as fine and bulk chemical, pharmaceutical, food and beverage, cosmetic, textile, and pulp and paper.5 Meanwhile, the needs for chiral compounds have risen continuously, especially in the pharmaceutical fields, due to the different biological activities and toxicities that may be exhibited by each enantiomer of the chiral compounds.6,7Asymmetric reduction of ketones by chemical and biological catalysts has been considered as a promising method to produce chiral alcohols.8–10 Enzymes catalyzing asymmetric reduction reactions have been widely studied due to their high specificity property, and the well-known enzyme used for this purpose is ADH.10 An acetophenone reductase from Geotrichum candidum NBRC 4597 (GcAPRD) is a novel ADH that reduces ketones to their corresponding (S)-alcohols.11 The GcAPRD has versatile and robust properties such as excellent enantioselectivity, broad substrate specificity, and non-aqueous solvents tolerance that make it potential for industrial applications.12–18 However, the free form of GcAPRD has limitations to recycle and operate in a flow process.
Enzyme immobilization is a method that improves stability and recyclability of the enzyme allowing the enzyme to operate under wide conditions.2 In most cases, the enzyme is confined with the immobilization supports that limits their mobility.19 Various techniques have been addressed to immobilize the enzyme, including the organic–inorganic nanocrystal method, first reported by Ge et al. in 2012.20–22 This method has been proven to be effective in immobilizing various biomolecules such as proteins, peptides, and antibodies.21,22 Moreover, limitations faced by other immobilization methods, such as insufficient binding of protein on support and severe loss of activities, have been overcome by the organic–inorganic nanocrystal method.19,23 Thereby, several studies have applied this method to immobilize broader kinds of biomolecules.21 Enzymes have been one of the most widespread biomolecules immobilized by this method for examples α-lactalbumin,22 laccase,22 carbonic anhydrase,22 lipases,22,24–26 α-amylases,27 papain,28,29 and α-acetolactate decarboxylases.30 However, only a few studies have focused on the immobilization of alcohol dehydrogenase (ADH) by this method,31 particularly for asymmetric synthesis,32 and there is no example yet of versatile ADH nanocrystals with broad substrate specificities.
Thus, this research aimed to immobilize the GcAPRD by using the organic–inorganic nanocrystal method. The preparation conditions of GcAPRD nanocrystal were optimized, and the properties of the GcAPRD nanocrystal were characterized. The GcAPRD nanocrystal showed improved stability and recyclability while retaining excellent enantioselectivity. To the best of our knowledge, this is the first study to report an ADH nanocrystal with broad substrate specificity.
2. Materials and methods
2.1 Reagents and materials
GcAPRD wild type was prepared according to the previous procedure.12 Chemicals for cell cultivation, enzyme purification, and preparation of the GcAPRD nanocrystal were supplied from Nacalai Tesque (Japan), except for 4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid (HEPES) and dithiothreitol (DTT), which were purchased from Sigma Aldrich (USA) and Wako (Japan), respectively. Protein concentration measurement reagent was supplied from Bio-Rad (USA). Commercial grade solvents, ketones, and alcohol standards (1b–3b) were purchased from Nacalai Tesque (Japan). Previously prepared alcohol (4b–7b) and chiral ester (2b–4b and 7b) standards were used in this study.33–352.2 Apparatus
Ultraviolet-visible spectroscopic analysis was conducted in a UV-1900-UV-Visible spectrophotometer from Shimadzu (Japan). Chiral gas chromatography (GC) was performed on a GC-14B equipped with flame ionization detector and a CP-Chirasil-Dex-CB column (Varian 0.32 mm × 0.25 μm × 50 m) using He carrier gas (5 mL min−1, head pressure: 274 kPa, injector: 180 °C, detector: 180 °C) (Shimadzu, Japan). The morphology and energy dispersive X-ray spectroscopy (EDX) analysis were studied by Bench-top Scanning Electron Microscope proX supplied by Phenom-World (Netherlands). Thermo Gravimetric Analysis (TGA) was performed on TG-DTA, DTG-60 (Shimadzu, Japan). The optical rotation value was measured by JASCO P-2200 Polarimeter with a 10 cm path-length cell. The 1H-NMR analysis was performed on a Bruker Biospin AVANCE III 400 spectrometer at 400 MHz in CDCl3.2.3 GcAPRD nanocrystal preparation
The phosphate-buffered saline (PBS) containing GcAPRD was mixed with a metal ion by pipetting for 5 times gently, incubated at 4 °C for 24 h, and centrifuged at 4 °C, 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min. The detailed experimental conditions are shown in the ESI (Table S1†). The supernatant was removed to measure the residual protein amount by Bradford method,36 and to calculate the immobilization yield (1). The precipitant was washed by distilled water and resuspended in PBS (GcAPRD nanocrystal).
000 rpm for 5 min. The detailed experimental conditions are shown in the ESI (Table S1†). The supernatant was removed to measure the residual protein amount by Bradford method,36 and to calculate the immobilization yield (1). The precipitant was washed by distilled water and resuspended in PBS (GcAPRD nanocrystal).
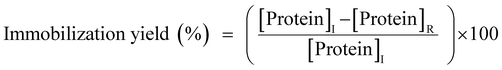 | (1) |
2.4 Relative reaction yield and activity measurement of enzyme
The relative reaction yield measurement to investigate the optimum immobilization conditions was conducted by mixing acetophenone 1a (5.0 mM), 2-propanol (15% v/v), free GcAPRD (8.30–16.50 μg of protein per mL) or GcAPRD nanocrystal (8.30–33.0 μg of protein per mL), NAD+ (0.20 mM), and HEPES–NaOH buffer (0.10 M, pH 7.2) up to 3 mL. For the investigation of the optimum immobilization conditions, the reaction was performed at 37 °C, 200 rpm for 10 min. The product formed (1b) was extracted with diethyl ether and analyzed by GC, using 3-methyl-1-butanol as an internal standard.Activity measurement to investigate the temperature and pH profile, and the stability of GcAPRD nanocrystal were assayed by using UV-spectrophotometer. All assays were performed at 37 °C in 1.0 mL HEPES–NaOH buffer (0.10 M, pH 7.2), consisting of 1a (5.0 mM) except for the pH profile experiment (5.6 mM), NADH (0.28 mM), and GcAPRD nanocrystal (0.80–4.00 μg of protein per mL). Initial velocity was determined by detecting the decrease of NADH at 340 nm for 132 s.
The recyclability was assessed by batch process. The reaction mixture consisted of 1a (3.0 mM), 2-propanol (15% v/v), GcAPRD nanocrystal (630 μg of protein), NAD+ (0.20 mM), and HEPES–NaOH buffer (0.10 M, pH 7.2) up to 1.5 mL. The reaction in each cycle was conducted at 30 °C, 200 rpm for up to 180 min. Afterward, the reaction mixture was centrifuged at 4 °C, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min. The supernatant was removed, extracted with diethyl ether, and analyzed by GC. The precipitant was washed by the reaction mixture with the absence of NAD+, and proceeded the next reaction cycle.
000 rpm for 5 min. The supernatant was removed, extracted with diethyl ether, and analyzed by GC. The precipitant was washed by the reaction mixture with the absence of NAD+, and proceeded the next reaction cycle.
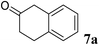
2.5 Micromole scale asymmetric reduction of ketones by GcAPRD nanocrystal
The experiment was conducted based on the previously reported procedure.13 Micromole scale reductions were performed in a 3.0 mL reaction mixture, which consists of 1a–7a (5.0 mM), 2-propanol (15% v/v), NAD+ (5.0 mM), HEPES–NaOH buffer (0.10 M, pH 7.2), and GcAPRD nanocrystal with the amount needed to achieve 1.0 μmol min−1 mL−1 reduction of each substrate to its corresponding alcohol for 1a–6a, and 350 μg of protein/mL for 7a. The mixture was incubated at 30 °C, 200 rpm for 3 h. A portion of the mixture was extracted with diethyl ether to determine the reaction yield by GC analysis. The enantioselectivity excess (ee) values and the absolute configurations of 1b, 5b, and 6b were determined by comparing their GC retention times with the corresponding authentic samples prepared before.35 Meanwhile, for 2b–4b and 7b, the reaction mixtures were extracted by dichloromethane, dried over MgSO4, and used for propionylation by propionyl chloride reaction. ee values and absolute configurations were determined by comparing the GC retention times of propionates of 2b–4b and 7b with the corresponding authentic samples prepared before.33,34 The chiral GC retention time of ketone, alcohol, and ester standards are given in Table S2.†2.6 Preparative scale asymmetric reduction of a ketone by GcAPRD nanocrystal
The reaction was performed in a 100 mL scale consisting of 6a (197 mg, 10.0 mM), 2-propanol (3.0% v/v), NAD+ (1.4 mM), HEPES–NaOH buffer (0.10 M, pH 7.2), and GcAPRD nanocrystal (2 mg of protein). The mixture was incubated at 30 °C, 130 rpm for 18 h, and extracted by diethyl ether for 3 times, dried over MgSO4, filtered, and evaporated. The corresponding alcohol was purified by silica gel column chromatography (hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 3
ethyl acetate, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford (S)-6b (165 mg, 0.87 mmol, 84%), and characterized by 1H-NMR analysis. The 1H-NMR spectra of alcohol (S)-6b was in agreement with that reported in the literature.37 ee was determined by chiral GC analysis, and the absolute configuration was determined by comparing the optical rotation value of product with the literature.38 [α]20D = −38.19 (c = 1.04, CHCl3, ee = >99%); lit38 (S)-6b: [α]20D = −33.70 (c = 1.44, CHCl3, ee = 91%). 1H-NMR (400 MHz, CDCl3, 25 °C, TMS): δ = 7.48 (d, J = 2.0 Hz, 1H, CH), 7.41 (d, J = 8.3 Hz, 1H, CH), 7.20 (dd, J = 8.2 Hz, 2.0 Hz, 1H, CH), 4.84–4.90 (m, 1H, CH), 1.82–1.83 (m, 1H, OH), 1.48 (d, J = 6.48 Hz, 3H, CH3).
1) to afford (S)-6b (165 mg, 0.87 mmol, 84%), and characterized by 1H-NMR analysis. The 1H-NMR spectra of alcohol (S)-6b was in agreement with that reported in the literature.37 ee was determined by chiral GC analysis, and the absolute configuration was determined by comparing the optical rotation value of product with the literature.38 [α]20D = −38.19 (c = 1.04, CHCl3, ee = >99%); lit38 (S)-6b: [α]20D = −33.70 (c = 1.44, CHCl3, ee = 91%). 1H-NMR (400 MHz, CDCl3, 25 °C, TMS): δ = 7.48 (d, J = 2.0 Hz, 1H, CH), 7.41 (d, J = 8.3 Hz, 1H, CH), 7.20 (dd, J = 8.2 Hz, 2.0 Hz, 1H, CH), 4.84–4.90 (m, 1H, CH), 1.82–1.83 (m, 1H, OH), 1.48 (d, J = 6.48 Hz, 3H, CH3).
2.7 GcAPRD nanocrystal characterization by scanning electron microscopy (SEM) and EDX analysis, and TGA
The prepared GcAPRD nanocrystal was washed by distilled water several times to remove the residual phosphate and metal ion prior to drying at room temperature. The dried GcAPRD nanocrystal was used for SEM and EDX analysis, and TGA.3. Results and discussions
3.1 Optimization of GcAPRD nanocrystal synthesis
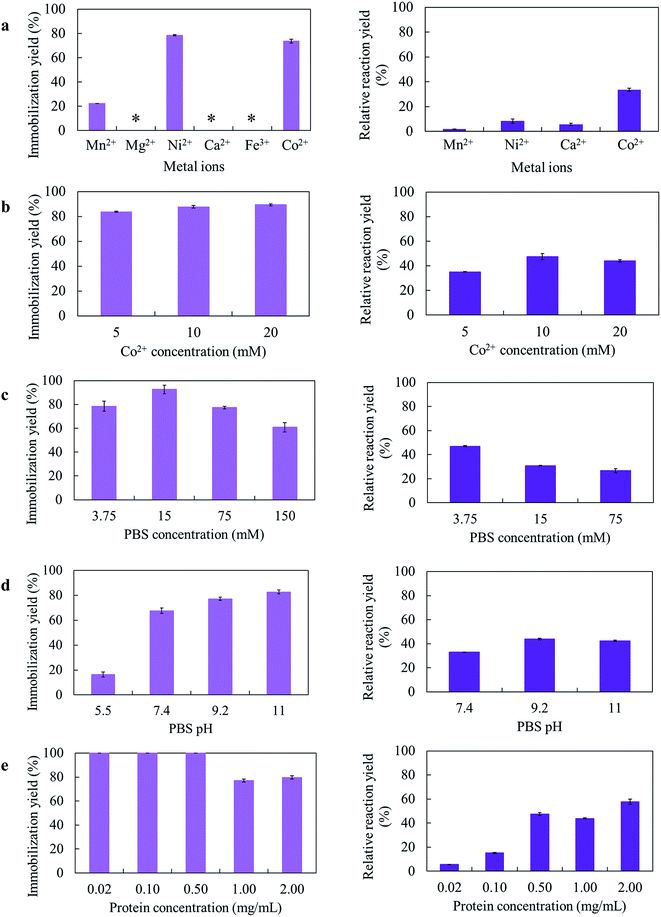
Several parameters for the GcAPRD nanocrystal preparation, such as kinds of metal ions, metal concentration, PBS concentration, PBS pH, and protein concentration, were varied. The immobilization yields and relative reaction yields are shown in Fig. 1.Different amino acid residues in each biomolecule present different affinities with various metal ions, and the appropriate metal ion concentration is necessary for the nucleation.21 In this research, 6 metal ions were used to prepare the GcAPRD nanocrystal (Fig. 1a). It was found that the immobilization yield was high when using Co2+ or Ni2+ as a metal ion. Nonetheless, only the GcAPRD nanocrystal synthesized by Co2+ retained sufficient activity. It was assumed that the His-tag located in the C-terminus of the GcAPRD acted as a binder with Co2+ or Ni2+ ion, which was comparable to the discovery of López-Gallego and Yate's.31 They reported that the presence of His-tagged protein is mandatory as the driving force for the crystal formation. The His-tag binds Co2+ ions via the coordination bond and forms a nucleation site, which initiates the crystal formation.31 Although Ni2+ could form the crystal with His-tagged protein, high concentration of nickel could cause the enzyme deactivation (Fig. S1†). To investigate the importance of Co2+ and His-tagged GcAPRD bonding, the formed GcAPRD nanocrystal was suspended in 500 mM imidazole in 5 mM PBS to break the Co2+ and His-tag bond, thus eluting the protein. After the elution, the GcAPRD nanocrystal was apparently detached, suggested that Co2+ and His-tag bonding played an important role in the GcAPRD nanocrystal formation (Fig. S2†). Additionally, the metal concentration was optimized to 10 mM (Fig. 1b), comparable with Kim et al.’s study (10 mM of cobalt for the formation of BSA–inorganic hybrid nanoflower).39
PBS concentration is another important parameter for the nanocrystal formation.28 Thereby, we investigated the effect of PBS concentration on the GcAPRD nanocrystal formation (Fig. 1c). The prepared GcAPRD nanocrystal performed the highest relative reaction yield at 3.75 mM PBS. However, the optimum PBS concentration is different depending on the biomolecule and other synthesis parameters.40,41 Synthesis pH is an essential parameter influencing the nanocrystal formation.29,41,42 Thus, this study investigated the effect of PBS pH on the GcAPRD nanocrystal formation (Fig. 1d). The immobilization yield of GcAPRD was low in acidic conditions (pH 5.5), and higher in higher pH (pH 7.4, 9.2, and 11.0). It was proposed that in acidic conditions, the biomolecule is highly protonated, causing strong repulsions with the positive charges of the transition metal (i.e., Cu2+and Co2+).43 Joshua et al., reported that protonating the imidazole nitrogen atom of the histidine residue (pKa 6.0) by lowering the pH to 6.0 can disrupt the coordination bond between histidine and Co2+.44 Altinkaynak et al. also reported an easier synthesis of lactoperoxidase hybrid nanoflower at pH 9.0 and 10.0.19 The GcAPRD nanocrystal showed the best relative reaction yield in pH 9.2, and pH 11.0.
The presence of biomolecule is mandatory for the hybrid nanocrystal formation.21,22 In this study, the protein concentration in the GcAPRD nanocrystal formation was varied to 0.02, 0.10, 0.05, 1.00, and 2.00 mg mL−1 (Fig. 1e). The experiments demonstrated that at the protein concentration of 0.05 mg mL−1 and below, the enzyme was entirely immobilized (immobilization yield >99%). On the other hand, the protein concentration of more than 1.00 mg mL−1 resulted in a decrease in immobilization yield, but protein immobilized per metal amount was high. Nevertheless, the relative reaction yield result showed that the reaction proceeded better when the synthesized protein concentrations were more than 0.50 mg mL−1, and the greatest relative reaction yield was found on the 2.00 mg mL−1 of protein. Ge et al. reported that different concentration of protein has a different nanocrystal formation mechanism. The lower concentration of protein causes fewer nucleation sites, leading to a larger nanocrystal with a more complex structure.22 At last, it was confirmed that the active GcAPRD was mandatory to the GcAPRD nanocrystal activity (ESI, section 4†). To understand this phenomenon clearly, SEM analysis on GcAPRD nanocrystal synthesized with various protein concentrations were performed as described in section 3.2.
3.2 Morphology analysis by SEM
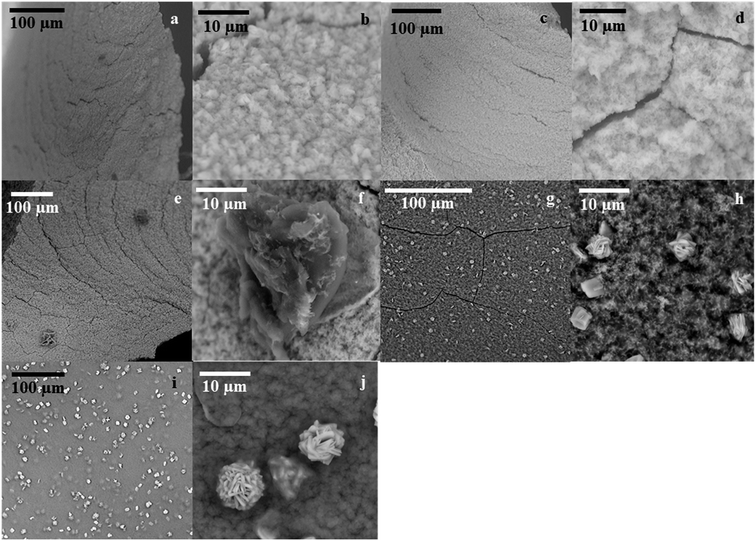
SEM analysis on GcAPRD nanocrystal synthesized with various protein concentrations was performed to understand its effect in the immobilization yield and relative reaction yield. As shown in Fig. 2, it was found that without enzyme, the cobalt-phosphate crystal was formed (Fig. 2a and b). In the presence of 0.02 mg mL−1 (Fig. 2c and d) and 0.10 mg mL−1 (Fig. 2e and f) of protein, the surfaces of the crystals were similar to the control, assuming that the small amount of protein was buried underneath the cobalt–phosphate crystals. Therefore, the mass transfer between the hindered enzyme and the substrate was limited, and low relative reaction yields were observed for the nanocrystals synthesized with 0.02 and 0.10 mg mL−1 of protein. The surfaces of GcAPRD nanocrystals synthesized with 0.50 mg mL−1 (Fig. 2g–h) and 2.00 mg mL−1 (Fig. 2i–j) protein were different from the control and displaying a parallel hexahedron structure assembled to a flower-like structure mixed with sponge-like structure. The proposed formation mechanism of the GcAPRD nanocrystal synthesized with 0.50 and 2.00 mg mL−1 of protein is presented in Fig. 3. There may be 3 steps, comprised of nucleation, aggregation, and anisotropic growth. A similar structure was reported previously by Wang et al. in the formation of nanocrystals with the presence of 2.00 mg mL−1 protein.27,31 The high relative reaction yield suggested that the GcAPRD was gathered on the surface of the nanostructure, and the enzymes were not buried. In addition, the GcAPRD nanocrystal was characterized by EDX analysis and TGA to examine the chemical composition and structure of the GcAPRD nanocrystal as in Fig. S3–S5 (ESI,† section 5).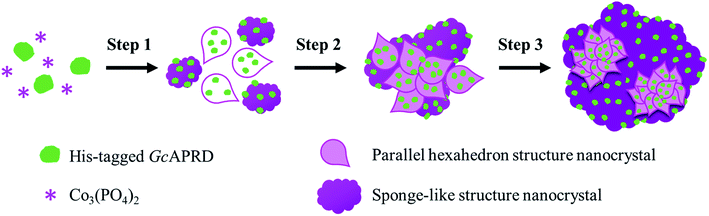 | ||
| Fig. 3 The proposed formation mechanism of GcAPRD nanocrystal, comprised of 3 steps: (1) nucleation, (2) aggregation, and (3) anisotropic growth. | ||
3.3 Temperature and pH profile, and stability of GcAPRD nanocrystal
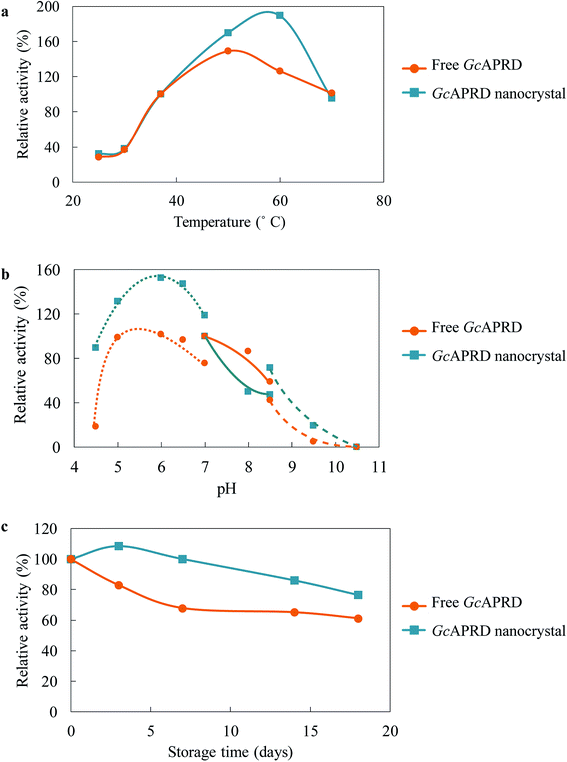
Immobilization has been reported to improve the temperature and pH profile, as well as the stability of the enzyme.2,31,40,41 The temperature profile of GcAPRD nanocrystal and free GcAPRD was examined (Fig. 4a). Both free GcAPRD and GcAPRD nanocrystal demonstrated poor activity at low temperatures (20–30 °C). The activities of both enzymes were increased in higher temperatures. The free GcAPRD performed the highest relative activity at 50 °C (170%), and the activity gradually decreased at the temperature above 60 °C. To our delight, the GcAPRD nanocrystal presented the improved temperature profile as the highest relative activity was observed at 60 °C (189%). At the same time, the effect of pH on the GcAPRD nanocrystal activity was investigated, and it showed less sensitivity than the free GcAPRD in acidic conditions (Fig. 4b). The relative activity of the free GcAPRD dramatically decreased from 99% to 18% when the pH was lowered from 5.0 to 4.5, while the GcAPRD nanocrystal's relative activity moderately decreased from 132% to 89%. The storage stability of the enzymes at 4 °C was investigated. As shown in Fig. 4c, after 7 days, the residual activity of the free GcAPRD was 68%, while the GcAPRD nanocrystal still retained 100% activity. Also, after 18 days, the free GcAPRD's residual activity was 61%, while the GcAPRD nanocrystal had 76% of residual activity.3.4 Recyclability of GcAPRD nanocrystal
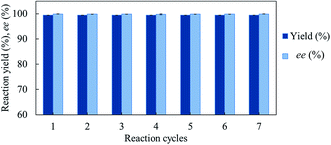
We investigated the recyclability of the GcAPRD nanocrystal by performing the reduction of 1a in batch process. The GcAPRD nanocrystal was able to be recycled with noticeable high reduction yield (>99%) and excellent enantioselectivity (>99%) up to 7 reaction cycles (Fig. 5). This result proved the superior recyclable properties of the GcAPRD nanocrystal, which is a promising approach to further applications.3.5 Substrate specificity of GcAPRD nanocrystal
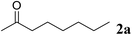
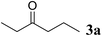
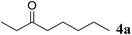
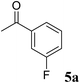
Excellent enantioselectivity and broad substrate specificity are one of the most crucial aspects of the enzyme utilization for industrial processes.10 The GcAPRD nanocrystal was tested to reduce broad kinds of ketones (Table 1). 1a, as a model substrate for the GcAPRD reduction, was reduced by GcAPRD nanocrystal with >99% yield and >99% ee (S). The GcAPRD nanocrystal also reduced challenging aliphatic ketones 2a–4a with high yields and enantioselectivities (up to >99% ee (S)). Moreover, the GcAPRD nanocrystal was able to enantioselectively reduce “difficult to resolve” ketone 3a with only one carbon difference between side chains adjacent to the carbonyl carbon. The enantioselectivities reported for the free enzyme were retained perfectly.9,12,13| Substrate | Yield (%) | ee (%) |
|---|---|---|
| a The reaction conditions are described in the section 2.5. | ||
 |
>99 | >99 (S) |
 |
>99 | >99 (S) |
 |
74 | 99 (S) |
 |
77 | >99 (S) |
 |
>99 | >99 (S) |
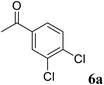 |
>99 | >99 (S) |
 |
91 | 96 (S) |
The GcAPRD nanocrystal produced beneficial chiral alcohols with excellent enantioselectivities (ee up to >99% (S)) such as (S)-5b (intermediate of EGFR and HER2 kinase inhibitors, a potential cancer treatment agent),45 (S)-6b (intermediate of sertraline, an obsessive-compulsive disorder medication),46 and (S)-7b (intermediate of aminotetralin, which can be used to stimulate dopamine, serotonin, and melatonin receptors).47,48 We were also able to isolate (S)-6b with 84% isolated yield, in a preparative scale reaction catalyzed by the GcAPRD nanocrystal. To the best of our knowledge, this is the first study to report an ADH nanocrystal with broad substrate specificity.
4. Conclusions
We successfully immobilized a novel ADH, GcAPRD, by the organic–inorganic nanocrystal method. The GcAPRD nanocrystal presented improved temperature and pH profile, and stability compared with those of the free GcAPRD. The GcAPRD nanocrystal overcame the recyclability limitation of the free GcAPRD. It could be recycled up to 7 cycles with remarkably high yield and excellent enantioselectivity. The GcAPRD nanocrystal reduced broad kinds of ketones to produce valuable chiral alcohols. The robust and versatile properties of the GcAPRD nanocrystal made it a promising and sustainable catalyst for industrial applications. Future studies will investigate the reaction of GcAPRD nanocrystal in non-aqueous solvent and perform cascade reactions to allow the production of broader kinds of beneficial compounds.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are grateful to the Open Research Facilities for Life Science and Technology, School of Life Science and Technology, Tokyo Institute of Technology, for the support of SEM and EDX analysis, and Open Facility Center, Materials Analysis Division, Tokyo Institute of Technology, for the support of TGA. This work was partially supported by the Japan Society for the Promotion of Science, Japan (grant number JP16K05864 and JP19K05560) to Tomoko Matsuda.Notes and references
- K. Nakamura, R. Yamanaka, T. Matsuda and T. Harada, Tetrahedron: Asymmetry, 2003, 14, 2659–2681 CrossRef CAS.
- A. Madhavan, R. Sindhu, P. Binod, R. K. Sukumaran and A. Pandey, Bioresour. Technol., 2017, 245, 1304–1313 CrossRef CAS PubMed.
- W. Stampfer, B. Kosjek, K. Faber and W. Kroutil, J. Org. Chem., 2003, 68, 402–406 CrossRef CAS PubMed.
- M. Giersberg, A. Degelmann, R. Bode, M. Piontek and G. Kunze, J. Ind. Microbiol. Biotechnol., 2012, 39, 1385–1396 CrossRef CAS PubMed.
- J.-M. Choi, S.-S. Han and H.-S. Kim, Biotechnol. Adv., 2015, 33, 1443–1454 CrossRef CAS PubMed.
- R. N. Patel, Bioorg. Med. Chem., 2018, 26, 1252–1274 CrossRef CAS PubMed.
- D. Ballesteros-vivas, G. Alvarez-rivera, M. Bueno and A. Cifuentes, TrAC, Trends Anal. Chem., 2020, 123, 115761 CrossRef.
- N. Itoh, Appl. Microbiol. Biotechnol., 2014, 98, 3889–3904 CrossRef CAS PubMed.
- A. A. Koesoema, D. M. Standley, T. Senda and T. Matsuda, Appl. Microbiol. Biotechnol., 2020, 104, 2897–2909 CrossRef CAS PubMed.
- H. Gröger, W. Hummel and R. Metzner, in Comprehensive Chirality, 2012 Search PubMed.
- T. Matsuda, S. Kanamaru, Y. Nakata, R. Kanamori and T. Fukae, Appl. Microbiol. Biotechnol., 2009, 86, 625–631 Search PubMed.
- T. Yamamoto, Y. Nakata, C. Cao, Y. Sugiyama, Y. Asanuma, S. Kanamaru and T. Matsuda, Appl. Microbiol. Biotechnol., 2013, 97, 10413–10421 CrossRef CAS PubMed.
- A. A. Koesoema, Y. Sugiyama, Z. Xu, D. M. Standley, M. Senda, T. Senda and T. Matsuda, Appl. Microbiol. Biotechnol., 2019, 103, 9543–9553 CrossRef CAS PubMed.
- K. Nakamura, Y. Inoue, T. Matsuda and I. Misawa, J. Chem. Soc., Perkin Trans. 1, 1999, 2397–2402 RSC.
- T. Matsuda, T. Harada and K. Nakamura, Chem. Commun., 2000, 1367–1368 RSC.
- T. Matsuda, K. Watanabe, T. Kamitanaka, T. Harada and K. Nakamura, Chem. Commun., 2003, 3, 1198–1199 RSC.
- T. Matsuda, R. Marukado, M. Mukouyama, T. Harada and K. Nakamura, Tetrahedron: Asymmetry, 2008, 19, 2272–2275 CrossRef CAS.
- T. Matsuda, Y. Yamagishi, S. Koguchi, N. Iwai and T. Kitazume, Tetrahedron Lett., 2006, 47, 4619–4622 CrossRef CAS.
- C. Altinkaynak, S. Tavlasoglu, N. Özdemir and I. Ocsoy, Enzyme Microb. Technol., 2016, 93–94, 105–112 CrossRef CAS PubMed.
- N. R. Mohamad, N. H. C. Marzuki, N. A. Buang, F. Huyop and R. A. Wahab, Biotechnol. Biotechnol. Equip., 2015, 29, 205–220 CrossRef CAS PubMed.
- J. Cui and S. Jia, Coord. Chem. Rev., 2017, 352, 249–263 CrossRef CAS.
- J. Ge, J. Lei and R. N. Zare, Nat. Nanotechnol., 2012, 7, 428–432 CrossRef CAS PubMed.
- C. Altinkaynak, S. Tavlasoglu, N. Özdemir and I. Ocsoy, Enzyme Microb. Technol., 2016, 93–94, 105–112 CrossRef CAS PubMed.
- S. Escobar, S. Velasco-Lozano, C. H. Lu, Y. F. Lin, M. Mesa, C. Bernal and F. López-Gallego, J. Mater. Chem. B, 2017, 5, 4478–4486 RSC.
- Y. Zhang, W. Sun, N. M. Elfeky, Y. Wang, D. Zhao, H. Zhou, J. Wang and Y. Bao, Enzyme Microb. Technol., 2020, 132, 109408 CrossRef CAS PubMed.
- S. S. Nadar and V. K. Rathod, Enzyme Microb. Technol., 2018, 108, 11–20 CrossRef CAS PubMed.
- L. B. Wang, Y. C. Wang, R. He, A. Zhuang, X. Wang, J. Zeng and J. G. Hou, J. Am. Chem. Soc., 2013, 135, 1272–1275 CrossRef CAS PubMed.
- J. Yu, X. Chen, M. Jiang, A. Wang, L. Yang, X. Pei, P. Zhang and S. G. Wu, RSC Adv., 2018, 8, 2357–2364 RSC.
- Y. Li, X. Fei, L. Liang, J. Tian, L. Xu, X. Wang and Y. Wang, J. Mol. Catal. B: Enzym., 2016, 133, 92–97 CrossRef CAS.
- F. Zhao, Q. Wang, J. Dong, M. Xian, J. Yu, H. Yin, Z. Chang, X. Mu, T. Hou and J. Wang, Process Biochem., 2017, 57, 87–94 CrossRef CAS.
- F. López-Gallego and L. Yate, Chem. Commun., 2015, 51, 8753–8756 RSC.
- X. Chen, L. Xu, A. Wang, H. Li, C. Wang, X. Pei, P. Zhang and S. G. Wu, J. Chem. Technol. Biotechnol., 2019, 94, 236–243 CrossRef CAS.
- A. A. Koesoema, Y. Sugiyama, Z. Xu, D. M. Standley, M. Senda, T. Senda and T. Matsuda, Appl. Microbiol. Biotechnol., 2019, 103, 9543–9553 CrossRef CAS PubMed.
- A. A. Koesoema, D. M. Standley, T. Kotchakorn, M. Tamura and T. Matsuda, Tetrahedron Lett., 2020, 61, 151682 CrossRef CAS.
- A. A. Koesoema, D. M. Standley, S. Ohshima, M. Tamura and T. Matsuda, Tetrahedron Lett., 2020, 151820 CrossRef CAS.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- R. Wang, J. Qi, Y. Yue, Z. Lian, H. Xiao, S. Zhuo and L. Xing, Tetrahedron, 2019, 75, 130463 CrossRef.
- Y. Li, S. Yu, X. Wu, J. Xiao, W. Shen, Z. Dong and J. Gao, J. Am. Chem. Soc., 2014, 136, 4031–4039 CrossRef CAS PubMed.
- K. H. Kim, J. M. Jeong, S. J. Lee, B. G. Choi and K. G. Lee, J. Colloid Interface Sci., 2016, 484, 44–50 CrossRef CAS PubMed.
- S. S. Nadar, S. D. Gawas and V. K. Rathod, Int. J. Biol. Macromol., 2016, 92, 660–669 CrossRef CAS PubMed.
- B. Somturk, M. Hancer, I. Ocsoy and N. Özdemir, Dalton Trans., 2015, 44, 13845–13852 RSC.
- B. Somturk, I. Yilmaz, C. Altinkaynak, A. Karatepe, N. Özdemir and I. Ocsoy, Enzyme Microb. Technol., 2016, 86, 134–142 CrossRef CAS PubMed.
- C. Altinkaynak, I. Yilmaz, Z. Koksal, H. Özdemir, I. Ocsoy and N. Özdemir, Int. J. Biol. Macromol., 2016, 84, 402–409 CrossRef CAS PubMed.
- J. A. Bornhorst and J. J. Falke, Methods Enzymol., 2000, 326, 245–254 CAS.
- H. Mastalerz, M. Chang, P. Chen, P. Dextraze, B. E. Fink, A. Gavai, B. Goyal, W. C. Han, W. Johnson, D. Langley, F. Y. Lee, P. Marathe, A. Mathur, S. Oppenheimer, E. Ruediger, J. Tarrant, J. S. Tokarski, G. D. Vite, D. M. Vyas, H. Wong, T. W. Wong, H. Zhang and G. Zhang, Bioorg. Med. Chem. Lett., 2007, 17, 2036–2042 CrossRef CAS PubMed.
- C. Barbieri, E. Caruso, P. D'Arrigo, G. Pedrocchi Fantoni and S. Servi, Tetrahedron: Asymmetry, 1999, 10, 3931–3937 CrossRef CAS.
- P. Bonomi, P. Cairoli, D. Ubiali, C. F. Morelli, M. Filice, I. Nieto, M. Pregnolato, P. Manitto, M. Terreni and G. Speranza, Tetrahedron: Asymmetry, 2009, 20, 467–472 CrossRef CAS.
- F. Orsini, G. Sello, E. Travaini and P. Di Gennaro, Tetrahedron: Asymmetry, 2002, 13, 253–259 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra03160g |
| This journal is © The Royal Society of Chemistry 2020 |