Open Access Article
Open Access ArticleMetabolism of insecticide diazinon by Cunninghamella elegans ATCC36112
Mei-ai Zhao†
a,
Hao Gu† b,
Chuan-Jie Zhang†c,
In-Hong Jeongd,
Jeong-Han Kime and
Yong-Zhe Zhu*b
b,
Chuan-Jie Zhang†c,
In-Hong Jeongd,
Jeong-Han Kime and
Yong-Zhe Zhu*b
aCollege of Life Sciences, Qingdao Agricultural University, Changcheng Rd, Chengyang, Qingdao City, Shandong Province 266-109, China
bCollege of Chemistry and Pharmacy, Qingdao Agricultural University, Changcheng Rd, Chengyang, Qingdao City, Shandong Province 266-109, China. E-mail: zhuyzh@qau.edu.cn; Fax: +86-532-8803-0220; Tel: +86-133-5532-5000
cCollege of Animal Science and Technology, Yangzhou University, Yangzhou, Jiangsu Province 225-009, China
dDivision of Crop Protection, National Institute of Agricultural Science, Rural Development Administration, Jeollabuk-do 55365, Republic of Korea
eDepartment of Agricultural Biotechnology, Seoul National University, 599 Gwanak-ro, Silim-dong, Gwanak-Gu, Seoul, 151-742, Republic of Korea
First published on 26th May 2020
Abstract
The fungal metabolism of diazinon was investigated and the microbial model (Cunninghamella elegans ATCC36112) could effectively degrade the organophosphorus pesticide (diazinon) mediated by cytochrome P450, which was mainly involved in oxidation and hydrolysis of phase I metabolism. Approximately 89% of diazinon was removed within 7 days and was not observed after 13 days with concomitant accumulation of eight metabolites. Structures of the metabolites were fully or tentatively identified with GC-MS and 1H, 13C NMR. The major metabolites of diazinon were diethyl (2-isopropyl-6-methylpyrimidin-4-yl) phosphate (diazoxon) and 2-isopropyl-6-methyl-4-pyrimidinol (pyrimidinol), and formation of minor metabolites was primarily the result of hydroxylation. To determine the responsible enzymes in diazinon metabolism, piperonyl butoxide and methimazole were treated, and the kinetic responses of diazinon and its metabolites by Cunninghamella elegans were measured. Results indirectly demonstrated that cytochrome P450 and flavin monooxygenase were involved in the metabolism of diazinon, but methimazole inhibited the metabolism less effectively. Based on the metabolic profiling, a possible metabolic pathway involved in phase I metabolism of diazinon was proposed, which would contribute to providing insight into understanding the toxicological effects of diazinon and the potential application of fungi on organophosphorus pesticides.
1. Introduction
Organophosphorus pesticides (OPs) have largely replaced the organochlorines from the mid-1960s,1 which constitutes an important aspect of modern agriculture. OPs are used to ensure better yield and quality of fruits and vegetables, but at the same time the contamination of food by OPs may increase their danger to humans.2 Due to the widespread use of OPs, residual OPs continue to accumulate in animal tissues and pass from one trophic level to another within food chains.3 So degradation of OPs with a more economical and pro-environmental strategy such as microorganisms is very important and urgent.Diazinon [O,O-diethyl O-2-isopropyl-6-methylpyrimidin-4-yl phosphorothioate] is an phosphorothioate insecticide and acaricide with contact, stomach, and respiratory action.4 It has been used in animal houses and households to control the ingestion and chewing of insects and mites on various crops, lawns, fruits and vegetables.5,6 Diazinon presents a water solubility of 60 mg L−1 and the partition coefficient (Koc) is 1000 mL g−1 in soil.5,7 Besides the solubility and adsorption of pesticides, the toxicity of pesticides to non-target species should also be considered.8 Based on the concern about non-targeted biotoxicity of diazinon, more and more studies indicated that diazinon caused oxidative damage through the generation of free radicals and induced lipid peroxidation and DNA fragmentation,2,9,10 and diazinon could damage the liver and kidneys, causing severe histopathological damage.2,13 Furthermore, diazinon will be oxidatively denatured to diazoxon when it enters the human body.11 This metabolite is more toxic than the parent compound, mainly in inhibiting acetylcholinesterase,12 leading to a cholinergic syndrome and associated neurotoxicity.2
Metabolism studies are very important for the understanding of pesticide toxicity and safety.14 At present, the research on diazinon metabolism mainly focuses on the metabolites, pathways and related metabolic toxicology of diazinon in mice, dogs and other mammals.15–17 In addition, the metabolism and toxicity of the pesticide in vitro metabolic model (human colon carcinoma cells and human liver microsomes, etc.)2,18 and bacteria (Acinetobacter and Pseudomonas sp, etc.)19 have also been reported. However, there are a few reports on fungal metabolism of diazinon.20
Cunninghamella elegans (C. elegans), a filamentous fungus, is widely used as a microbial model of the mammalian metabolism of different xenobiotics, including pesticides, drugs and other pollutants.21–24 Previous studies have shown that the fungal biotransformation of many pesticides is mainly mediated by the cytochrome P450 (CYP450s),24–28 which has similar metabolic systems and processes as human metabolism. Diverse chemical reaction are catalyzed by P450 monooxygenases including classical reaction of hydroxylation, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C double bond epoxidation reaction, and aromatic ring hydroxylation reaction, even the cleavage of the C–C bond subjected to multiple substrate oxidations.29 So, the application of this microbial model and enzyme inhibitor can be speculated or assisted to confirm the pesticide metabolism pathway in mammals, therefore greatly reducing the cost of experimental animals and experiments. The main purpose of this study is to use C. elegans to biotransform diazinon and identify the metabolites with gas chromatography mass spectrometry (GC-MS) and nuclear magnetic resonance (NMR). Under the action of enzyme inhibitors, oxidases involved in metabolism have also been studied.
C double bond epoxidation reaction, and aromatic ring hydroxylation reaction, even the cleavage of the C–C bond subjected to multiple substrate oxidations.29 So, the application of this microbial model and enzyme inhibitor can be speculated or assisted to confirm the pesticide metabolism pathway in mammals, therefore greatly reducing the cost of experimental animals and experiments. The main purpose of this study is to use C. elegans to biotransform diazinon and identify the metabolites with gas chromatography mass spectrometry (GC-MS) and nuclear magnetic resonance (NMR). Under the action of enzyme inhibitors, oxidases involved in metabolism have also been studied.
2. Materials and methods
2.1 Chemicals and reagents
Diazinon, diazinon-O-analog (diazoxon) and 2-isopropyl-6-methyl-4-pyrimidinol (pyrimidinol) standards (purity > 98%) purchased from Chem service (West Chester, PA, U.S.A.). Methimazole (MZ) and piperonyl butoxide (PB) were from Macklin® (Shanghai, China). Ethyl acetate and methanol were of HPLC grade and from Burdick and Jackson® (Seoul, Korea). Anhydrous sodium sulfate and sodium chloride were obtained from Junsei (Tokyo, Japan). Potato dextrose agar (PDA) and potato dextrose broth (PDB) were supplied by BD Korea (Seoul, Korea). All other chemicals were of reagent grade and of the highest purity available.2.2 Microorganism
Cunninghamella elegans ATCC36112 was obtained from American Type Culture Collection (Manassas, VA, U.S.A.). Stock cultures of C. elegans ATCC36112 were maintained on PDA plates at 28 °C. Spores and mycelia from several plates were used to inoculate on PDA medium. At 28 °C, C. elegans ATCC36112 grew for 48 h.2.3 Metabolic reaction
Approximately 2 g mycelia was transferred into fresh PDB medium (500 mL), followed by the addition of diazinon (5 mg in 1 mL acetonitrile), which were incubated at 28 °C with shaking at 170 rpm for 13 days. The final pH value was adjusted to 7.0 with 0.1 M phosphate buffer.30 Control experiments were conducted in the absence of either diazinon (blank control) or fungi (negative control). According to the sterilization standard of PDA and PDB, all the media were sterilized at 121 °C for 15 min. The natural pH value of the medium was 5.1 ± 0.2. The pH was measured by Orion Star™ A211 pH Benchtop Meter.2.4 Enzyme inhibitor reaction
Additionally, the effects of enzyme inhibitors (PB and MZ) on the biodegradation of C. elegans were studied. Different concentrations (2, 10, 50 mg L−1) of enzyme inhibitor stock solutions were added to the inoculated PDB medium (500 mL, pH 7.0), followed by adding diazinon (5 mg in 1 mL acetonitrile) and pre-incubated for 12 hours. Sample was collected at 2 hours, 1, 3, 5, 7, 10, and 13 days, respectively.2.5 Extraction of metabolites
To detect parent pesticide and its biotransformation products, the culture medium was extracted. At each metabolic reaction time (2 hours, 1, 3, 5, 7, 10, and 13 days), 50 mL of the culture medium was transferred to a 1000 mL separatory funnel with 20 g of NaCl, and afterward extracted twice with ethyl acetate (100 and 50 mL). The combined ethyl acetate phases were passed through anhydrous Na2SO4 and evaporated at 35 °C. Subsequently, the obtained residue was dissolved in methanol (2 mL) and filtered through a 0.22 μm polytetrafluoroethylene (PTFE) filter for HPLC and GC-MS analysis. Before GC-MS analysis, the extract was dried by nitrogen and dissolved in dry pyridine, derivatized with BSTFA + TMCS (99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) at 70 °C for 37 min.
1) at 70 °C for 37 min.
2.6 Enrichment and isolation of metabolites by large scale culture
To collect enough metabolites for subsequent analysis, 9 bottles (800 mL per each) culture medium were set up. The culture medium was extracted in multiple portions in a separatory funnel, then concentrated and dissolved in methanol. Isolation of the metabolites was carried out using a previously reported method.25 Separation of metabolites was through silica gel (20 g) column chromatography (1.5 cm i.d. × 40 cm) with 50 mL of hexane/ethyl acetate by stepwise gradient elution (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 90
0, 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 80
10, 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, 70
20, 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, 60
30, 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40, 50
40, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, 40
50, 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60, 30
60, 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70, 20
70, 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80, 10
80, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90 and 0
90 and 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, v/v). Each eluted component was detected by HPLC. M1 was eluted in a fraction of 30
100, v/v). Each eluted component was detected by HPLC. M1 was eluted in a fraction of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 (hexane/ethyl acetate), and M2 was collected in a fraction of 0
70 (hexane/ethyl acetate), and M2 was collected in a fraction of 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (hexane/ethyl acetate). After drying, M1 and M2 were then dissolved in CDCl3 (99.8%, Merck) for NMR.
100 (hexane/ethyl acetate). After drying, M1 and M2 were then dissolved in CDCl3 (99.8%, Merck) for NMR.
2.7 Instrumental analysis condition
Quantitative analysis of diazinon was conducted using an Ultimate 3000 HPLC system (Thermo Scientific, Sunnyvale, CA, U.S.A.) equipped with Luna C18 column (250 mm × 4.6 mm, 5 μm, Phenomenex®, Torrance, CA, U.S.A.) at 40 °C. The mobile phase consisted water (A) and methanol (B), while the flow rate was 1 mL min−1 and the volume of injection was 10 μL. The gradient condition was as follows: 5% B at 0 to 1 min, 90% B at 20–25 min, 95% B at 30 min, 5% B at 32–40 min. Samples were detected by UV absorption at 245 nm.GC-MS analysis was performed with a Bruker SCION TQ equipped with an CP-8400 autosampler. A fused-silica capillary column (Rxi-5Sil MS, 30 m × 0.25 mm i.d., 0.25 μm film thickness) was used for GC separation. The oven program was 1 min at 80 °C, 7 °C min−1 to 240 °C, and 5 °C min−1 to 295 °C (30 min). In the full-scan mode, electron ionization (EI) mass spectra in the range of 50–600 (m/z) were recorded at 70 eV electron energy. Helium was used as carrier gas at 1.0 mL min−1. Splitless injections of 2 μL sample were carried out. The injector temperature was 260 °C, the interface temperature was set at 280 °C, and the solvent delay time was set to 5 min. Data analysis was performed with Bruker MS workstation software (version 8.0, Germany) and ACD/MS Fragmenter 2017 software (ACD Labs, Canada).
NMR spectroscopy was used to confirm the structure of metabolites. 1H and 13C NMR spectra were recorded on a 400 MHz NMR spectrometer (Jeol JNM-LA400, JEOL Ltd., Tokyo, Japan) in CDCl3 (99.8%, Merck) at 298 K.
3. Results and discussion
3.1 Metabolism of diazinon by C. elegans
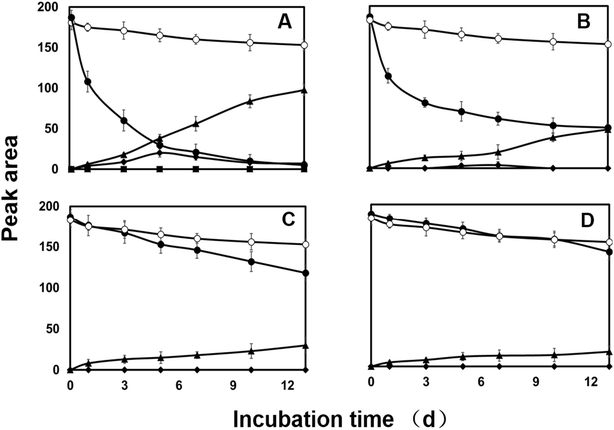
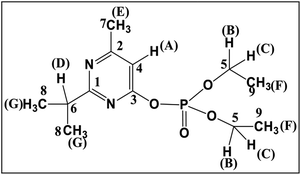
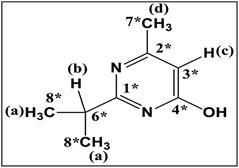
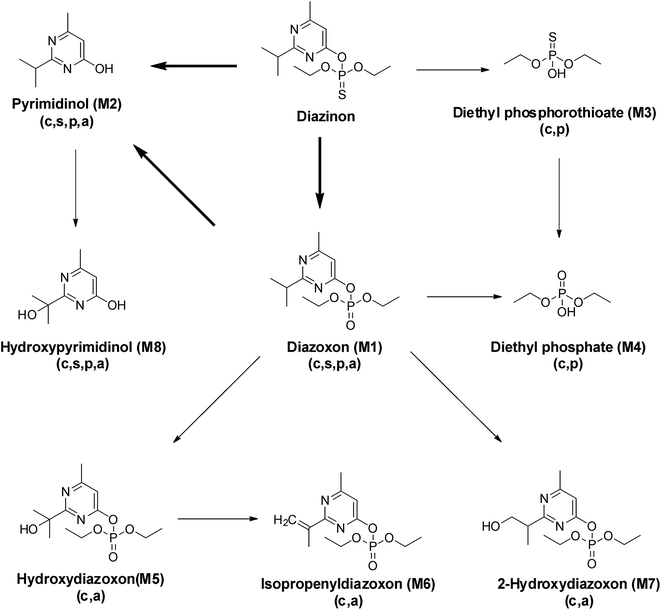
HPLC analysis of the culture extract indicated that diazinon was transformed to two metabolites (M1 and M2). In the range of 0.1 to 100.0 mg L−1, there is a linear relationship (Y = 6.2191x − 1.3314, R2 = 0.9999) between the concentrations of diazinon standard and the peak area. LOD (S/N > 3) was calculated as 0.1 mg L−1. The structure of metabolites was shown in Fig. 5. For example, diazinon was detected at approximately 50% degraded in the culture at 2 days after treatment and was undetectable after 13 days (Fig. 1A), M1 and M2 were observed as major metabolites. With the production of metabolites, the accumulation of M1 reached a maximum at 5 days and gradually decreased to trace level after 13 days, while M2 showed a trend of continuous growth. Then the identification of those was carried out through large scale culture. Approximately, eight metabolites of diazinon were observed in culture supernatants including two major metabolites (M1 and M2) by GC-MS (Fig. 3). Minor metabolites (M3–M8) were hardly located on the chromatogram because of the trace levels, so their formation pattern could not be established. In a sterilized control experiments, no appreciable degradation was observed.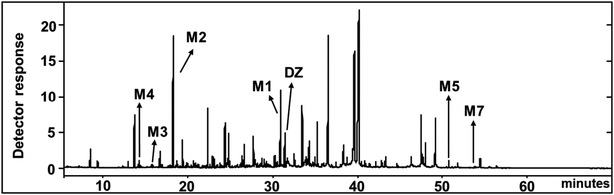 | ||
| Fig. 3 Total ion chromatograms (TIC) of diazinon (DZ) and its TMS-derivatized metabolites by C. elegans at 3rd day. | ||
In GC-MS, mass spectral details of those metabolites were described in Table 1. Molecular ion of M1 was observed at m/z 288, which was 16 Da lower than that of diazinon (M+, 304), suggesting that metabolic oxidative desulfuration took place in diazinon (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S→P
S→P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). The molecular ion and the fragment ions of M1 could be speculated as diazoxon. In the case of TMS-derivatized M2, gave a molecular ion of m/z 224 from GC-MS analysis, it was 72 Da higher than pyrimidinol (M+, 152), which would be fragmented from diazinon (Fig. 4). M2 could be speculated as pyrimidinol. To verify this hypothesis, diazoxon and pyrimidinol were purchased and analyzed by GC-MS and NMR. GC-MS analysis showed the retention times of two standards at 30.9 and 18.2 min, and their molecular ion peaks at m/z 288 (16) and m/z 224 (17), respectively. 1H and 13C NMR spectra of metabolites matched with the structures of standards (Table 2). On the basis of these results, M1 and M2 were identified as diazoxon and pyrimidinol, respectively.
O). The molecular ion and the fragment ions of M1 could be speculated as diazoxon. In the case of TMS-derivatized M2, gave a molecular ion of m/z 224 from GC-MS analysis, it was 72 Da higher than pyrimidinol (M+, 152), which would be fragmented from diazinon (Fig. 4). M2 could be speculated as pyrimidinol. To verify this hypothesis, diazoxon and pyrimidinol were purchased and analyzed by GC-MS and NMR. GC-MS analysis showed the retention times of two standards at 30.9 and 18.2 min, and their molecular ion peaks at m/z 288 (16) and m/z 224 (17), respectively. 1H and 13C NMR spectra of metabolites matched with the structures of standards (Table 2). On the basis of these results, M1 and M2 were identified as diazoxon and pyrimidinol, respectively.
| Compound | Retention time (min) | Molecular weight and fragment ionsa (m/z) |
|---|---|---|
| a Values in parentheses are the relative abundance of specified fragment ions. | ||
| Diazinon (parent) | 31.4 | 304 (M+, 21), 276 (15), 248 (14), 216 (11), 199 (42), 179 (86), 152 (53), 137 (100), 93 (31) |
| Diazoxon (M1) | 30.9 | 288 (M+, 16), 273 (75), 260 (16), 217 (20), 151 (26), 137 (100) |
| Pyrimidinol (M2) | 18.2 | 224 (M+, 17), 209 (100), 196 (15), 181 (9), 126 (7) |
| Diethyl phosphorothioate (M3) | 15.9 | 242 (M+, 38), 227 (29), 199 (38), 171 (100), 165 (64), 153 (56), 137 (43), 121 (46) |
| Diethyl phosphate (M4) | 14.3 | 226 (M+, 4), 211 (9), 199 (14), 183 (12), 171 (4), 155 (100), 139 (7) |
| Hydroxydiazoxon (M5) | 50.7 | 376 (M+, 27), 361 (4), 303 (10), 251 (100), 236 (18), 197 (37), 155 (47), 105 (38) |
| Isopropenyl diazoxon (M6) | 33.0 | 286 (M+, 4), 271 (13), 192 (27), 179 (20), 149 (9), 147 (18), 137 (16), 73 (100) |
| 2-Hydroxylated diazoxon (M7) | 53.8 | 376 (M+, 17), 361 (5), 343 (4), 296 (9), 251 (100), 223 (10), 209 (24), 195 (24), 181 (12), 169 (12), 155 (14) |
| Hydroxypyrimidinol (M8) | 30.7 | 312 (M+, 37), 297 (79), 282 (38), 267 (74), 253 (47), 223 (53), 193 (22), 126 (25), 73 (100) |
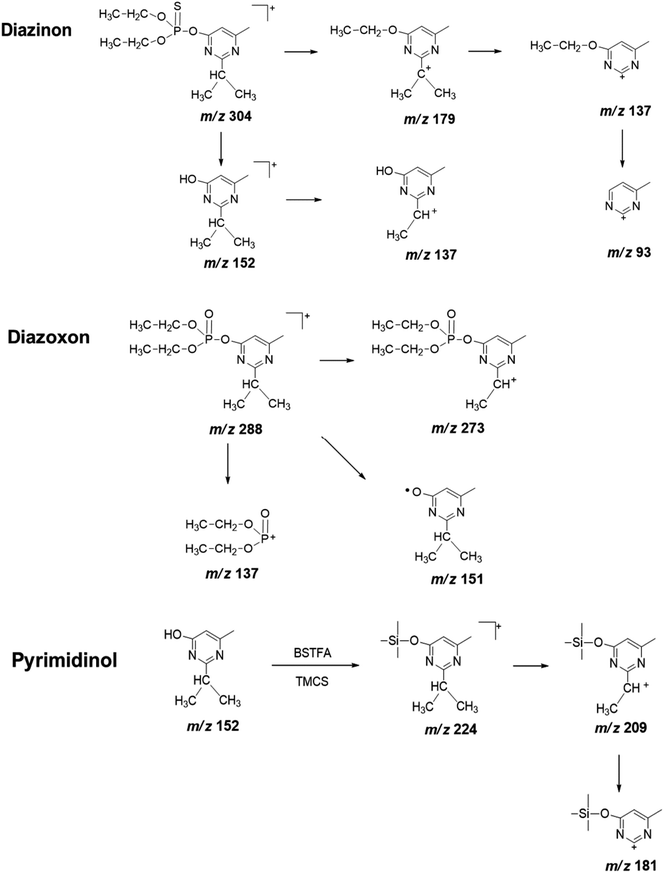 | ||
| Fig. 4 Proposed fragmentation pathway of diazinon, diazoxon, and pyrimidinol standard under electron ionization conditions. | ||
To obtain useful information for explaining the ion spectrum of unknown degradation products, we continued to study the MS2 fragment ions of diazinon, diazoxon and pyrimidinol (Fig. 4, Table 1). Diazinon showed its molecular ion at m/z 137 ([M + H]+) (100), which was the most abundant fragment ion. This may be associated with the cleavage of the P–O-pyrimidine group. Similar reactions have been observed in diazoxon at m/z 137. The ion peaks at m/z 248 and 216 from parent represent consecutive eliminations of ethylene molecules, which may be formed by a classical elimination reaction involving a four-member transition state.31 The ion at m/z 179 represents the cleavage of the P–O bond, accompanied by ethyl rearrangement or migration to an aromatic bonded oxygen atom.32 As for pyrimidinol derivatized, it has a stable structure and fewer fragment ions are obtained under the condition of electron ionization, its fragment ion m/z 209 as the base peak may be formed by losing a methyl from isopropyl.
Other metabolites (M3–M8) were also observed but at trace levels. M3 had shown its ion peak at m/z 242, suggesting that it would be a derivative of diethyl phosphorothioate. Other fragment ion observed at m/z 227, was corresponded to the loss of methyl groups from the precursor ion. The characteristic fragment at m/z 171 might be resulted from m/z 227 after losing two ethyl groups. Molecular ion of M4 was observed at m/z 226, which was 16 Da lower than that of diethyl phosphorothioate (M3, m/z 242), indicating that P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S was oxidized to P
S was oxidized to P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O. The most abundant fragment ion m/z 155 might be resulted from m/z 211 after losing two ethyl groups. In addition, M3 and M4 can be matched by library search and they had also been confirmed in some animal experiments.33,34 Therefore, M3 and M4 were tentatively identified as diethyl phosphorothioate and diethyl phosphate, respectively. Metabolites M5 and M7 gave similar fragment ions, and their molecular ions and base peaks were m/z 376 and m/z 251, respectively. The mass/charge m/z 251 might be formed by the cleavage of the aryl and alkyl group or ethyl rearrangement, and the fragment at m/z 223 was observed, indicating that an ethyl group was removed from the precursor ion m/z 251. Hydroxylation is an important way of pesticide metabolism by Cunninghamella elegans.25,31,35 Similarly, hydroxylation may occur in M5 and M7, both of them had been reported in the previous studies.31,36,37 So M5 and M7 were tentatively identified as hydroxydiazoxon and 2-hydroxylated diazoxon, respectively. Molecular ion of M6 was observed at m/z 286, which was 2 Da lower than that of metabolite M1 (m/z 288). Fragment ion m/z 271 was corresponded to the loss of a methyl from the ion at m/z 286. The ion m/z 192 would be formed by the cleavage of the P–O-pyrimidine group and ethyl rearrangement. Furthermore, fragment ion m/z 149 and m/z 137 were obtained by the cleavage of the P–O-pyrimidine group. It was worth noting that m/z 149 was 2 Da lower than m/z 151 of diazoxon, indicating that two hydrogen atoms were removed from the isopropyl group of the nitrogen-containing heterocycle. Thus, metabolite M6 was tentatively identified as isopropenyl diazoxon, which has also been reported before.36 M8 gave molecular ion at m/z 312, which was 88 Da (-OTMS) higher than that of metabolite M2 (m/z 224). The characteristic fragment at m/z 297 may be obtained after losing a methyl, which has a similar fragmentation mechanism with pyrimidinol. It had been reported that Müecke et al. (1970) treated rats with diazinon l4C-labeled in the pyrimidine ring and ethoxy groups, and hydroxypyrimidinol was identified in the urine.38 So metabolite M8 was tentatively identified as hydroxypyrimidinol (Fig. 5).
O. The most abundant fragment ion m/z 155 might be resulted from m/z 211 after losing two ethyl groups. In addition, M3 and M4 can be matched by library search and they had also been confirmed in some animal experiments.33,34 Therefore, M3 and M4 were tentatively identified as diethyl phosphorothioate and diethyl phosphate, respectively. Metabolites M5 and M7 gave similar fragment ions, and their molecular ions and base peaks were m/z 376 and m/z 251, respectively. The mass/charge m/z 251 might be formed by the cleavage of the aryl and alkyl group or ethyl rearrangement, and the fragment at m/z 223 was observed, indicating that an ethyl group was removed from the precursor ion m/z 251. Hydroxylation is an important way of pesticide metabolism by Cunninghamella elegans.25,31,35 Similarly, hydroxylation may occur in M5 and M7, both of them had been reported in the previous studies.31,36,37 So M5 and M7 were tentatively identified as hydroxydiazoxon and 2-hydroxylated diazoxon, respectively. Molecular ion of M6 was observed at m/z 286, which was 2 Da lower than that of metabolite M1 (m/z 288). Fragment ion m/z 271 was corresponded to the loss of a methyl from the ion at m/z 286. The ion m/z 192 would be formed by the cleavage of the P–O-pyrimidine group and ethyl rearrangement. Furthermore, fragment ion m/z 149 and m/z 137 were obtained by the cleavage of the P–O-pyrimidine group. It was worth noting that m/z 149 was 2 Da lower than m/z 151 of diazoxon, indicating that two hydrogen atoms were removed from the isopropyl group of the nitrogen-containing heterocycle. Thus, metabolite M6 was tentatively identified as isopropenyl diazoxon, which has also been reported before.36 M8 gave molecular ion at m/z 312, which was 88 Da (-OTMS) higher than that of metabolite M2 (m/z 224). The characteristic fragment at m/z 297 may be obtained after losing a methyl, which has a similar fragmentation mechanism with pyrimidinol. It had been reported that Müecke et al. (1970) treated rats with diazinon l4C-labeled in the pyrimidine ring and ethoxy groups, and hydroxypyrimidinol was identified in the urine.38 So metabolite M8 was tentatively identified as hydroxypyrimidinol (Fig. 5).
3.2 Effects of enzyme inhibitor on the degradation of diazinon
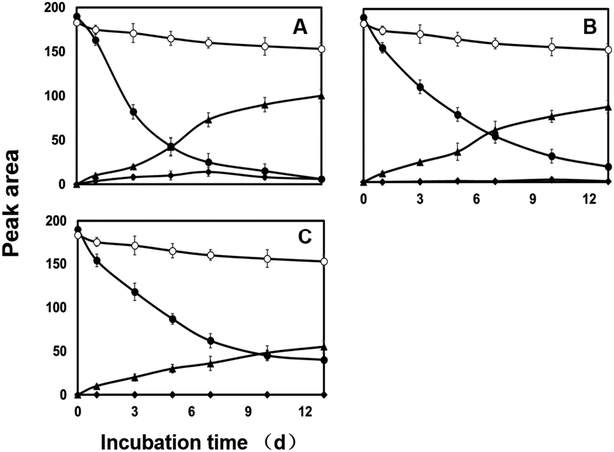
Both CYP450s and flavin monooxygenases (FMO) are phase I metabolic enzymes that catalyze the oxidation of sulfur and phosphorus compounds of xenobiotic compounds.39 The enzyme inhibitor piperonyl butoxide (PB) and methimazole (MZ) were introduced to explore the response in diazinon metabolism.25In PB-treated cultures, a large portion of diazinon still persisted compared to control. For example, residual diazinon was approximately 25–70% of the initial dose (Fig. 1B–D) after 13 days. M2, as a major metabolite, its concentration gradually increased until the end of the experiment in all cultures. However, the concentrations of M2 were far less than those of the control. M1 was only at trace level (2 mg L−1, Fig. 1B) or was not observed (10, 50 mg L−1, Fig. 1C and D) in PB-treatment experiment. Compared with the control, the degradation of diazinon was inhibited in the presence of PB, indicating that CYP450s may be involved in the biotransformation of diazinon. Previously, inhibition of CYP450s by metyrapone or carbon monoxide resulting in the attenuation of biotransformation reaction has been reported.40,41 In MZ-treated cultures, diazinon rapidly dissipated in the low level culture (2 mg L−1 of MZ) and was undetectable at 13th day, while significant amounts of diazinon (5–20% of initial dose) still persisted in the high level cultures (10, 50 mg L−1 of MZ) (Fig. 2). Overall profiles of diazinon and metabolites with the lowest concentration of MZ (2 mg L−1) were not distinguishable from those of the control (Fig. 2A). However, a less amount but similar response has been observed at higher concentrations of inhibitor (10 and 50 mg L−1) (Fig. 2B and C). For example, M1 was almost not observed during the whole experiment at high concentrations, while M2 gave the similar kinetic responses with those of the control or PB-treatment experiments. In this study, the residue of diazinon in MZ-treated culture was lower than that in PB-treated culture, which indicated that CYP450s were the major contributors to biotransformation. But the role of FMO in the oxidative metabolism was also not negligible,25,36,42 which was mainly reflected in the inhibitory effect of MZ on biotransformation. Lim et al. (2017) have previously reported that CYP450s were the main catabolic enzymes in xenobiotic biotransformation and contribution of FMO was limited.43 The change in M1 was more pronounced only at the lowest concentration of inhibitor (2 mg L−1 of PB). This may be due to the inhibition of the activity of the responsible enzyme at higher dose, which affects the oxidative desulfurization process. In conclusion, CYP450s play a major role in the oxidative metabolism of diazinon.
On the basis of these results, the metabolic pathway of diazinon was proposed (Fig. 5). In the presence of C. elegans, two major pathways of biotransformation are proposed. The first pathway is oxidative desulfurization and hydrolysis reactions, and the second pathway is hydroxylation reaction. Two reactions in the first pathway were common in some animal experiments44 and human liver microsome experiments.11,45,46 Under the catalysis of CYP450s, diazinon was directly transformed into three different metabolites (M1, M2 and M3). M1 (diazoxon) was formed with the oxidative desulfurization pathway, M2 and M3 were formed with the hydrolysis reaction, which is consistent with the almost immediate onset of M2 and M3 at 2 h after treatment and fragment ion analysis. The low-toxic pyrimidinol (M2) was also obtained from diazinon via M1 with oxidative desulfurization, hydrolysis pathways, which is the main degradation product in some soil,47,48 plant49 and animal44 experiments, as well as documented in other pesticides.50–52 For example, chlorpyrifos can form diethyl phosphate, diethyl phosphorothioate and 2,3,5-trichloro-4-pyridinyl alcohol through the above reactions.53 In this study, M1 is more toxic than diazinon, and there may be four ways to continue to decompose it. It will be degraded to M2 and M4 with the first pathway, and it will also be degraded to M5 and M7 with the second pathway. By observing the structure of minor metabolites (M5, M7, M8), we speculate on the occurrence of hydroxylation reactions, which may be related to CYP450s.36
One of the pathway for M4 is the same as that of M2, it could be formed from diazinon via M1 (diazinon → M1 → M4). Here, we also detected diethyl phosphate (M4) and diethyl phosphorothioate (M3) except pyrimidinol, which indicated the occurrence of dearylation. We captured the trend that M3 increased firstly and decreased gradually, while M4 continued to increase within 13 days. This may be the result of partial or complete conversion of M3 to M4, so another way of M4 is formed from diazinon via M3 with hydrolysis and oxidative desulfurization pathways (diazinon → M3 → M4). The rate of dearylation and desulfurization was related to the chemical structure of each pesticide and the CYP450s that catalyze the reactions.11,38 Previous study using microsome from human liver showed that diazinon was more readily detoxified (dearylation) than bioactivated (desulphuration) under the catalysis of CYP450s,45 which was necessary to fully understand the toxicological effects of pesticides on human body.
Furthermore, Casida (2011) had proved that CYP450s have situ selectivity in hydroxylation.54 M5 was firstly observed on the 5th day and M7 on the 7th day. By comparing the structure of these two metabolites, we speculated that a hydroxylation reaction had occurred. M5 was further metabolized through a dehydration reaction to give the respective isopropenyl substituted compounds M6, which was detected on the 7th day. M8 was also found except M1, M2, M3, and M4 on the first day. Based on the metabolic profiling, M8 can only come from M2, which may be the result of hydroxylation reaction.
4. Conclusions
This study has demonstrated the biodegradation of diazinon in C. elegans and described its metabolic pathways. Eight metabolites were fully or tentatively identified with GC-MS and 1H, 13C NMR. CYP450s and FMO were involved in the phase I metabolism of diazinon. In addition, to fully understand the toxicological effects of pesticides on the human body, it is necessary to determine the relative importance of different pathways in future experiments.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This study was carried out with the support of the “Research Program for Agricultural Science & Technology Development (Project PJ0140182018)”, National Institute of Agricultural Sciences, and Rural Development Administration, Republic of Korea.References
- B. D. Siegfried and M. E. Scharf, Mechanisms of organophosphate resistance in insects, in Biochemical sites of insecticide action and resistance, Springer, Berlin, Heidelberg, 2001, pp. 269–291 Search PubMed.
- M. Boussabbeh, I. B. Salem, M. Hamdi, S. B. Fradj, S. Abid-Essefi and H. Bacha, Environ. Sci. Pollut. Res., 2016, 23, 2882–2889 CrossRef CAS PubMed.
- R. L. Camacho-Morales and J. E. Sánchez, Mushroom Biotechnology, Academic Press., 2016, pp. 203–214 Search PubMed.
- T. M. A. M. Thabit and M. A. H. El-Naggar, Soil Environ., 2013, 32, 96–102 Search PubMed.
- C. D. S. Tomlin, The pesticide manual, BCPC Publications, Alton, Hampshire, UK, 14th edn, 2006, pp. 526–527 Search PubMed.
- V. Aggarwal, X. Deng, A. Tuli and K. S. Goh, Diazinon—chemistry and environmental fate: a California perspective, in Reviews of Environmental Contamination and Toxicology, Springer, New York, NY, 2013, vol. 223, pp. 107–140 Search PubMed.
- G. Briceño, M. S. Fuentes, O. Rubilar, M. Jorquera, G. Tortella, G. Palma, M. J. Amoroso and M. C. Diez, J. Basic Microbiol., 2015, 55, 293–302 CrossRef PubMed.
- P. S. C. Rao and A. G. Hornsby, Behavior of pesticides in soils and water, Circular E-Oklahoma State University, Cooperative Extension Service, USA, 1991, p. 142 Search PubMed.
- O. Akturk, H. Demirin, R. Sutcu, N. Yilmaz, H. Koylu and I. Altuntas, Cell Biol. Toxicol., 2006, 22, 455–461 CrossRef CAS PubMed.
- W. Wang, S. M. Luo, J. Y. Ma, W. Shen and S. Yin, J. Agric. Food Chem., 2018, 67, 19–31 CrossRef PubMed.
- W. A. Kappers, R. J. Edwards, S. Murray and A. R. Boobis, Toxicol. Appl. Pharmacol., 2001, 177, 68–76 CrossRef CAS PubMed.
- R. Khaghani and M. R. Zare, Health Scope, 2019, 9(1), e83067 CrossRef.
- M. D. Shah and M. Iqbal, Food Chem. Toxicol., 2010, 48, 3345–3353 CrossRef CAS PubMed.
- H. Lee, E. Kim, Y. Shin, J. H. Lee, H. G. Hur and J. H. Kim, J. Korean Soc. Appl. Biol. Chem., 2016, 59, 9–14 CrossRef CAS.
- A. F. Machin, H. Rogers, A. J. Cross, M. P. Quick, L. C. Howell and N. F. Janes, Pest Manag. Sci., 1975, 6, 461–473 CrossRef CAS.
- L. Ezzi, Z. Haouas, I. B. Salah, A. Sakly, I. Grissa, S. Chakroun, E. Kerkeni, M. Hassine, M. Mehdi and H. B. Cheikh, Environ. Sci. Pollut. Res., 2016, 23, 11163–11170 CrossRef CAS PubMed.
- F. Iverson, D. L. Grant and J. Lacroix, Bull. Environ. Contam. Toxicol., 1975, 13, 611–618 CrossRef CAS PubMed.
- S. Watanabe, U. Kuzhiumparambil and S. Fu, AAPS J., 2018, 20, 42 CrossRef PubMed.
- F. Amani, A. A. S. Sinegani, F. Ebrahimi and S. Nazarian, Biological Journal of Microorganism, 2019, 7, 27–39 CrossRef PubMed.
- A. El-Ghany and I. A. Masmali, J. Plant Pathol. Microbiol., 2016, 7, 1–7 Search PubMed.
- L. G. Sultatos, J. Toxicol. Environ. Health, 1994, 43, 271–289 CrossRef CAS PubMed.
- D. Zhang, Y. Yang, J. E. A. Leakey and C. E. Cerniglia, FEMS Microbiol. Lett., 1996, 138, 221–226 CrossRef CAS PubMed.
- J. D. Moody, J. P. Freeman and C. E. Cerniglia, Drug Metab. Dispos., 1999, 27, 1157–1164 CAS.
- C. J. Cha, D. R. Doerge and C. E. Cerniglia, Appl. Environ. Microbiol., 2001, 67, 4358–4360 CrossRef CAS PubMed.
- Y. Z. Zhu, Y. S. Keum, L. Yang, H. Lee, H. Park and J. H. Kim, J. Agric. Food Chem., 2010, 58, 12379–12384 CrossRef CAS PubMed.
- W. Palmer-Brown, P. L. de Melo Souza and C. D. Murphy, Environ. Sci. Pollut. Res., 2019, 26, 1414–1421 CrossRef CAS PubMed.
- R. F. Wang, W. W. Cao, A. A. Khan and C. E. Cerniglia, FEMS Microbiol. Lett., 2000, 188, 55–61 CrossRef CAS PubMed.
- J. Amadio, K. Gordon and C. D. Murphy, Appl. Environ. Microbiol., 2010, 76, 6299–6303 CrossRef CAS PubMed.
- R. Hussain, M. Ahmed, T. A. Khan and Y. Akhter, Appl. Microbiol. Biotechnol., 2020, 104, 989–999 CrossRef CAS PubMed.
- M. Forrest, K. A. Lord, N. Walker and H. C. Woodville, Environ. Pollut., Ser. A, 1981, 24, 93–104 CrossRef CAS.
- V. N. Kouloumbos, D. F. Tsipi, A. E. Hiskia, D. Nikolic and R. B. Breemen, J. Am. Soc. Mass Spectrom., 2003, 14, 803–817 CrossRef CAS PubMed.
- T. Carins, E. G. Siegmund and J. E. Froberg, Bull. Environ. Contam. Toxicol., 1985, 35, 291–295 CrossRef PubMed.
- R. S. H. Yang, E. Hodgson and W. C. Dauterman, J. Agric. Food Chem., 1971, 19, 10–13 CrossRef CAS PubMed.
- R. S. H. Yang, E. Hodgson and W. C. Dauterman, J. Agric. Food Chem., 1971, 19, 14–19 CrossRef CAS.
- Y. Z. Zhu, M. Fu, I. H. Jeong, J. H. Kim and C. J. Zhang, J. Agric. Food Chem., 2017, 65, 10711–10718 CrossRef CAS PubMed.
- T. R. Roberts, P. J. Jewess, P. W. Lee, P. H. Nicholls and J. R. Plimmer, Part 2: fungicides. Metabolic pathways of agrochemicals, The Royal Society of Chemistry, Cambridge, UK, 1999, pp. 966–969 Search PubMed.
- M. Ibáñez, J. V. Sancho, Ó. J. Pozo and F. Hernández, Anal. Bioanal. Chem., 2006, 384, 448–457 CrossRef PubMed.
- W. Müecke, K. O. Alt and H. E. Esser, J. Agric. Food Chem., 1970, 18, 208–212 CrossRef PubMed.
- Z. C. Wang, Z. J. Kang, X. Y. Shi and X. W. Gao, Chin. J. Pestic. Sci., 2015, 17, 1–14 CAS.
- W. Yang, T. Jing, D. Acosta and P. J. Davis, Xenobiotica, 1993, 23, 973–982 CrossRef CAS PubMed.
- D. Zhang, E. B. Hansen, J. Deck, T. M. Heinze, A. Henderson, W. A. Korfmacher and C. E. Cerniglia, Xenobiotica, 1997, 27, 301–315 CrossRef CAS PubMed.
- S. K. Krueger and D. E. Williams, Pharmacol. Ther., 2005, 106, 357–387 CrossRef CAS PubMed.
- D. S. Lim, D. H. Lim, J. H. Lee, E. T. Oh and Y. S. Keum, J. Agric. Food Chem., 2017, 65, 3056–3064 CrossRef CAS PubMed.
- A. W. Abu-Qare and M. B. Abou-Donia, Fresenius’ J. Anal. Chem., 2001, 370, 403–407 CrossRef CAS PubMed.
- C. Sams, J. Cocker and M. S. Lennard, Xenobiotica, 2004, 34, 861–873 CrossRef CAS PubMed.
- C. A. Ellison, Y. Tian, J. B. Knaak, P. J. Kostyniak and J. R. Olson, Drug Metab. Dispos., 2012, 40, 1–5 CrossRef CAS PubMed.
- M. S. Díaz-Cruz and D. Barceló, J. Chromatogr. A, 2006, 1132, 21–27 CrossRef PubMed.
- H. Shemer and K. G. Linden, J. Hazard. Mater., 2006, 136, 553–559 CrossRef CAS PubMed.
- S. K. Lee, K. Kim, C. K. Park and E. C. Hwang, Agric. Res. Seoul Natl. Univ., 1985, 69–76 CAS.
- T. Ma and J. E. Chambers, Food Chem. Toxicol., 1994, 32, 763–767 CrossRef CAS PubMed.
- J. E. Chambers, T. Ma, J. S. Boone and H. W. Chambers, Life Sci., 1994, 54, 1357–1364 CrossRef CAS PubMed.
- K. A. Y. A. Ptashne, R. M. Wolcott and R. A. Neal, J. Pharmacol. Exp. Ther., 1971, 179, 380–385 CAS.
- T. S. Poet, H. Wu, A. A. Kousba and C. Timchalk, Toxicol. Sci., 2003, 72, 193–200 CrossRef CAS PubMed.
- J. E. Casida, J. Agric. Food Chem., 2011, 59, 2923–2931 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this paper. |
| This journal is © The Royal Society of Chemistry 2020 |