Open Access Article
Open Access ArticleInvestigating partitioning of free versus macrocycle bound guest into a model POPC lipid bilayer†
Harshita Kumari *a,
Andrew Eisenhartb,
Jinnipha Pajoubponga,
Frank Heinrich*cd and
Thomas L. Beck*b
*a,
Andrew Eisenhartb,
Jinnipha Pajoubponga,
Frank Heinrich*cd and
Thomas L. Beck*b
aJames L. Winkle College of Pharmacy, University of Cincinnati, 231 Albert Sabin Way, MSB #3109 C, Cincinnati, OH 45267-0514, USA. E-mail: kumariha@ucmail.uc.edu
bDepartment of Chemistry, University of Cincinnati, Cincinnati, Ohio 45221, USA. E-mail: becktl@ucmail.cu.edu
cDepartment of Physics, Carnegie Mellon University, Pittsburgh, Pennsylvania 15213, USA. E-mail: fheinrich@cmu.edu
dCenter for Neutron Research, National Institute of Standards and Technology, Gaithersburg, Maryland 20988, USA
First published on 17th April 2020
Abstract
We report on the permeation of free and macrocycle-bound avobenzone across a POPC lipid bilayer through combined neutron reflectometry experiments and molecular dynamics simulations. Results indicate that the p-phosphonated calix[8]arene macrocycle limits the avobenzone penetration into the upper leaflet of the membrane. Hence, it could serve as a useful vehicle for safer formulations.
Introduction
Supramolecular chemistry is transitioning from synthesizing unusable host–guest complexes to designing and developing molecular machines.1–3 Stoddart used molecules on the nanometric scale as switches in electronic devices and linear motor-molecules in nanoelectromechanical systems.4,5 Investigating molecular movements and controlling them will define the next decade of supramolecular chemistry. This new approach has motivated us to investigate the mechanism of penetration of a novel supramolecular host–guest complex across lipid bilayers/biomembranes, which is potentially useful to control UV radiation exposure, a potent cause of skin cancer.The prevalence of malignant melanoma (a type of skin cancer), despite the extensive use of sunscreens, is a global concern.6,7 UVA and UVB penetration across the epidermis leads to generation of reactive oxidative species, DNA/protein/lipid damage and activation of varying signal transduction pathways that compromises the skin's defence systems.8,9 The use of sunscreens is continuously on the rise; however, the toxic effect of some organic sunscreens10,11 on coral reefs12–14 and challenges with formulation of inorganic sunscreen agents, such as, zinc oxide and titanium dioxide,15–17 has limited the number of available molecules that provide effective UV protection.
The photo-stability of sunscreen actives, toxicological impacts of photo-degradation products and controlling skin penetration are three of the major challenges in this area. Sunscreens, such as, oxybenzone/benzophenone-3 are emerging as environmental and human contaminants as they have been detected in human urine (97% of the population) or impacting coral reefs.18 Thus, skin penetration across deeper layers (dermis) is a major concern with organic photoactives. Although macromolecular quantitative estimation of skin penetration of sunscreen is possible via radiolabelled 14C tape stripping assays on human/pig skin,19 a nanometric molecular-level understanding is still lacking. In this work, we study the interaction of an organic active (avobenzone, Fig. 1) with model lipid bilayer membranes, and we examine how complexing it with a macrocycle could potentially impact/restrain its pathway across the bilayer membrane.
For this study, we chose p-phosphonated calix[8]arene (calix[8]-PO3H2)20 as the host molecule and avobenzone as the sunscreen agent (Fig. 1). Calixarenes are cyclic oligomers of several phenolic units connected with methylene bridges.21–24 Although sparingly soluble in water, the base molecule has been modified with functional groups, such as, phosphonate, sulphonate or amine to induce aqueous solubility. In addition, the cavity size can be modulated to accommodate a suitably sized guest. Our recent rat and human cell toxicology study on calix[8]-PO3H2 revealed its non-toxic behaviour that renders it as a useful biomedical macrocycle for potential application as a nanocarrier.25 Complexation of calix[8]-PO3H2 with avobenzone26 was chosen for three reasons: (a) water solubility of the macrocycle; (b) large internal cavity and upper rim H-bonding functionality of the host and (c) non-toxic behaviour towards rat and human cell lines-.25
Avobenzone (1-(4-methoxyphenyl)-3-(4-tertbutylphenyl)-propane-1,3-dione) also known as Parsol 1789 is a broad-spectrum sunscreen agent. Its ability to absorb both UVA and UVB radiations makes it one of the most useful sunscreen agents that prevents photodamage of skin. However, once exposed to sun, avobenzone offers only 30 minutes of photo-protection. Hence, it is formulated with photostabilizers, such as octocrylene, which is a known endocrine disrupter and releases free radicals.27–29 Experimental and theoretical studies suggest that the chelated enol form is the ground state. Upon UV irradiation in polar nonprotic solvents, the enol form converts to the keto tautomer.30 Herein, we are exploring a rather inert macrocycle that could provide a confined environment for photostability and/or controlled penetration across bilayer. In this study, we have limited our scope to the interaction of free versus macrocycle-complexed avobenzone with an artificial bilayer.
Neutron reflectometry
We tested the interaction of avobenzone and calix[8]-PO3H2 with single-membrane POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) tethered lipid bilayer membranes (tBLMs).31 Solvent-immersed tBLMs allow for a characterization of the membrane before and after addition of avobenzone and calix[8]-PO3H2 from solution, thus mimicking the encounter of those substances by the membrane in the body. This differentiates tBLMs from conventionally used stacked lipid bilayer model systems32,33 in air. The chemical structure of the glycerophospholipid POPC consists of a zwitterionic phosphatidylcholine headgroup linked to a glycerol, which is ester-bound to two fatty acid groups – palmitic acid (C16:0) and oleic acid (C18:1). This fatty acid composition mimics that of phospholipids mainly found in the eukaryotic cell membrane,34 including keratinocytes.35 In addition to phospholipids, native eukaryotic membranes also contain glycolipids, sterols and various proteins, which are not included in the current simplified model system.Using a POPC model membrane constitutes the first step in establishing more complex SC lipid membrane model systems. Specifically, in the cornification process, keratinocytes, which are living cells containing phospholipid bilayer membranes, a nucleus and cytoplasm, are transformed to corneocytes via apoptosis. Consequently, the digestion of the nucleus and the loss of the cytoplasm and all intracellular organelles occur, whereas the cell membrane is replaced by the cornified cell envelope (CE). The CE consists of crosslinked structural proteins (e.g. loricrin, involucrin), which are also covalently linked to ceramides on the exterior surface providing a hydrophobic interface between the CE and the intercellular lipid lamellae.36,37
The simplified biomimetic lipid bilayer model has allowed us to investigate its fundamental interaction with molecules of interest between the molecular and the micrometre scale using neutron reflectometry (NR). Because neutrons are uncharged and highly penetrating particles that have wavelengths comparable to molecular sizes and inter-molecular distances, neutrons are an ideal probe for characterizing the structure and dynamics of complex materials such as lipid bilayers. Moreover, neutrons interact with hydrogen and deuterium differently allowing one to use of the scattering contrast between H2O and D2O to investigate hydrogen-rich biological membranes.38 There are three different neutron scattering techniques commonly applied for different membrane models: (a) neutron diffraction for stacked bilayers (b) small angle neutron scattering for vesicles and bicelles, and (c) NR for single bilayers such as the POPC used in this work.38,39
NR is a flexible tool in structural biology due to its ability to discern the biomolecular architectures of lipid membranes and membrane-associated proteins without destroying the sample. It allows to mimic biological processes and to measure their structural responses. However, it does require specific deuteration to resolve individual components of interest. For this study, we conducted NR experiments with hydrogenated and deuterated POPC bilayers, and simultaneously analysed both data sets to obtain one structural model.
NR measurements were performed at the CGD-Magik reflectometer40 at the NIST Center for Neutron Research (NCNR). Reflectivity curves were recorded for momentum transfer values 0.01 ≤ qz ≤ 0.25 Å−1. For each measurement, adequate counting statistics were obtained after 5–7 h. The NCNR fluids cell41 allows for in situ solvent exchange; therefore, subsequent measurements were performed on the same sample area. The entire flow cell was maintained at room temperature (RT). Solvent exchange was accomplished by rinsing ∼10 ml of water through the cell (volume ∼ 1.3 ml) using a syringe.
We conducted a set of NR experiments with a hydrogenated POPC bilayer and another identical set of experiments with a deuterated POPC-d31 bilayer. After measurement of the as-prepared bilayer in H2O and D2O, avobenzone was added to the sample cell at a concentration of 100 μM dissolved in H2O and D2O. NR measurements were conducted while the bilayer was in contact with either solution. The sample was measured again after rinsing with H2O and D2O, respectively. Data for both bilayers were co-refined sharing conserved model parameters across data sets, in particular those associated with the volume profile of avobenzone. The co-refinement of data from two lipid bilayers with differently labelled hydrocarbon chains significantly boosted the resolution of the avobenzone profile, in this region. We performed an identical set of measurements with a 100 μM complex of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 avobenzone
1 avobenzone![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) calix[8]-PO3H2 instead of pure avobenzone.
calix[8]-PO3H2 instead of pure avobenzone.
1D-component volume occupancy (CVO) profiles along the lipid bilayer normal were obtained as previously described.42–44 Bilayer fit parameters were the hydrocarbon thickness for each bilayer subsection, the bilayer completeness, and the thickness of the sub-membrane space. One roughness parameter was applied to all distributions. The thin layer of native silicon oxide was modelled by a single distribution with individual roughness and thickness parameters. Hermite splines defined by control points that were on average 15 Å apart were used to model the CVO profiles of avobenzone and calix[8]-PO3H2. The number of control points was iteratively refined during model optimization for each CVO profile. Fit parameters associated with each control point were a volume occupancy, a deviation from equidistant separation, and (for the CVO profile of the complex) a nSLD (neutron scattering length density) value between those of avobenzone and calix[8]-PO3H2. Such a variable nSLD per control point allowed us to separate individual CVO profiles for avobenzone (average nSLD ρ = 1.37 × 10−6 Å−2) and calix[8]-PO3H2 (ρ = 2.73 × 10−6 Å−2). The exchange of labile protons in isotopically different buffers affects the nSLD of molecular components and was taken into account during data analysis. Optimization of model parameters was performed using the ga_refl and Refl1D software packages developed at the NCNR.45 A Monte Carlo Markov chain-based global optimizer45 was used to determine fit parameter confidence limits.
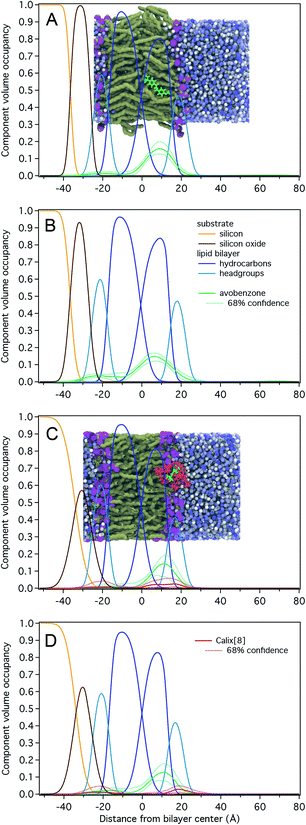
Results for NR measurements with avobenzone without calix[8] are shown in Fig. 2A and B and Table S1.† The substrate-supported POPC and POPC-d31 lipid bilayers were complete (surface coverage 1.00 ± 0.01) and had total hydrocarbon thicknesses of 30 ± 1 Å, which is ∼3 Å larger than those determined for stacked lipid bilayer membranes and vesicles.46 Avobenzone is observed interacting with the lipid bilayer at the hydrocarbon/headgroup interface of the substrate-distal lipid leaflet or solvent-side region without penetrating the bilayer to the substrate-proximal leaflet. The observed surface volume density of avobenzone is 3.0 ± 0.5 Å3/Å2 during bilayer incubation and 3.8 ± 0.4 Å3/Å2 after solvent rinse, showing a stable bilayer-association with 1 avobenzone molecule per ∼2.7 lipid molecules. Adding avobenzone does not affect the structural integrity of the lipid bilayer as the bilayer completeness remains at 100 ± 1%. A substantial thickening of the lipid bilayer of ∼0.5 Å per leaflet upon avobenzone addition is observed.
Fig. 2C, D and Table S2† show NR results for the measurements involving the avobenzone![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) calix[8]-PO3H2 complex. The POPC and POPC-d31 lipid bilayers were structurally identical to those prepared for the avobenzone measurements, albeit the hydrocarbon region was on average 2 Å thinner, and therefore closer to literature values.46 While a comparable surface volume density of bilayer-associated avobenzone of 2.3 ± 0.6 Å3/Å2 (1 avobenzone per ∼3.1 lipid molecules) was observed, the density of calix[8]-PO3H2 arene was barely significant with 0.7 ± 0.6 Å3/Å2. Since the solvent-excluded volume of avobenzone is roughly a third of that of calix[8]-PO3H2, the molar ratio of calix[8]-PO3H2 to avobenzone at the membrane is substantially less than 1. This indicates a dissociation of avobenzone from calix[8]-PO3H2 before or upon bilayer interaction, which is consistent with the -weak hydrogen bonding of avobenzone to calix[8]-PO3H2 at the upper rim.26 Post-rinsing, both the amount of bilayer-associated avobenzone and macrocycle remains unchanged. The lipid bilayer remains structurally intact throughout the experiment.
calix[8]-PO3H2 complex. The POPC and POPC-d31 lipid bilayers were structurally identical to those prepared for the avobenzone measurements, albeit the hydrocarbon region was on average 2 Å thinner, and therefore closer to literature values.46 While a comparable surface volume density of bilayer-associated avobenzone of 2.3 ± 0.6 Å3/Å2 (1 avobenzone per ∼3.1 lipid molecules) was observed, the density of calix[8]-PO3H2 arene was barely significant with 0.7 ± 0.6 Å3/Å2. Since the solvent-excluded volume of avobenzone is roughly a third of that of calix[8]-PO3H2, the molar ratio of calix[8]-PO3H2 to avobenzone at the membrane is substantially less than 1. This indicates a dissociation of avobenzone from calix[8]-PO3H2 before or upon bilayer interaction, which is consistent with the -weak hydrogen bonding of avobenzone to calix[8]-PO3H2 at the upper rim.26 Post-rinsing, both the amount of bilayer-associated avobenzone and macrocycle remains unchanged. The lipid bilayer remains structurally intact throughout the experiment.
Molecular dynamics simulations
Molecular dynamics simulations were conducted using the GROMACS/4.2.1 suite of programs. The system was set up by conjoining an equilibrated box containing a POPC lipid bilayer with two equilibrated water boxes. The combined configuration forms a rectangular prism of dimensions 6 nm × 6 nm × 12 nm. Forcefields used were CHARMM27 for the lipids, the TIP3P water model, and the CHARMM general forcefield for avobenzone and calix[8]-PO3H2. Initial positions for the avobenzone molecule and the avobenzone![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) calix[8]-PO3H2 complex were chosen to be 2 nm from the lipid surface, to prevent premature interaction with the POPC surface. The initial configuration of the avobenzone
calix[8]-PO3H2 complex were chosen to be 2 nm from the lipid surface, to prevent premature interaction with the POPC surface. The initial configuration of the avobenzone![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) calix[8]-PO3H2 complex was chosen following an unrestrained MD equilibration of the two species in an aqueous environment, this resulting structure represents a low energy configuration of the two molecules interacting in a “bowl–solute” conformation. The systems were equilibrated using the NVT ensemble followed by the NPT ensemble for 1 ns and 5 ns, respectively, using the Nose–Hoover thermostat and the Parrinello–Rahman barostat. After equilibration, umbrella pulling simulations were conducted in the negative z-direction to generate configuration windows for the potential-of-mean-force (PMF) calculations. 300 configurations were extracted from the pull trajectories to ensure proper sampling. Each simulation window was re-equilibrated for a short period 0.5 ns, then production runs were conducted. During production, the center of mass of the “pulled” molecule(s) (avobenzone or avobenzone–calix[8]-PO3H2) are restrained, but rotation around their centre of mass and molecule flexing is allowed to prevent unrealistic configurations. Each production run was sampled for 6 ns with a 2 fs timestep and coupling times of 2 and 4 fs for the thermostat and barostat respectively. Samples from each production window were analysed using the GROMACS implementation of the weighted histogram analysis (gmx WHAM).47 The PMFs were shifted so that zero energy corresponds to the solutes in bulk water.
calix[8]-PO3H2 complex was chosen following an unrestrained MD equilibration of the two species in an aqueous environment, this resulting structure represents a low energy configuration of the two molecules interacting in a “bowl–solute” conformation. The systems were equilibrated using the NVT ensemble followed by the NPT ensemble for 1 ns and 5 ns, respectively, using the Nose–Hoover thermostat and the Parrinello–Rahman barostat. After equilibration, umbrella pulling simulations were conducted in the negative z-direction to generate configuration windows for the potential-of-mean-force (PMF) calculations. 300 configurations were extracted from the pull trajectories to ensure proper sampling. Each simulation window was re-equilibrated for a short period 0.5 ns, then production runs were conducted. During production, the center of mass of the “pulled” molecule(s) (avobenzone or avobenzone–calix[8]-PO3H2) are restrained, but rotation around their centre of mass and molecule flexing is allowed to prevent unrealistic configurations. Each production run was sampled for 6 ns with a 2 fs timestep and coupling times of 2 and 4 fs for the thermostat and barostat respectively. Samples from each production window were analysed using the GROMACS implementation of the weighted histogram analysis (gmx WHAM).47 The PMFs were shifted so that zero energy corresponds to the solutes in bulk water.
Fig. 3 (top) shows the free energy change as a function of the distance of avobenzone from the POPC lipid bilayer's centre of mass. A negative change in free energy is observed as avobenzone partitions into the lipid bilayer. The local minimum of the free energy profile is located at ∼0.7 nm, which indicates that avobenzone after partitioning into the membrane will settle near this position and which is in agreement with the peak of the CVO profiles of avobenzone obtained from the NR experiments (Fig. 2; Tables S1 and S2†).
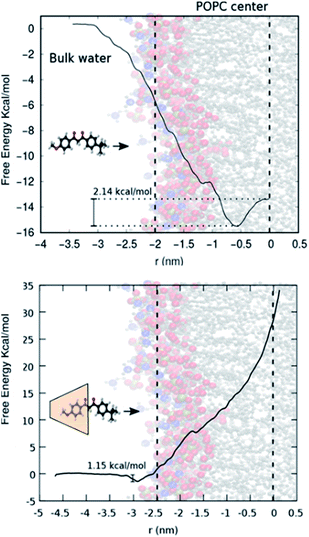 | ||
| Fig. 3 (Top) Schematic showing POPC bilayer and avobenzone. (Bottom) Calix[8]-PO3H2 + avobenzone with POPC bilayer. | ||
Fig. 3 (bottom) shows the free energy change as the avobenzone![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) calix[8]-PO3H2 complex approaches the bilayer. In contrast to the result with avobenzone only, a shallow local minimum is located at the surface of the bilayer (3 nm from the bilayer center). Moving into membrane the free energy rises sharply not exhibiting an energetically favourable configuration. This indicates the difficulty the avobenzone
calix[8]-PO3H2 complex approaches the bilayer. In contrast to the result with avobenzone only, a shallow local minimum is located at the surface of the bilayer (3 nm from the bilayer center). Moving into membrane the free energy rises sharply not exhibiting an energetically favourable configuration. This indicates the difficulty the avobenzone![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) calix[8] complex faces attempting to permeate even the outer layers of the bilayer. The free energy continues to rise after crossing the center of mass of the POPC lipid bilayer further indicating that the forced permeation in our simulations is disrupting the bilayer structure. Error bars in Fig. 3 are omitted for clarity, but standard deviation of each point on the curve is estimated to be between 0.3 and 0.5 kcal mol−1 using the bootstrap analysis method.47 The local free energy minimum of −1.5 kcal mol−1 is too shallow to stably bind a significant amount of the complex at a solution concentration of 100 μM.48 Simulations therefore agree with the absence of a significant amount of calix[8]-PO3H2 detected in NR.
calix[8] complex faces attempting to permeate even the outer layers of the bilayer. The free energy continues to rise after crossing the center of mass of the POPC lipid bilayer further indicating that the forced permeation in our simulations is disrupting the bilayer structure. Error bars in Fig. 3 are omitted for clarity, but standard deviation of each point on the curve is estimated to be between 0.3 and 0.5 kcal mol−1 using the bootstrap analysis method.47 The local free energy minimum of −1.5 kcal mol−1 is too shallow to stably bind a significant amount of the complex at a solution concentration of 100 μM.48 Simulations therefore agree with the absence of a significant amount of calix[8]-PO3H2 detected in NR.
Conclusions
Combined NR and simulation results from the present study reveal the interaction of avobenzone with the bulk solvent-side region (outer leaflet) of a POPC membrane. Un-complexed avobenzone is shown to strongly penetrate into the membrane and associate with 5 lipid molecules; however, in the presence of calix[8]-PO3H2 the complex encounters a large free energy barrier to membrane entry. These energetically favourable positions lead to the penetration of a single avobenzone to the first hydrophobic region of the POPC lipid bilayer, while the avobenzone![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) calix[8]-PO3H2 complex instead enjoys an energetically favourable position at the surface of the bilayer. These preferred positions agree with the NR results showing calix[8]-PO3H2 accumulating at the lipid bilayer surface and avobenzone settling in the first hydrophobic region. The avobenzone by itself displays a 2.1 kcal mol−1 energy well in the first inner leaflet, higher than the energy well of the avobenzone
calix[8]-PO3H2 complex instead enjoys an energetically favourable position at the surface of the bilayer. These preferred positions agree with the NR results showing calix[8]-PO3H2 accumulating at the lipid bilayer surface and avobenzone settling in the first hydrophobic region. The avobenzone by itself displays a 2.1 kcal mol−1 energy well in the first inner leaflet, higher than the energy well of the avobenzone![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) calix[8]-PO3H2 complex at the lipid bilayer surface (1.2 kcal mol−1). In addition, simulations have identified a free energy minimum for membrane-bound avobenzone 6 Å from the membrane centre, inside the first hydrophobic region of the bilayer. This is in agreement with the experimentally determined peak positions of the avobenzone CVO profiles of 10 ± 3 Å and 7 ± 3 Å (Table S1†). The free energy difference between membrane bound avobenzone and avobenzone at infinite dilution of −16 kcal mol−1 estimates to an affinity in the nM range.48 This calculated value being lower than the experimental concentration of 100 μM may suggest that the overall free energy difference between bound and solvated states should be smaller than the calculated −16 kcal mol−1 if the concentration in the simulated bulk phase matched that of the experiments. These results showcase the ability of the calix[8]-PO3H2 macrocycle to reduce the penetration of the avobenzone into a simple lipid bilayer, providing a useful way of anchoring targeted molecules to the surface of a membrane and reducing molecular permeation.
calix[8]-PO3H2 complex at the lipid bilayer surface (1.2 kcal mol−1). In addition, simulations have identified a free energy minimum for membrane-bound avobenzone 6 Å from the membrane centre, inside the first hydrophobic region of the bilayer. This is in agreement with the experimentally determined peak positions of the avobenzone CVO profiles of 10 ± 3 Å and 7 ± 3 Å (Table S1†). The free energy difference between membrane bound avobenzone and avobenzone at infinite dilution of −16 kcal mol−1 estimates to an affinity in the nM range.48 This calculated value being lower than the experimental concentration of 100 μM may suggest that the overall free energy difference between bound and solvated states should be smaller than the calculated −16 kcal mol−1 if the concentration in the simulated bulk phase matched that of the experiments. These results showcase the ability of the calix[8]-PO3H2 macrocycle to reduce the penetration of the avobenzone into a simple lipid bilayer, providing a useful way of anchoring targeted molecules to the surface of a membrane and reducing molecular permeation.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research is funded by Start-up funds (HK) and NSF grants CHE-1565632 and CHE-1955161 (TLB). FH acknowledges support from the U.S. Department of Commerce through an MSE grant (70NANB17H299). Certain commercial materials, equipment, and instruments are identified in this work to describe the experimental procedure as completely as possible. In no case does such an identification imply a recommendation or endorsement by NIST, nor does it imply that the materials, equipment, or instrument identified are necessarily the best available for the purpose.Notes and references
- V. Balzani, A. Credi, F. M. Raymo and J. F. Stoddart, Angew. Chem., 2000, 39, 3348–3391 CrossRef CAS.
- H. Huang, T. Orlova, B. Matt and N. Katsonis, Macromol. Rapid Commun., 2018, 39, 1700387 CrossRef PubMed.
- D. Zhang, A. Martinez and J.-P. Dutasta, Chem. Rev., 2017, 117, 4900–4942 CrossRef CAS PubMed.
- P. M. Mendes, A. H. Flood and J. F. Stoddart, Appl. Phys. A: Mater. Sci. Process., 2005, 80, 1197–1209 CrossRef CAS.
- R. A. Bissell, E. Córdova, A. E. Kaifer and J. F. Stoddart, Nature, 1994, 369, 133–137 CrossRef CAS.
- B. Kütting and H. Drexler, Int. Arch. Occup. Environ. Health, 2010, 83, 843–854 CrossRef PubMed.
- T. S. Poon, R. S. Barnetson and G. M. Halliday, J. Invest. Dermatol., 2003, 121, 184–190 CrossRef CAS PubMed.
- F. H. M. de Melo, F. Molognoni and M. Galvonas, in Recent Advances in the Biology, Therapy and Management of Melanoma, 2013, ch. 5, DOI:10.5772/54937.
- J. A. Ruszkiewicz, A. Pinkas, B. Ferrer, T. V. Peres, A. Tsatsakis and M. Aschner, Toxicol. Rep., 2017, 4, 245–259 CrossRef CAS PubMed.
- M. S. Díaz-Cruz and D. Barceló, TrAC, Trends Anal. Chem., 2009, 28, 708–717 CrossRef.
- S. Kim and K. Choi, Environ. Int., 2014, 70, 143–157 CrossRef CAS PubMed.
- R. Danovaro, L. Bongiorni, C. Corinaldesi, D. Giovannelli, E. Damiani, P. Astolfi, L. Greci and A. Pusceddu, Environ. Health Perspect., 2008, 116, 441–447 CrossRef CAS PubMed.
- S. L. Schneider and H. W. Lim, J. Am. Acad. Dermatol., 2019, 80, 266–271 CrossRef CAS PubMed.
- M. M. P. Tsui, J. C. W. Lam, T. Y. Ng, P. O. Ang, M. B. Murphy and P. K. S. Lam, Environ. Sci. Technol., 2017, 51, 4182–4190 CrossRef CAS PubMed.
- C. Botta, J. Labille, M. Auffan, D. Borschneck, H. Miche, M. Cabié, A. Masion, J. Rose and J.-Y. Bottero, Environ. Pollut., 2011, 159, 1543–1550 CrossRef CAS PubMed.
- K. Schilling, B. Bradford, D. Castelli, E. Dufour, J. F. Nash, W. Pape, S. Schulte, I. Tooley, J. van den Bosch and F. Schellauf, Photochem. Photobiol. Sci., 2010, 9, 495–509 RSC.
- R. Dunford, A. Salinaro, L. Cai, N. Serpone, S. Horikoshi, H. Hidaka and J. Knowland, FEBS Lett., 1997, 418, 87–90 CrossRef CAS PubMed.
- J. C. DiNardo and C. A. Downs, J. Cosmet. Dermatol., 2018, 17, 15–19 CrossRef PubMed.
- C. Jacques-Jamin, H. Duplan, H. Rothe, O. Vaillant, J. Eilstein, S. Grégoire, R. Cubberley, D. Lange, C. Ellison, M. Klaric, N. Hewitt and A. Schepky, Skin Pharmacol. Physiol., 2017, 30, 234–245 CrossRef CAS PubMed.
- A. D. Martin and C. L. Raston, Chem. Commun., 2011, 47, 9764–9772 RSC.
- K. D. Shimizu and J. Rebek, Jr., Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 12403–12407 CrossRef CAS PubMed.
- R. H. Vreekamp, J. P. M. van Duynhoven, M. Hubert, W. Verboom and D. N. Reinhoudt, Angew. Chem., Int. Ed., 1996, 35, 1215–1218 CrossRef CAS.
- Z. Zhang, A. Drapailo, Y. Matvieiev, L. Wojtas and M. J. Zaworotko, Chem. Commun., 2013, 49, 8353 RSC.
- Y. Zhou, C. Liu, H. Xu, H. Yu, Q. Lu and L. Wang, Spectrochim. Acta, Part A, 2007, 66, 919–923 CrossRef PubMed.
- A. Dawn, X. Yao, Y. Yu, J. Jiang and H. Kumari, Supramol. Chem., 2019, 31, 425–431 CrossRef CAS PubMed.
- A. Das, M. Mirzamani, H. Kumari and A. Gudmundsdottir, 2020, manuscript in preparation.
- C. M. Kawakami and L. R. Gaspar, J. Photochem. Photobiol., B, 2015, 151, 239–247 CrossRef CAS PubMed.
- J. Wang, L. Pan, S. Wu, L. Lu, Y. Xu, Y. Zhu, M. Guo and S. Zhuang, Int. J. Environ. Res. Public Health, 2016, 13, 782 CrossRef CAS PubMed.
- M. Lodén, H. Beitner, H. Gonzalez, D. W. Edström, U. Åkerström, J. Austad, I. Buraczewska-Norin, M. Matsson and H. C. Wulf, Br. J. Dermatol., 2011, 165, 255–262 CrossRef PubMed.
- G. J. Mturi and B. S. Martincigh, J. Photochem. Photobiol., A, 2008, 200, 410–420 CrossRef CAS.
- R. Budvytyte, G. Valincius, G. Niaura, V. Voiciuk, M. Mickevicius, H. Chapman, H.-Z. Goh, P. Shekhar, F. Heinrich, S. Shenoy, M. Lösche and D. J. Vanderah, Langmuir, 2013, 29, 8645–8656 CrossRef CAS PubMed.
- G. S. Gooris, M. Kamran, A. Kros, D. J. Moore and J. A. Bouwstra, Biochim. Biophys. Acta, Biomembr., 2018, 1860, 1272–1281 CrossRef CAS PubMed.
- D. Groen, F. Berthaud, J. A. Bouwstra, C. Chapuis, G. S. Gooris and M. Boncheva, Biochim. Biophys. Acta, Biomembr., 2014, 1838, 310–318 CrossRef CAS PubMed.
- N. R. Yepuri, T. A. Darwish, A. M. Krause-Heuer, A. E. Leung, R. Delhom, H. P. Wacklin and P. J. Holden, ChemPlusChem, 2016, 81, 315–321 CrossRef CAS PubMed.
- A. Kalinin, L. N. Marekov and P. M. Steinert, J. Cell Sci., 2001, 114, 3069–3070 CAS.
- C. R. Harding, Dermatol. Ther., 2004, 17, 6–15 CrossRef PubMed.
- E. Candi, R. Schmidt and G. Melino, Nat. Rev. Mol. Cell Biol., 2005, 6, 328–340 CrossRef CAS PubMed.
- G. Fragneto, Eur. Phys. J. Spec. Top., 2012, 213, 327–342 CrossRef CAS.
- G. Fragneto, R. Delhom, L. Joly and E. Scoppola, Curr. Opin. Colloid Interface Sci., 2018, 38, 108–121 CrossRef CAS.
- J. A. Dura, D. J. Pierce, C. F. Majkrzak, N. C. Maliszewskyj, D. J. McGillivray, M. Lösche, K. V. O’Donovan, M. Mihailescu, U. Perez-Salas, D. L. Worcester and S. H. White, Rev. Sci. Instrum., 2006, 77(7), 74301 CrossRef PubMed.
- B. J. Kirby, P. A. Kienzle, B. B. Maranville, N. F. Berk, J. Krycka, F. Heinrich and C. F. Majkrzak, Curr. Opin. Colloid Interface Sci., 2012, 17, 44–53 CrossRef CAS.
- A. Benedetto, F. Heinrich, M. A. Gonzalez, G. Fragneto, E. Watkins and P. Ballone, J. Phys. Chem. B, 2014, 118, 12192–12206 CrossRef CAS PubMed.
- F. Heinrich and M. Lösche, Biochim. Biophys. Acta, Biomembr., 2014, 1838, 2341–2349 CrossRef CAS PubMed.
- P. Shekhar, H. Nanda, M. Lösche and F. Heinrich, J. Appl. Phys., 2011, 110(10), 102216 CrossRef PubMed.
- B. J. Kirby, P. A. Kienzle, B. B. Maranville, N. F. Berk, J. Krycka, F. Heinrich and C. F. Majkrzak, ýCurr. Opin. Colloid Interface Sci, 2012, 17, 44–53 CrossRef CAS.
- N. Kučerka, S. Tristram-Nagle and J. F. Nagle, J. Membr. Biol., 2006, 208, 193–202 CrossRef PubMed.
- J. S. Hub, B. L. de Groot and D. van der Spoel, J. Chem. Theory Comput., 2010, 6, 3713–3720 CrossRef CAS.
- M. Barros, F. Heinrich, S. A. K. Datta, A. Rein, I. Karageorgos, H. Nanda, M. Lösche and W. I. Sundquist, J. Virol., 2016, 90, 4544–4555 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra02850a |
| This journal is © The Royal Society of Chemistry 2020 |