Open Access Article
Open Access ArticleEffects of lipid membrane composition on the distribution of biocidal guanidine oligomer with solid supported lipid membranes†
Yeonjeong Ha and
Jung-Hwan Kwon
and
Jung-Hwan Kwon *
*
Division of Environmental Science and Ecological Engineering, Korea University, 145 Anam-ro, Seongbuk-gu, Seoul 02841, Republic of Korea. E-mail: junghwankwon@korea.ac.kr; Tel: +82 2 3290 3041
First published on 10th June 2020
Abstract
Polyhexamethylene guanidine (PHMG) is a cationic antimicrobial oligomer that has been used prevalently over the past few decades. However, due to the lack of inhalation toxicity assessment of PHMG, it has caused severe health damage, including fatal lung fibrosis, after being used as one of the major active ingredients of humidifier disinfectants in Korea. Because the first step of the entry of PHMG into airway is its association with cell membranes, the distribution of PHMG between lipid membranes and water is very important to know the depositional flux in the respiratory systems and related toxic mechanisms. We developed a quantitative method to determine the distribution constant (Klipw) of PHMG between solid supported lipid membranes and water and evaluated the effects of lipid membrane compositions on the Klipw of PHMG. PHMG accumulated into anionic lipid membranes rapidly compared to into cationic or zwitterionic lipid membranes, suggesting fast adsorption of PHMG onto anionic lipid head groups. Klipw values with anionic/zwitterionic lipid mixtures were higher than Klipw values with anionic lipids only, potentially due to the later phase separation after preferential interaction between PHMG and anionic lipids in lipid mixtures. In addition, Klipw values increased with increasing single acyl chain lipid content in unsaturated lipids and decreasing cholesterol content. These results imply that changes in lipid spontaneous curvature and lipid bilayer packing density also affect the membrane distribution of PHMG.
1. Introduction
Biocides are inorganic or synthetic organic chemicals that inhibit or destroy microorganisms and are generally used for disinfection, sterilization, and preservation of products, such as food and cosmetics.1,2 Among a broad range of biocide chemicals, the polymeric guanidine group (e.g., polyhexamethylene biguanide (PHMB), polyhexamethylene guanidine (PHMG) and oligo(2-(2-ethoxy)ethoxyethyl) guanidine chloride (PGH)), which is classified as a cationic antimicrobial oligomer group, has been most widely used to inactivate bacteria, fungi, and parasites for many decades.3 Because all guanidine-based antimicrobial polymers are positively charged at physiological pH, guanidine groups strongly interact with bacterial cell membranes, which are primarily composed of negatively charged bilayers made up of phosphatidylglycerol (PG).4,5 Many previous studies have shown that lipid membrane damage and disruption caused by electrostatic interaction between guanidine-based biocides and negatively charged lipid head groups are the primary mechanisms of antimicrobial activity.6–10 In contrast, the guanidine-based biocides have negligible effects on zwitterion lipid membranes, such as phosphatidylcholine (PC) and phosphatidylethanolamine (PE),4,5,11 and zwitterion lipids are the primary components of fish and mammalian lipid membranes.12 Therefore, it has generally been believed that the guanidine family destroys microorganisms without damaging the host (e.g., fish and human) cells.However, recent studies have found that the guanidine group has toxic effects on aquatic organisms. For example, pulmonary toxicity of gills, including inflammation and fibrosis, was observed for zebrafish exposed to 0.229 mg L−1 of PHMG in saline solution,13 and sub-lethal concentrations (below 0.47 mg L−1) of PHMG significantly increased reactive oxygen species generation and caused thyroid endocrine disruption in zebrafish larvae.14 The EC50 value of PHMB for zebrafish liver cells (ZFL) was also reported as 11.02 μg mL−1.15 Furthermore, it was found that PHMB can penetrate through mammalian cell membranes and strongly interact with nucleic acids.16 Additionally, Lim et al.17 found that PHMG strongly interacts with the zwitterion lipid monolayer, 1,2-dipalmitoyl-sn-glycero-3-phosphatidylcholine (DPPC), which is a primary component of human lung surfactant. Shim et al., also reported that when aerosol particles with radioactive In-labeled PHMG were exposed to rats, they were transported and accumulated into the lungs.18 These studies raise concerns regarding the potential harmful effects of the guanidine group on human as well as aquatic organism health. Indeed, fatal misuse of PHMG and PGH as active ingredients of household humidifier disinfectants has resulted in the death of hundreds of people, including pregnant women and children, in Republic of Korea.19,20
Although investigating the interactions between cationic antimicrobial oligomers and lipid membranes that act as the primary barrier for living organisms is the foremost step toward assessing the toxicity and bioavailability of these chemicals, only a few studies have investigated the interaction mechanisms,4,5,10,17,21 and the effects of lipid composition on the membrane insertion of oligomers.22,23 To the best of our knowledge, the distribution constants of guanidine oligomers between lipid membranes and water (Klipw) and the effects of lipid membrane characteristics on Klipw have not been investigated in previous studies.
In this study, we used solid supported lipid membranes (SSLMs) to quantitatively determine the Klipw of PHMG. PHMG was selected as a representative of the guanidine family because it is one of the primary chemicals used as humidifier disinfectants that have caused unprecedented tragedy in Korea. SSLMs, solids that are uniformly coated with lipid membranes, are recently used in various applications such as imaging techniques for medical diagnostics and biochemical analysis.24,25 Since SSLMs are stable and maintain the fluidity of lipid membranes,26–28 SSLMs have successfully used to evaluate the lipid membrane distribution of a broad range of chemicals, including hydrophilic and hydrophobic, from a molecular to nanoparticle level.27,29–34 The main objective of this study is to assess the effects of lipid membrane composition on the distribution. Various lipid membranes (e.g., different hydrophilic head charge and lipid packing parameters) were employed for the quantitative method. We also suggest the probable distribution mechanism of PHMG into the lipid membranes.
2. Experimental
2.1 Materials
Powdered PHMG was supplied by SK Chemicals Co., Ltd. (Seongnam, Republic of Korea) in 2011, and this product is no longer commercially available in Korea after the humidifier disinfectant incident mentioned above. All lipid membrane components used in this study, except cholesterol, were purchased from Avanti Polar lipids (Alabaster, AL, USA) and cholesterol was obtained from Sigma-Aldrich (St. Louis, MO, USA). Table 1 summarizes the characteristics of each lipid used in this study, including the lipid head group charge, acyl chain number, main transition temperature, and physical state at 25 °C. Eosin Y solution (5 wt% in water) and non-porous silica microspheres (mean diameter: 3 μm) were also purchased from Sigma-Aldrich (St. Louis, MO, USA) to measure PHMG concentration in water and synthesize SSLMs, respectively.| Lipid componenta | Carbon chain:double bond | Main transition temperature (Tm, °C)b | Physical state at room temperature | Lipid head charge |
|---|---|---|---|---|
| a Full names of the lipid components used in this study are listed below: DOTAP: 1,2-dioleoyl-3-trimethylammonium-propane, 14:1 PC: 1,2-dimyristoleoyl-sn-glycero-3-phosphocholine, 16:1 PC: 1,2-dipalmitoleoyl-sn-glycero-3-phosphocholine, DOPC: 1,2-dioleoyl-sn-glycero-3-phosphocholine, POPC: 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine, DOPG: 1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol), POPG: 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol), DOPS: 1,2-dioleoyl-sn-glycero-3-phospho-L-serine, Lyso PG: 1-oleoyl-2-hydroxy-sn-glycero-3-phospho-(1′-rac-glycerol).b Avanti Polar Lipids, Inc. provides the main transition temperature for glycerophospholipids. Information found at https://avantilipids.com/tech-support/physical-properties/phase-transition-temps. | ||||
| DOTAP | (C 18:1, 18:1) | ∼−0 | Liquid crystalline | Positive |
| 14:1 PC | (C 14:1, 14:1) | <−30 | Liquid crystalline | Zwitterion |
| 16:1 PC | (C 16:1, 16:1) | −36 | Liquid crystalline | Zwitterion |
| DOPC (18:1 PC) | (C 18:1, 18:1) | −17 | Liquid crystalline | Zwitterion |
| POPC | (C 16:0, 18:1) | −9 | Liquid crystalline | Zwitterion |
| DOPG | (C 18:1, 18:1) | −18 | Liquid crystalline | Negative |
| POPG | (C 16:0, 18:1) | −2 | Liquid crystalline | Negative |
| DOPS | (C 18:1, 18:1) | −11 | Liquid crystalline | Negative |
| Lyso PG | (C 18:1) | — | — | Negative |
2.2 SSLMs
The detailed procedure for preparing SSLMs with lipid membranes has been described in previous studies.29–31 First, lipids dissolved in chloroform were transferred to glass vials and a thin film was formed on the glass vial walls via nitrogen purging. The lipid membrane vesicles were synthesized by the rapid extrusion process using 0.8 μm polycarbonate membrane filters. Then, the lipid vesicle dispersions were mixed with silica microspheres with 1–2 min of vigorous vortex mixing followed by gentle mixing in an incubator overnight at 25 °C, 150 rpm. After mixing, supernatants containing excess lipid vesicles that were not adsorbed onto microspheres were removed. The mass of the lipids (m) adsorbed onto the silica microspheres were calculated from the mass difference between the initial and supernatant lipids. A total organic carbon analyzer was used to measure the concentrations of lipids. A stable and uniform coating of silica microspheres with lipid membranes was confirmed from images obtained through confocal fluorescence microscopy with the aid of a fluorescent-labeled lipid membrane, as reported previously.302.3 Determination of the distribution constant of PHMG between water and lipid membranes (Klipw)
SSLMs and 1.5 mL PHMG aqueous solution were placed into polypropylene (PP) tubes. The tubes were incubated in an incubator at 150 rpm and 25 °C. After incubation, the tubes sat quiescently over 60 min to settle the SSLMs. Then, 1 mL of supernatant was transferred into a clean PP tube and excess PHMG that did not adsorb onto the SSLMs was measured.The initial (C0) and supernatant (Cf) concentrations of PHMG were determined by a spectrophotometric method with 0.05% Eosin Y solution. 1 mL of pH 3.6 glycine buffer solution and 200 μL of 0.05% Eosin Y solution were added sequentially to 1 mL PHMG aqueous solution followed by vortex mixing. Then, the mixtures were left for 5–10 min for color development. Finally, the PHMG concentrations in the mixtures were quantified by measuring the visible light absorbance at 549 nm using a DR/4000U UV/Vis spectrophotometer (Hach Co., Loveland, CO, USA). The absorbance at 549 nm was confirmed as being proportional to the concentration of PHMG in the mixtures (r2 > 0.998). In aqueous PHMG, the concentration range was 0–10 mg L−1 (Fig. S1, ESI†).
The mass of PHMG that accumulated in the lipid membranes was calculated using eqn (1), and the lipid–water distribution constant (Klipw) was calculated using eqn (2):
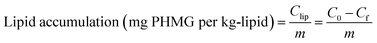 | (1) |
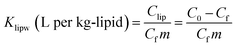 | (2) |
3. Results and discussion
3.1 Determination of apparent time to reach equilibrium and concentration dependence
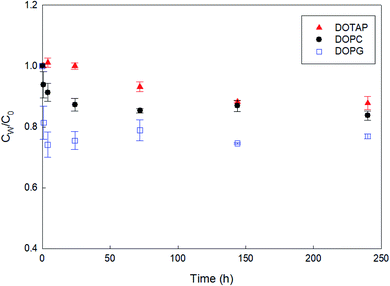
As a preliminary experiment, we used glass vials to contain PHMG aqueous solution and attempted to determine the apparent time taken to reach the distribution equilibrium of PHMG between water and SSLMs. However, we found that the PHMG concentration significantly decreased due to the electrostatic interaction between negatively charged glass vial walls and cationic PHMG oligomers (Fig. S2(a), ESI†). In addition, when glass vials were used, PHMG interacted with non-porous bare silica microspheres that have negative charges on their surface (48% removal after 3 h of incubation), and the rate of interactions between PHMG and bare silica beads was very similar to the PHMG interaction rates with DOPG lipids, which have negative lipid head charges (Fig. S2(b), ESI†). In contrast, there was a negligible interaction between PHMG and PP centrifuge tubes for 72 h (Fig. S2(a)†). Therefore, PP centrifuge tubes were used for all experiments in this study.Fig. 1 shows the rate of PHMG distribution with SSLMs with three different head group charges. DOTAP, DOPC, and DOPG unsaturated lipid membranes with the same acyl chain length (C18:1) but with positive, zwitterion, and negative charges in their head group, respectively, were utilized. As shown in Fig. 1, the PHMG distribution equilibrium with DOPG lipids was attained very quickly (within 1 h) due to the fast electrostatic interactions between the positive PHMG oligomer and anionic DOPG lipids. For DOPC lipids with a zwitterion head group and cationic DOTAP, the distribution equilibrium occurred within 24 and 144 h, respectively. Therefore, 72 h was selected as the apparent equilibrium time for zwitterion and negative lipid membranes, except Klipw values shown in Fig. 3(b). (In Fig. 3(b), 240 h was used as the equilibrium time to investigate the effects of the head group charge on Klipw values). The different apparent times taken to reach equilibrium implies the different lipid–water distribution mechanisms. Detailed discussion on the distribution mechanisms of PHMG with different lipid membranes is described in Section 3.2 and 3.3.
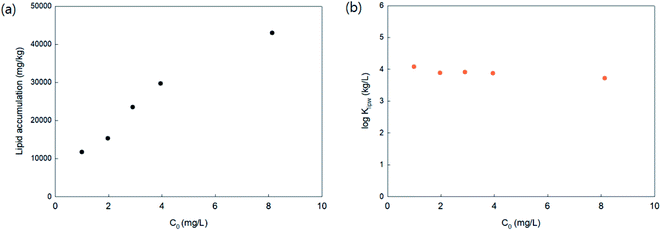
To investigate the PHMG concentration dependence on lipid accumulation, lipid membrane accumulations with 16:1 PC lipids were calculated using five different initial concentrations of PHMG (1.0, 2.0, 3.0, 4.0, and 8.1 mg L−1). As shown in Fig. 2(a), in the range of 2.0–4.0 mg L−1, the amount of PHMG distributed with 16:1 PC lipid membranes increased proportionally with an increase in initial PHMG aqueous concentration, resulting in concentration independent values of lipid–water distribution constants (Klipw) (Fig. 2(b)). Klipw values obtained using 1.0 and 8.1 mg L−1 deviated slightly compared with Klipw values in the range of 2.0–4.0 mg L−1 (Fig. 2(b)), however, the differences were below 5%. This confirms that the PHMG concentration range of 2.0–4.0 mg L−1 is lower than the concentration limit, above which the capacity of SSLMs for PHMG is saturated. Therefore, the PHMG initial concentration ranging from 2.0 to 6.0 mg L−1 was used to determine lipid–water partitioning constants (Klipw).
3.2 Effects of lipid head group charge
To assess the effects of membrane composition on the Klipw of PHMG, we first calculated time-dependent lipid accumulation using three different lipid membrane head charges (Fig. 3(a)). At all incubation times shown in Fig. 3 (from 1 to 240 h), lipid accumulation was the highest with DOPG, followed by DOPC, and the lowest with DOTAP. This indicates that the guanidine group of PHMG strongly interacts with the lipid membranes with negatively charged heads. A previous study31 found that negatively charged fullerene nanoparticles are strongly accumulated with oppositely charged lipid membranes (DOTAP) compared to other lipid membranes (e.g., DOPC and DOPG). Therefore, it can be concluded that if the target chemicals have charges, interactions between the adsorbate and oppositely charged lipid membranes are critical mechanisms for lipid membrane distribution. The Klipw obtained using eqn (2) between DOPG lipid membranes and water is the highest among three lipid membranes (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Klipw,DOPG = 4.25 ± 0.03), followed by DOPC (log
Klipw,DOPG = 4.25 ± 0.03), followed by DOPC (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Klipw,DOPC = 4.02 ± 0.06), and the lowest with DOTAP (log
Klipw,DOPC = 4.02 ± 0.06), and the lowest with DOTAP (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Klipw,DOTAP = 3.86 ± 0.10) (Fig. 3(b)).
Klipw,DOTAP = 3.86 ± 0.10) (Fig. 3(b)).
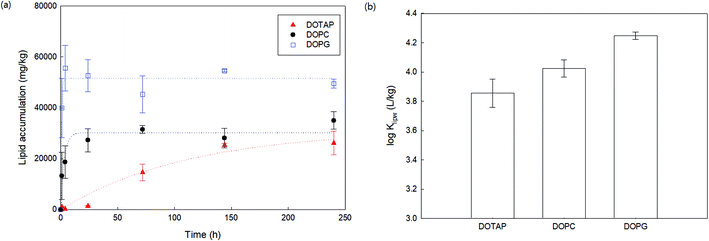 | ||
| Fig. 3 (a) Lipid accumulation rates of PHMG (C0 = 4.04 mg L−1) using three different lipid membranes with different head group charges. Dotted lines indicate that the Langmuir adsorption model fits using eqn (3). (b) The lipid–water distribution constant (Klipw) of PHMG between water and three lipid membranes calculated after 240 h of incubation. Error bars represent standard deviations of triplicates. | ||
To obtain the rate parameters and investigate the effects of the lipid head group charge on accumulation rates, we employed the Langmuir adsorption model (eqn (3)) and fit the model to the experimental data (Fig. 3(a)). This Langmuir adsorption model was first developed by Wilhelm et al.35 to describe the interaction rates of iron nanoparticles with cells, and has successfully been applied to previous studies investigating the lipid membrane interaction kinetics of charged nanoparticles, such as gold36 and fullerene nanoparticles.31 The Langmuir adsorption model is:
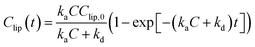 | (3) |
| Lipid membrane | ka (L mg−1 h−1) | kd (h−1) | Kb (L mg−1) | τc (h) | r2 |
|---|---|---|---|---|---|
| a Langmuir adsorption model (eqn (3)) was used to obtain kinetic parameters.b K is the affinity constant and defined as ka/kd.c τ (= 1/(kaC + kd)) is characteristic time, and small τ indicates a faster interaction between lipid membranes and PHMG. | |||||
| DOPG | 9.51 × 10−3 | 1.47 | 6.48 × 10−3 | 0.66 | 0.76 |
| DOPC | 1.19 × 10−3 | 0.32 | 3.76 × 10−3 | 3.11 | 0.71 |
| DOTAP | 3.41 × 10−5 | 8.75 × 10−3 | 3.90 × 10−3 | 112.36 | 0.92 |
As shown in Table 2, the affinity constant (K) with the DOPG lipid membrane is significantly higher than that with DOPC and DOTAP lipids due to the PHMG adsorption via the strong electrostatic interaction between the positive oligomer and negative lipid membranes. The characteristic times (τ) of DOPG and DOPC are 0.66 and 3.11 h, respectively, which are similar levels to the overall interaction rates between gold nanoparticles and egg phosphatidylcholine lipid bilayers (0.19 < τ < 3.76), and between fullerene nanoparticles and DOTAP, DOPC, and DOPG lipid membranes (0.12 < τ < 15.03). As both gold and fullerene nanoparticles have charges on their surfaces, the first step of the interactions between these nanoparticles with lipid membranes is adsorption onto the lipid membrane surface via electrostatic interactions. Therefore, for DOPG and DOPC lipid membranes, the rapid adsorption of PHMG to the lipid membrane head group could be the primary mechanism of lipid membrane distribution. However, the characteristic time (τ) of DOTAP lipids (τ = 112.36 h) is much longer compared to that of other lipid membranes, indicating that surface adsorption might not be the primary mechanism for cationic lipid membranes.
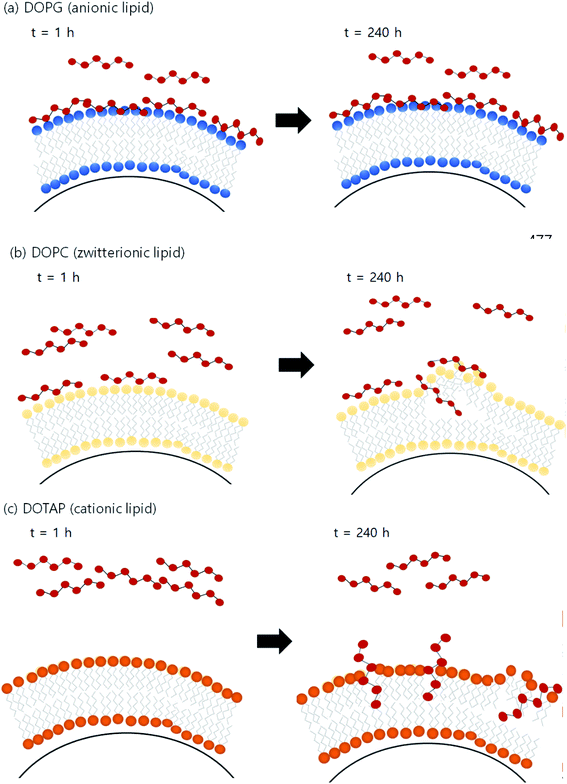
Fig. 4 illustrates the proposed mechanism of PHMG distribution into three different lipid membranes. For DOPG lipid membranes, PHMG rapidly bound to the lipid head groups and the amount of PHMG distributed in the lipid membrane stayed the same (Fig. 4(a)). Similarly, PHMG adsorbs onto the head group of the zwitterionic lipid membrane and it is also possible that PHMG slowly enters the lipid membrane, causing membrane disruption (Fig. 4(b)). For the DOTAP lipid membranes, initially PHMG rarely interacted with them due to electrostatic repulsion; however, lipid accumulation significantly increased after 72 h of incubation. One possible mechanism of PHMG distribution into the DOTAP lipid membrane is that positively charged guanidine group of PHMG may pretend to the cationic head of the DOTAP lipid, which allows PHMG to act as one molecule of DOTAP lipid. This possibly aligns PHMG with DOTAP lipids in the lipid bilayer (Fig. 4(c)). Once PHMG is located in the DOTAP membrane, membrane disruption and pore formation are also possible.
We prepared SSLMs with two different anionic lipids (POPG and DOPS) as well as zwitterionic/anionic lipid mixtures (POPC/POPG and POPC/DOPS) to assess the effects of lipid mixtures on the membrane distribution of PHMG (Fig. 5(a)). Klipw values of two anionic lipids (POPG and DOPS) were higher than the Klipw of zwitterionic lipids (POPC) due to the strong electrostatic interactions. Interestingly, Klipw values of zwitterionic/anionic lipid mixtures (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio) were even higher than Klipw values of anionic lipids only. This could be attributed to lipid lateral separation in a membrane bilayer composed with two different head groups. Recent studies21,37–39 have proposed that the lipid rearrangement and lateral separation of zwitterionic lipids from anionic lipids can occur when cationic antimicrobial peptides (AMPs) preferentially interact with anionic lipids in zwitterionic/anionic lipid mixtures (Fig. 5(b)). Lateral separation creates higher negative charge density on the unit surface area of lipid membranes, which can make more PHMG interact with lipid membranes with zwitterionic/anionic lipid mixtures than anionic lipids only. This phase separation also promotes membrane disruption and can be a significant cause of higher Klipw values of zwitterionic/anionic lipid mixtures than anionic lipids. Indeed, species with membranes composed of zwitterionic and anionic lipids may be more susceptible to positive antimicrobial agents than other species composed of primarily anionic lipids due to this lipid segregation mechanism.21
1 molar ratio) were even higher than Klipw values of anionic lipids only. This could be attributed to lipid lateral separation in a membrane bilayer composed with two different head groups. Recent studies21,37–39 have proposed that the lipid rearrangement and lateral separation of zwitterionic lipids from anionic lipids can occur when cationic antimicrobial peptides (AMPs) preferentially interact with anionic lipids in zwitterionic/anionic lipid mixtures (Fig. 5(b)). Lateral separation creates higher negative charge density on the unit surface area of lipid membranes, which can make more PHMG interact with lipid membranes with zwitterionic/anionic lipid mixtures than anionic lipids only. This phase separation also promotes membrane disruption and can be a significant cause of higher Klipw values of zwitterionic/anionic lipid mixtures than anionic lipids. Indeed, species with membranes composed of zwitterionic and anionic lipids may be more susceptible to positive antimicrobial agents than other species composed of primarily anionic lipids due to this lipid segregation mechanism.21
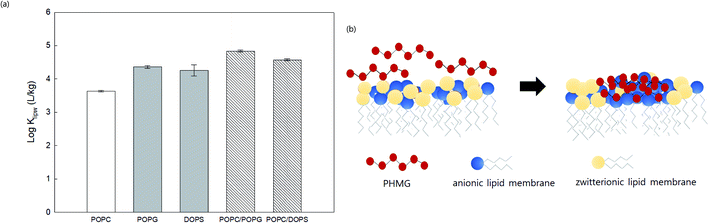 | ||
Fig. 5 (a) Lipid–water distribution constant (Klipw) of PHMG (C0 = 5.0 mg L−1) between water and lipid membranes with different compositions after 72 h of incubation. POPC has a zwitterion head group, and POPG and DOPS are anionic lipid membranes. POPC/POPG and POPC/DOPS are zwitterionic/anionic lipid mixtures prepared at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio. Error bars represent the standard deviations of three replicates. (b) Illustration showing the lipid lateral separation caused by binding PHMG to the lipid membrane composed with zwitterionic/anionic lipid mixtures. Strong interactions between PHMG and anionic lipid membranes promote the clustering of anionic lipid membranes around PHMG, which causes lipid rearrangement/segmentation. This figure was inspired by Teixeira et al.21 1 molar ratio. Error bars represent the standard deviations of three replicates. (b) Illustration showing the lipid lateral separation caused by binding PHMG to the lipid membrane composed with zwitterionic/anionic lipid mixtures. Strong interactions between PHMG and anionic lipid membranes promote the clustering of anionic lipid membranes around PHMG, which causes lipid rearrangement/segmentation. This figure was inspired by Teixeira et al.21 | ||
3.3 Effects of lipid packing parameters
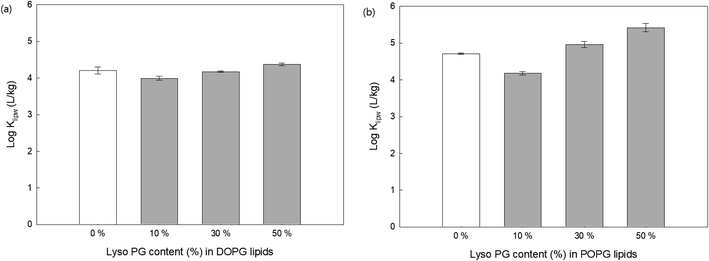
In addition to the electrical potential of membrane head groups, lipid membrane packing parameters are also considered important factors. Lipid packing parameters directly affect the hydrophobic interactions between the hydrocarbon acyl chain and antimicrobial oligomers that control the membrane insertion of them. The insertion of antimicrobial agents into the hydrophobic region of lipid membranes induces lipid packing frustration and membrane disruption,21,40 which are the critical mechanisms for antibacterial activity of the guanidine oligomers. Here, lipid lacking was modulated by (1) adding lipids with a single acyl chain to anionic lipid membranes (DOPG and POPG), (2) adding cholesterols to 16:1 PC (zwitterionic lipids), and (3) using different acyl chain lengths (14:1, 16:1, and 18:1 PC).To change lipid membrane packing, we first altered the spontaneous curvature of the bilayer lipid membranes by adding a lyso PG, which has a single acyl chain to the DOPG (Fig. 6(a)) and POPG (Fig. 6(b)) with different content. Membrane curvature depends on the size of the head group and acyl chain composition.41 For example, lipids with small polar head groups impose a negative radius of curvature, where the head group spacing is smaller. In contrast, lipids with a single acyl chain have a positive radius of curvature where the distance between the head groups is larger.40,41 In addition, adding single acyl chain lipids thins the bilayer and induces disorder due to the curvature changes.42 Recent studies have shown that the positive radius of the curvature created by lyso-lipids promotes more AMP insertion into the lipid membranes followed by pore formation inside the bilayer, whereas AMPs are only adsorbed onto lipid bilayer surfaces when the lipid membranes are composed of unsaturated PC lipids.42–44 Fig. 6 shows that for both DOPG and POPG lipid membranes, the addition of 10% lyso PG content decreased the Klipw values compared with PG lipids without lyso PG. One possible explanation for this is that adding lyso PG lipids decreases the negative charge density on the lipid membrane surfaces, leading to less PHMG being adsorbed onto the lipid surfaces. On the other hand, Klipw values increased upon increasing the lyso PG content by up to 50%. Since adding more lyso PG induces head group space enlargement and lipid membrane thinning, a greater amount of PHMG enters the lipid membranes and forms pores followed by membrane disruption. Therefore, lipid packing change caused by spontaneous curvature alteration is a critical factor that impacts PHMG distribution.
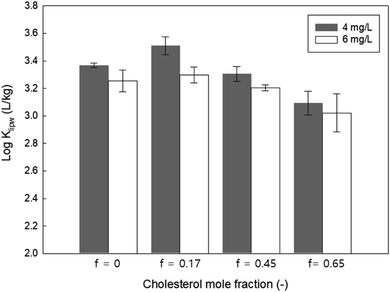
We also modulated the membrane packing parameters by containing different mole fractions of cholesterol in the zwitterionic lipid membrane (14:1 PC). Cholesterol creates more densely packed lipid bilayers and decreases the fluidity of the lipid membranes.45 As shown in Fig. 7, the Klipw of PHMG decreased with increasing cholesterol content for both the initial PHMG concentration of 4 and 6 mg L−1. This suggests that decreasing the membrane fluidity with increasing cholesterol content may prevent the insertion of PHMG into the hydrophobic region of lipid membranes. Thus, as Paliienko et al.46 suggested, living organisms with less or lack of cholesterol content in the lipid membranes are more susceptible than organisms with cholesterol containing membranes.
Finally, the effects of acyl chain lengths on the PHMG distribution were investigated. We measured the lipid accumulation rates of PHMG using three different unsaturated lipid membranes (Fig. S3, ESI†). However, we were unable to identify significant differences among the three lipids. Previous studies have reported that Klipw values of hydrophobic organic pollutants and fullerene nanoparticles are influenced by acyl chain length due to membrane fluidity changes or changes in hydrophobic interactions between lipid membranes and chemicals.30,47,48 However, the impact of acyl chain length on the lipid water distribution of PHMG was negligible in this study.
3.4 Environmental and toxicological implications
This study elucidates the quantitative measurements of the distribution constants (Klipw) of PHMG between the lipid membranes and water using SSLMs. We focused on the lipid accumulation of PHMG, and proposed distribution mechanisms for various membrane compositions. PHMG strongly and rapidly interacted with the negative and zwitterion lipid head groups, indicating that PHMG likely possesses cytotoxicity not only for microorganisms but also higher level organisms, such as fish and mammals, including humans. Klipw with zwitterionic/anionic lipid mixtures was higher than that with anionic lipid membranes, indicating that tissues with higher zwitterionic/anionic lipid content in their lipid membranes are more susceptible to its cytotoxicity. Changes in lipid packing parameters (i.e., lipid curvature and cholesterol content) also affect the distribution of PHMG in the lipid membranes. Considering cells of living organisms have a variety of cell membrane compositions, the results of this study are useful for identifying susceptible species among microbes and pathogens, and for assessing the potential cytotoxicity.Although PHMG is one of the lethal ingredients of humidifier disinfectants, which are responsible for many deaths from pulmonary disease,49–51 PHMG distribution into lung surfactant has not previously been quantitatively investigated. Klipw values of ca. 104 L kg−1 imply that the removal of PHMG from the respiratory tracts would be difficult via ciliary motion because lung surfactant is a monolayer consisting of zwitterionic and anionic lipid mixtures.52 However, considering the primary lipid component of lung surfactant is DPPC (C16:0/C16:0), which is in the gel-phase at room temperature (the main transition temperature, Tm = 41 °C), SSLMs are not suitable for evaluating the exact distribution through lung surfactant due to their limitations.53,54 It is worthy of investigating the distribution of PHMG between cell membranes and water considering the major components of lung surfactants and mixed lipid compositions that mimic human respiratory systems.
4. Conclusions
In this study, a quantitative method was developed to investigate the lipid–water distribution constant (Klipw) of PHMG using solid supported lipid membranes (SSLMs). The rate of PHMG distribution with lipids depends on lipid head group charges. This indicates that there are significant effects of head charges on the lipid membrane distribution mechanisms of PHMG. Moreover, we found that Klipw values were significantly influenced by lipid membrane compositions such as head group charges and lipid packing parameters. Thus, the results of this study provide useful information for evaluating cytotoxicity of PHMG towards living organisms including fish and human.Conflicts of interest
The authors declare no competing interests.Acknowledgements
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2019R1I1A1A01059970) and by the Ministry of Environment (MOE), Republic of Korea as “Technology Program for establishing biocide safety management” (2018002490001). The authors thank Mr Sang-Hyun Choi for assisting with the spectrophotometric analysis of PHMG. The authors also thank Editage (www.editage.co.kr) for providing English language editing guidance.References
- K. Rasmussen, P. Chemin and P. Haastrup, J. Hazard. Mater., 1999, 67, 237–251 CrossRef CAS PubMed.
- J. S. Chapman, Int. Biodeterior. Biodegrad., 2003, 51, 133–138 CrossRef CAS.
- K. Chindera, M. Mahato, A. K. Sharma, H. Horsley, K. Kloc-Muniak, N. F. Kamaruzzaman, S. Kumar, A. McFarlane, J. Stach, T. Bentin and L. Good, Sci. Rep., 2016, 6, 23121 CrossRef CAS PubMed.
- T. Ikeda, S. Tazuke and M. Watanabe, Biochim. Biophys. Acta, 1983, 735, 380–386 CrossRef CAS.
- T. Ikeda, A. Ledwith, C. H. Bamford and R. A. Hann, Biochim. Biophys. Acta, 1984, 769, 57–66 CrossRef CAS.
- P. Broxton, P. M. Woodcock and P. Gilbert, J. Appl. Bacteriol., 1983, 54, 345–353 CrossRef CAS PubMed.
- P. Broxton, P. M. Woodcock, F. Heatley and P. Gilbert, J. Appl. Bacteriol., 1984, 57, 115–124 CrossRef CAS PubMed.
- Z. X. Zhou, A. Zheng and J. J. Zhong, Acta Biochim. Biophys. Sin., 2011, 43, 729–737 CrossRef CAS PubMed.
- A. M. Carmona-Ribeiro and L. D. D. Carrasco, Int. J. Mol. Sci., 2013, 14, 9906–9946 CrossRef PubMed.
- P. Gilbert and L. E. Moore, J. Appl. Microbiol., 2005, 99, 703–715 CrossRef CAS PubMed.
- X. L. Luo, Z. R. Jiang, N. Y. Zhang, Z. X. Yang and Z. X. Zhou, Polymers, 2017, 9, 517 CrossRef PubMed.
- S. A. Zabelinskii, N. B. Brovtsyna, M. A. Chebotareva, O. B. Gorbunova and A. I. Krivchenko, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 1995, 111, 127–140 CrossRef CAS.
- H. Oh, C. Y. Kim, B. Ryu, U. Kim, J. Kim, J. M. Lee, B. H. Lee, J. Moon, C. R. Jung and J. H. Park, Zebrafish, 2018, 15, 460–472 CrossRef CAS PubMed.
- H. Kim and K. Ji, Ecotoxicol. Environ. Saf., 2019, 184, 109663 CrossRef CAS PubMed.
- V. Christen, S. Faltermann, N. R. Brun, P. Y. Kunz and K. Fent, Sci. Total Environ., 2017, 586, 1204–1218 CrossRef CAS PubMed.
- M. J. Allen, A. P. Morby and G. F. White, Biochem. Biophys. Res. Commun., 2004, 318, 397–404 CrossRef CAS PubMed.
- C. Lim, S. Park, J. Park, J. Ko, D. W. Lee and D. S. Hwang, J. Hazard. Mater., 2018, 353, 271–279 CrossRef CAS PubMed.
- H. E. Shim, J. Y. Lee, C. H. Lee, S. Mushtaq, H. Y. Song, L. Song, S. J. Choi, K. Lee and J. Jeon, Chemosphere, 2018, 207, 649–654 CrossRef CAS PubMed.
- Y. J. Park, M. H. Jeong, I. J. Bang, H. R. Kim and K. H. Chung, Inhalation Toxicol., 2019, 31, 161–166 CrossRef CAS PubMed.
- J. H. Lee, Y. H. Kim and J. H. Kwon, Environ. Sci. Technol., 2012, 46, 2498–2500 CrossRef CAS PubMed.
- V. Teixeira, M. J. Feio and M. Bastos, Prog. Lipid Res., 2012, 51, 149–177 CrossRef CAS PubMed.
- V. Vasquez-Montes, J. Gerhart, D. Thevenin and A. S. Ladokhin, J. Mol. Biol., 2019, 431, 5004–5018 CrossRef CAS PubMed.
- A. G. Dos Santos, J. T. Marques, A. C. Carreira, I. R. Castro, A. S. Viana, M. P. Mingeot-Leclercq, R. F. M. de Almeida and L. C. Silva, Phys. Chem. Chem. Phys., 2017, 19, 30078–30088 RSC.
- R. Mei, Y. Wang, W. Liu and L. Chen, ACS Appl. Mater. Interfaces, 2018, 10, 23605–23616 CrossRef CAS PubMed.
- X. Su, Y. Wang, W. Wang, K. Sun and L. Chen, ACS Appl. Mater. Interfaces, 2016, 8, 10201–10211 CrossRef CAS PubMed.
- T. M. Bayerl and M. Bloom, Biophys. J., 1990, 58, 357–362 CrossRef CAS PubMed.
- M. M. Baksh, M. Jaros and J. T. Groves, Nature, 2004, 427, 139–141 CrossRef CAS PubMed.
- E. T. Castellana and P. S. Cremer, Surf. Sci. Rep., 2006, 61, 429–444 CrossRef CAS PubMed.
- Y. Ha, L. E. Katz and H. M. Liljestrand, Environ. Sci. Technol., 2015, 49, 14546–14553 CrossRef CAS PubMed.
- Y. Ha, H. M. Liljestrand and L. E. Katz, Water Sci. Technol., 2013, 68, 290–295 CrossRef CAS PubMed.
- Y. Ha, X. Z. Wang, H. M. Liljestrand, J. A. Maynard and L. E. Katz, Environ. Sci. Technol., 2016, 50, 6717–6727 CrossRef CAS PubMed.
- A. Loidl-Stahlhofen, A. Eckert, T. Hartmann and M. Schottner, J. Pharm. Sci., 2001, 90, 599–606 CrossRef CAS PubMed.
- J. J. Garcia-Celma, L. Hatahet, W. Kunz and K. Fendler, Langmuir, 2007, 23, 10074–10080 CrossRef CAS PubMed.
- F. Zhao, J. P. Holmberg, Z. Abbas, R. Frost, T. Sirkka, B. Kasemo, M. Hassellöv and S. Svedhem, RSC Adv., 2016, 6, 91102–91110 RSC.
- C. Wilhelm, F. Gazeau, J. Roger, J. Pons and J.-C. Bacri, Langmuir, 2002, 18, 8148–8155 CrossRef CAS.
- W. C. Hou, B. Y. Moghadam, C. Corredor, P. Westerhoff and J. D. Posner, Environ. Sci. Technol., 2012, 46, 1869–1876 CrossRef CAS PubMed.
- R. M. Epand, S. Rotem, A. Mor, B. Berno and R. F. Epand, J. Am. Chem. Soc., 2008, 130, 14346–14352 CrossRef CAS PubMed.
- R. M. Epand and R. F. Epand, Biochim. Biophys. Acta, Biomembr., 2009, 1788, 289–294 CrossRef CAS PubMed.
- A. Arouri, M. Dathe and A. Blume, Biochim. Biophys. Acta, Biomembr., 2009, 1788, 650–659 CrossRef CAS PubMed.
- B. D. van Rooijen, M. M. Claessens and V. Subramaniam, Biochim. Biophys. Acta, 2009, 1788, 1271–1278 CrossRef CAS PubMed.
- H. T. McMahon and E. Boucrot, J. Cell Sci., 2015, 128, 1065–1070 CrossRef CAS PubMed.
- S. Y. Woo and H. Lee, Phys. Chem. Chem. Phys., 2017, 19, 21340–21349 RSC.
- M. T. Lee, F. Y. Chen and H. W. Huang, Biochemistry, 2004, 43, 3590–3599 CrossRef CAS PubMed.
- M. T. Lee, W. C. Hung, F. Y. Chen and H. W. Huang, Biophys. J., 2005, 89, 4006–4016 CrossRef CAS PubMed.
- I. Ermilova and A. P. Lyubartsev, RSC Adv., 2020, 10, 3902–3915 RSC.
- K. O. Paliienko, T. O. Veklich, O. Y. Shatursky, O. A. Shkrabak, A. O. Pastukhov, M. O. Galkin, N. V. Krisanova, A. J. Chunikhin, A. V. Rebriev, A. V. Lysytsya, T. A. Borisova and S. O. Kosterin, Toxicol. In Vitro, 2019, 60, 389–399 CrossRef CAS PubMed.
- H. Yamamoto and H. M. Liljestrand, Environ. Sci. Technol., 2004, 38, 1139–1147 CrossRef CAS PubMed.
- M. C. Antunes-Madeira and V. M. Madeira, Biochim. Biophys. Acta, 1987, 901, 61–66 CrossRef CAS.
- H. N. Jung, T. Zerin, B. Podder, H. Y. Song and Y. S. Kim, Toxicol. In Vitro, 2014, 28, 684–692 CrossRef CAS PubMed.
- H. R. Kim, G. W. Hwang, A. Naganuma and K. H. Chung, J. Toxicol. Sci., 2016, 41, 711–717 CrossRef CAS PubMed.
- S. H. Choi, S. K. Park, H. J. Kang, K. Lee and J. H. Kwon, Bull. Korean Chem. Soc., 2015, 36, 1819–1823 CrossRef CAS.
- R. Veldhuizen, K. Nag, S. Orgeig and F. Possmayer, Biochim. Biophys. Acta, 1998, 1408, 90–108 CrossRef CAS.
- Z. V. Feng, T. A. Spurlin and A. A. Gewirth, Biophys. J., 2005, 88, 2154–2164 CrossRef CAS PubMed.
- F. Tokumasu, A. J. Jin, G. W. Feigenson and J. A. Dvorak, Ultramicroscopy, 2003, 97, 217–227 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra03108a |
| This journal is © The Royal Society of Chemistry 2020 |