Open Access Article
Open Access ArticleConstruction of gold-siRNANPR1 nanoparticles for effective and quick silencing of NPR1 in Arabidopsis thaliana†
Wen-Xue Lei‡
ab,
Zi-Shuai An‡ab,
Bai-Hong Zhang‡ab,
Qian Wu ab,
Wen-Jun Gongab,
Jin-Ming Li
ab,
Wen-Jun Gongab,
Jin-Ming Li *ab and
Wen-Li Chen
*ab and
Wen-Li Chen *ab
*ab
aMOE Key Laboratory of Laser Life Science & Institute of Laser Life Science, College of Biophotonics, South China Normal University, Guangzhou 510631, China. E-mail: chenwl@scnu.edu.cn; lijinm@scnu.edu.cn; Fax: +86-20-85216052; Tel: +86-20-85211436-8611
bGuangdong Provincial Key Laboratory of Laser Life Science, College of Biophotonics, South China Normal University, Guangzhou 510631, China
First published on 20th May 2020
Abstract
In recent years, gold nanoparticles (AuNPs) have been widely used as gene silencing agents and therapeutics for treatment of cancers due to their high transfection efficiency and lack of cytotoxicity, but their roles in gene silencing in plants have not yet been reported. Here, we report synthesis of AuNPs-branched polyethylenimine and its integration with the small interfering RNAs (siRNA) of NPR1 to form a AuNPs-siRNANPR1 compound. Our results showed that AuNPs-siRNANPR1 was capable of infiltrating into Arabidopsis cells. AuNPs-siRNANPR1 silenced 80% of the NPR1 gene in Arabidopsis. Bacteriostatic and ion leakage experiments suggest that the NPR1 gene in Arabidopsis leaves was silenced by AuNPs-siRNANPR1. In Columbia-0 plants, compared with the control group treated with buffer solution, the AuNPs-siRNANPR1 treatment significantly increased the number of colonies and cell death, and the leaves turned yellow, similar to the phenotype of the npr1 leaves. These results indicated this AuNPs-siRNANPR1 silencing the NPR1 gene method is simple, effective and quick (3 days), and a powerful tool to study gene functions in plants.
Introduction
Research on plant gene function heavily relies on agrobacterium-mediated gene gun bombardment and PEG-transformed protoplast regeneration. However, these experiments are time-consuming and sometime not effective.1 Thus, development of a rapid and effective method for silencing genes will be essential to accelerate research on plant gene regulation. Inorganic nanoparticles (INPs) as gene vectors, have recently attracted great attention for their non-immunogenicity, immunity from pathogens, and low toxicity, although their transfection efficiency is lower than that of viral vectors. INPs have advantages of large loading capacity, high stability, and easy preparation and storage.2–6 Gold nanoparticles (AuNPs) are especially useful for delivery of small interfering RNA (siRNA) and protect RNAs from nuclease degradation.7 The multilayer AuNPs integrated with siRNA effectively silenced the genes, as shown by the assay of luciferase activity.8 In addition, the PEGylated AuNPs containing SH-siRNA showed significant RNA inhibition (RNAi) in HuH-7 cells.2 However, there is no report on usage of AuNPs in plant gene silencing.RNAi induced by siRNA does not completely silence the targeted gene9 and is thus specially suitable for functional studies of lethal phenotype genes such as ATG6 in Arabidopsis. In this study, we reported the construction of AuNPs-siRNANPR1, and demonstrated for the first time its effective silencing the NPR1 gene (nonexpressor of pathogenesis-related gene 1) in Arabidopsis.
Materials and methods
Preparation of AuNPs
AuNPs were prepared by chemical reduction.10–12 The solution of 25 mL 0.056% chloroauric acid (Aladdin, Shanghai, China) in a 50 mL conical bottle was added by 0.8 mL 1% branched polyethylenimine (Shyuanye, Shanghai, China) 25 kD (AuNPs-PEI) and stirred at 500 rpm continuously for 24 hours at room temperature (25 °C). When the color of the solution no longer change, the reaction is completed and the solution is filtered using a 0.2 μm microporous filter (Biofil, Guangzhou, China). The AuNPs modified by PEI were stored in the original volume of distilled water and in a refrigerator at 4 °C. We modified the preparation procedure of AuNPs-PEI, used a 30 kDa dialysis bag (MYM, Beijing, China) for dialysis to remove PEI which was not combined with AuNPs to allow minimization of PEI interference.13 The size and zeta potential of the nanomedicines were measured using ZEN3690 zetasizer (Malvern, USA).The actual amount of chloroauric acid in preparation: 0.056% g mL−1 × 25 mL = 0.014 g; the actual mass of AuNPs: mAuNPs/mchlorogold acid = 197/394, mAuNPs = 0.007 g; so we estimated the concentration of nano gold as follows: 0.007 g/25.8 mL = 2.71 × 10−4 g mL−1 ≈ 1 mM.
Preparation of AuNPs-siRNANPR1
In the pre experiment, we found that the NPR1 gene could be silenced by mixing siRNA (20 μM) and AuNPs (1 mM) in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 and diluting 15 times with a infiltration buffer (10 mM MgCl2 and 10 mM MES (2-(N-morpholino) ethanesulfonic acid) pH 5.7) for infiltration into the leaves of Arabidopsis thaliana.
9 and diluting 15 times with a infiltration buffer (10 mM MgCl2 and 10 mM MES (2-(N-morpholino) ethanesulfonic acid) pH 5.7) for infiltration into the leaves of Arabidopsis thaliana.
Infiltrating leaves
In this study, the leaves collected for the experiment were all infiltrated by different solutions (the leaves without infiltration were not used in the experiment). Leaves of 3 weeks-old Arabidopsis were selected for infiltration of AuNPs. AuNPs infiltration was performed by pressure infiltration with a 1 mL syringe through the abaxial leaf surface. In this process, a small amount (about 10 μL) of liquid will gently and slowly be penetrated into the leaves. Infiltration is best performed when the plant stomata are opened in the morning. We chose to infiltrate at 8–10 in the morning. The cultivation conditions of plant materials were 16 h light (6 a.m. to 10 p.m.), 8 h dark (10 p.m. to 6 a.m.), the light intensity is 120 μmol L−1 m−2 s, the ambient temperature was controlled at 23 °C, and the relative humidity of the environment is 82%. Infiltration is a time-consuming process. Through many experiments, we found that it is easy to infiltrate at 8 to 10 a.m. (plants get light from 6 a.m.). The infiltration of tobacco (Nicotiana benthamiana) leaves is the same as that of Arabidopsis, but because the Nicotiana benthamiana leaves are more tender, the infiltration needs to be as gentle as possible to avoid mechanical damage. The standard for successful infiltration is to see the wetting of entire leaves.Extraction of Arabidopsis protoplasts
The healthy leaves of Arabidopsis thaliana (Col-0) before flowering were selected. SiRNA (20 μM) and AuNPs (1 mM) were mixed in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 and diluted 15 times with a infiltration buffer. The protoplasts were extracted after 1–2 hours of infiltration. The enzymatic hydrolysate was prepared with cellulose R-10 (1.5%) (Yakult Honsha, Tokyo, Japan), macrozyme R-10 (0.75%) (Yakult Honsha, Tokyo, Japan), CaCl2 (10 mM), BSA (0.1%), mannitol (0.5 M) and MES (10 mM, pH = 5.7). The epidermis was peeled off with transparent tape, and several parts of the leaves were cut off and placed in 5 mL enzymatic hydrolysate. After 3 hours of the enzymatic reaction at 26 °C in darkness, under 40–50 rpm oscillation, the solution turned from yellow to green (strictly controlling the temperature of enzymatic hydrolysis). The enzymatic hydrolysate passes through 200 mesh sieve holes, the filtrate is placed in centrifugal tube, and the residue in beaker is washed with W5 that is prepared with NaCl (154 mM), CaCl2 (125 mM), KCl (50 mM), MES (2 mM) and glucose (5 mM). The mixed was centrifuged at 4 °C, at 60 × g for 5 minutes, and remove supernatant. The protoplast was washed with 5 mL W5 and centrifuged at 100 × g, for 3 min. After three times of W5 wash, add 5 mL W5 and gently pipet it for use.14,15
9 and diluted 15 times with a infiltration buffer. The protoplasts were extracted after 1–2 hours of infiltration. The enzymatic hydrolysate was prepared with cellulose R-10 (1.5%) (Yakult Honsha, Tokyo, Japan), macrozyme R-10 (0.75%) (Yakult Honsha, Tokyo, Japan), CaCl2 (10 mM), BSA (0.1%), mannitol (0.5 M) and MES (10 mM, pH = 5.7). The epidermis was peeled off with transparent tape, and several parts of the leaves were cut off and placed in 5 mL enzymatic hydrolysate. After 3 hours of the enzymatic reaction at 26 °C in darkness, under 40–50 rpm oscillation, the solution turned from yellow to green (strictly controlling the temperature of enzymatic hydrolysis). The enzymatic hydrolysate passes through 200 mesh sieve holes, the filtrate is placed in centrifugal tube, and the residue in beaker is washed with W5 that is prepared with NaCl (154 mM), CaCl2 (125 mM), KCl (50 mM), MES (2 mM) and glucose (5 mM). The mixed was centrifuged at 4 °C, at 60 × g for 5 minutes, and remove supernatant. The protoplast was washed with 5 mL W5 and centrifuged at 100 × g, for 3 min. After three times of W5 wash, add 5 mL W5 and gently pipet it for use.14,15
Confocal microscopy imaging
The images were obtained in Zeiss LSM 880 confocal laser-scanning microscope (Carl Zeiss Microscopy GmbH, Jena, Germany) and analyzed with ZEN software (Carl Zeiss)16 to observe infiltration of AuNPs-siRNANPR1 into plant cells. FAM-siRNANPR1 (20 μM) and AuNPs (1 mM) were mixed in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 and diluted 15 times with a infiltration buffer. After infiltrating this compound into Nicotiana benthamiana leaves or Arabidopsis leaves for 1 hour, images or movies were collected with excitation at 492 and 518 nm wavelengths (green is the fluorescence of FAM; red is the spontaneous fluorescence of chlorophyll).
9 and diluted 15 times with a infiltration buffer. After infiltrating this compound into Nicotiana benthamiana leaves or Arabidopsis leaves for 1 hour, images or movies were collected with excitation at 492 and 518 nm wavelengths (green is the fluorescence of FAM; red is the spontaneous fluorescence of chlorophyll).
Western blot (WB)
Leaves were collected at different time points, after the AuNPs-siRNANPR1 are infiltrated into the Col-0 and NPR1-GFP. We improved WB method on the basis of the protocols of Yan17 and Chen:18 leaves from 4 weeks-old plants were treated with AuNPs-siRNANPR1. 0.4 g samples were collected, frozen in liquid nitrogen, and homogenized in protein extraction buffer (5 mM Tris–HCl, 15 mM NaCl, 0.5 mM EDTA, 1% (v/v) β-ME, 0.2% (v/v) Triton X-100, 0.5% (v/v) Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, and 40 μM MG-115). Homogenates were centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 17 min at 4 °C. Proteins were then denatured at 75 °C with 5× SDS-PAGE loading buffer supplemented with DL-dithiothreitol (100 mM DTT) for 10 min. Total proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) with a constant current of 100 mA and then transferred onto polyvinylidene difluoride membranes at a constant voltage 100 V (Bio-Rad, CA, USA). Skimmed milk (5% (w/v)) was prepared by dissolving milk powder in Tris-buffered Saline Tween-20 buffer and used to block the resulting membrane (Merck, Darmstadt, Germany) and to dilute antibodies for immunoblotting. Antibodies of green fluorescent protein (GFP) (JL-8 Monoclonal Antibody, Fisher Scientific, 632381) and β-actin were purchased from the Beyotime Institute of Biotechnology (Shanghai, China); antibodies of NPR1 were purchased from the Agrisera (Swedish). The β-actin antibody was used as control for analysis.
000 g for 17 min at 4 °C. Proteins were then denatured at 75 °C with 5× SDS-PAGE loading buffer supplemented with DL-dithiothreitol (100 mM DTT) for 10 min. Total proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) with a constant current of 100 mA and then transferred onto polyvinylidene difluoride membranes at a constant voltage 100 V (Bio-Rad, CA, USA). Skimmed milk (5% (w/v)) was prepared by dissolving milk powder in Tris-buffered Saline Tween-20 buffer and used to block the resulting membrane (Merck, Darmstadt, Germany) and to dilute antibodies for immunoblotting. Antibodies of green fluorescent protein (GFP) (JL-8 Monoclonal Antibody, Fisher Scientific, 632381) and β-actin were purchased from the Beyotime Institute of Biotechnology (Shanghai, China); antibodies of NPR1 were purchased from the Agrisera (Swedish). The β-actin antibody was used as control for analysis.
qPCR analysis
After AuNPs-siRNANPR1 or AuNPs-siRNAn.c. (negative control) were infiltrated into Col-0 and NPR1-GFP, leave samples were collected at different time points for evaluation of the silencing effect on NPR1 gene. Leaves (0.05 g) were collected and frozen in liquid nitrogen and stored at −80 °C. RNA was extracted by plant total RNA isolation kit (Sangon biotech, Shanghai, China) and then treated with primeScript RT Master Mix (Takara, Tokyo, Japan) according to the manufacturer's instructions, to synthesize the cDNA.19,20 Refer to Yan's procedure for real time PCR (qPCR).17 The primers used for NPR1 are listed as follows: F: GATCGCAAAACAAGCCACTATGG and R: ATCGAGCAGCGTCATCTTCAATT.Growth and inoculation of strains
This study used the P. syringae strain (Pst DC3000 (AvrRps4)) provided by Dr Yang of South China Normal University. Pst DC3000 (AvrRps4) was cultured in King's B medium containing 50 μg mL−1 rifampicin and 100 μg mL−1 kanamycin for 18 hours at 28 °C, centrifuged (4000 rpm min−1, twice), washed with 10 mM MgCl2 twice, and then resuspended in 10 mM MgCl2 and diluted to the required concentration (OD600 = 1.0).15Infection experiment of pathogenic bacteria
After 3 days of AuNPs-siRNANPR1 infiltration, bacterial solution of Pst DC3000 (AvrRps4) (OD600 = 0.001 diluted from the stock solution of OD = 1.0) was infiltrated and sampled after 3 days. Two small round leaves were beaten with sterilized puncher (different plants need to be wiped with sterilized water soaked paper before punching), placed in 1.5 mL EP tube, smashed with a grinding rod, and then added with 500 μL MgCl2 (10 mM) and mashed as fully as possible. In 96-well plate, dilute the sample to 6 concentrations, and then coat on King's B media containing 50 μg mL−1 rifampicin and 100 μg mL−1 kanamycin at 28 °C. The colony count was observed and recorded 2 days later.21,22Conductivity experiment
After 3 days of AuNPs-siRNANPR1 infiltration, bacterial solution of Pst DC3000 (AvrRps4) with OD600 = 0.02 (diluted from stock solution of OD = 1.0) was infiltrated and sampled after 3 days. Six small round leaves with a diameter of 8 mm were obtained with a perforator (75% alcohol disinfection). Then the leaves were placed in a 50 mL centrifugal tube, washed twice with 50 mL ddH2O, and added 15 mL ddH2O 10 minutes later. Measure and record data with conductivity meter (Mettler Toledo FE30-FiveEasy, Shanghai, China) every 2 hours.21,22Design of different siRNAs
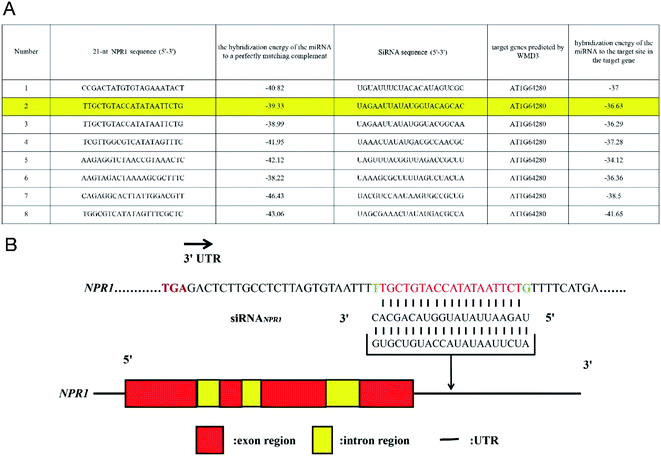
On the basis of Arabidopsis Information Resource (TAIR) genome annotation, we designed siRNA for silencing NPR1 gene by using the Web MicroRNA Designer platform (WMD3, http://wmd3.weigelworld.org), sequence is: UAGAAUUAUAUGGUACAGCAC (Fig. 1A). SiRNA was inserted into 3′ untranslated region (UTR) of NPR1 gene (Fig. 1B).23,24 We added the FAM fluorescent sequence to the 5′ end of the siRNA to construct FAM-siRNANPR1. The sequence of “UUCUCCGAACGUGUCACGUTT” was used as a negative control. SiRNAn.c. is a universal negative control without species specificity. It has been used in mice, humans and other species as a negative control.25–27Statistics of plant resistance to Pst DC3000 (AvrRps4)
Count the number of colonies with corresponding dilution on the solid medium, and then calculate the bacterial growth by the following formula:| Colony-forming units (cfu)/leaf disc = ((colonies × 10dilution/10) × 500)/2 |
Plant material
Arabidopsis wild type (ecotype Columbia, Col) comes from Professor Kohki Yoshimoto of Japan Plant Science Center. NPR1-GFP (with the npr1-2 mutant as the background) comes from the laboratory of Xinnian Dong of Duke University was obtained by the floral dip method.28 Nicotiana benthamiana wild type comes from the laboratory of Xinnian Dong of Duke University also.Statistical analysis
All tests were repeated at least three times and the results were analyzed by GraphPad Prism 8.16 Statistical analyses were performed using Student's t-test (*, p < 0.05, **, p < 0.01, and***, p < 0.001). Each value was the mean ± S.D. of three independent replicates.Result & discussion
Synthesis of AuNPs and siRNA-AuNPs
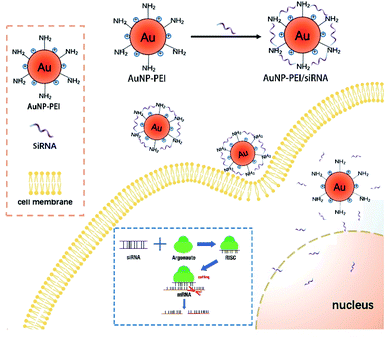
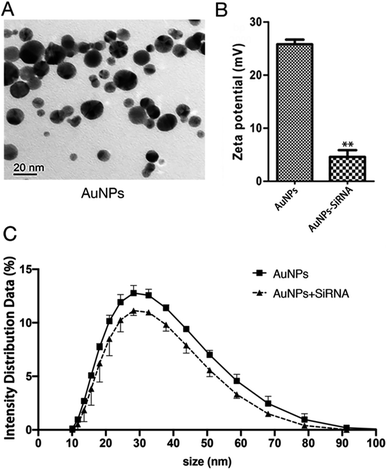
AuNPs have been used for delivery of siRNA, DNAs and proteins.29–31 The following mechanism has been proposed for the function of AuNPs: (1) oligonucleotides are covalently linked to the mercaptan sulfide to AuNPs to make functional surface; (2) the siRNA-AuNPs are recognized by the surface receptors of cells through endocytosis and penetrate cell membrane; and (3) siRNA are released from the particles and entered the nucleus (Fig. 2).8The concentration of AuNPs is about 1 mM and the size of the synthesized AuNPs was in range of 3–45 nm (Fig. 3A) and similar to that reported in ref. 32, and might thus be easily absorbed by cells. In order to explore the optimal binding ratio of AuNPs and siRNA, we tested different combination of AuNPs with siRNA.
We mixed siRNA (20 μM) and AuNPs (1 mM) in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 and diluting 15 times with a infiltration buffer (10 mM MgCl2 and 10 mM MES, pH 5.7) for measure particle size and potential. The same infiltration buffer and AuNPs were mixed at a ratio of 1
9 and diluting 15 times with a infiltration buffer (10 mM MgCl2 and 10 mM MES, pH 5.7) for measure particle size and potential. The same infiltration buffer and AuNPs were mixed at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 and diluted 15 times with this infiltration buffer to measure the particle size and potential of AuNPs before binding to siRNA. The average size of AuNPs before binding siRNA is about 34 nm and the zeta potential is 25 mV. The particle size and zeta potential of AuNPs changed after binding with siRNA (Fig. 3B, C, Tables S2 and S3†). The particle size of AuNPs increased by about 4 nm (Fig. 3C and Table S3†). Because the surface of AuNPs is positively charged and the molecular surface of siRNA is negatively charged, the zeta potential reduces from about 25 mV to about 5 mV (Fig. 3B and Table S2†). These changes in particle size and zeta potential can explain the successful formation of the AuNPs-siRNA compounds.33 In addition, only slight change in particle size was found before and after bonding, which indicated that the combination of AuNPs and siRNA will not cause the AuNPs to become too large and thus affect the infiltration effect.
9 and diluted 15 times with this infiltration buffer to measure the particle size and potential of AuNPs before binding to siRNA. The average size of AuNPs before binding siRNA is about 34 nm and the zeta potential is 25 mV. The particle size and zeta potential of AuNPs changed after binding with siRNA (Fig. 3B, C, Tables S2 and S3†). The particle size of AuNPs increased by about 4 nm (Fig. 3C and Table S3†). Because the surface of AuNPs is positively charged and the molecular surface of siRNA is negatively charged, the zeta potential reduces from about 25 mV to about 5 mV (Fig. 3B and Table S2†). These changes in particle size and zeta potential can explain the successful formation of the AuNPs-siRNA compounds.33 In addition, only slight change in particle size was found before and after bonding, which indicated that the combination of AuNPs and siRNA will not cause the AuNPs to become too large and thus affect the infiltration effect.
For their inert and non-toxic properties and good biocompatibility, AuNPs were considered as a good carrier of nucleotides.34 In addition, AuNPs are easily prepared for desired size and dispersibility of biomolecules in complex with oligopeptides or nucleic acids, and may thus be integrated into biological systems.35 The large ratio of surface area/volume of the AuNPs will provide space for loading RNA or DNA.36 As a nucleic acid carrier, AuNPs can improve the survival time of DNA and siRNA in cellular environment, the efficiency of cell uptake from endoplasmic vesicles, and the subsequent release.30,37 Positively charged AuNPs bind efficiently to negatively charged nucleic acid and also prevent them from being digested by enzymes.38 Through the “proton sponge” effect, the combination of branched PEI and AuNPs can transfer plasmid DNA to cells approximately 15 times more efficiently than unmodified PEI.39 We modified the preparation procedure of AuNPs-PEI, and dialyze the suspension to remove PEI that is not combined with AuNPs to minimize PEI interference.13
AuNPs-FAM-siRNANPR1 was observed to enter plant cells
The FAM-siRNANPR1 was mixed with AuNPs (AuNPs-siRNANPR1) in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
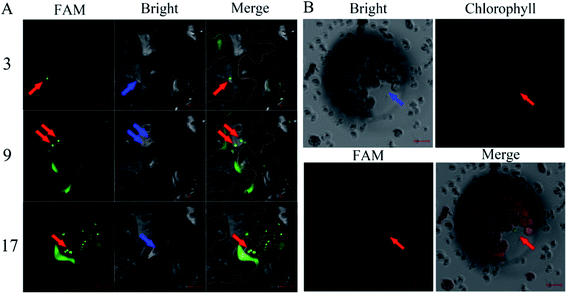
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, diluted three times with the buffer of a buffer of 10 mM MgCl2 and 10 mM MES, incubated at 25 °C under 60 rpm shaking for half an hour, and then diluted five times. In order to investigate whether the silencing complex can enter plant cells, 5 weeks-old leaves of Nicotiana benthamiana were selected for infiltration of AuNPs-FAM-siRNANPR1. AuNPs-FAM-siRNANPR1 infiltration was performed by pressure infiltration with a 1 mL syringe through the abaxial leaf surface. The criterion for successful infiltration was that the whole leaf was wet without obvious mechanical damage. Considering the quenching of fluorescence signal, we only observed the fluorescence of 1 h injection. In a relatively short period of time, only part of the AuNPs-FAM-siRNANPR1 can enter the cell through endocytosis. So fluorescent signals can be detected in the cytoplasm and extracellular matrix. In order to determine the position of AuNPs-FAM-siRNANPR1 in the cell and eliminate the possibility that the FAM fluorescence of the upper layer overlaps with that of the lower layer, the mesophyll cells were scanned 30 layers from top to bottom, and the images of each layer were decomposed. AuNPs-FAM-siRNANPR1 are small spherical bright spots. FAM fluorescence exists in different positions of different layers, which can effectively explain that AuNPs-FAM-siRNANPR1 are in the inner part of Nicotiana benthamiana mesophyll cells. At the same time, we provide the decomposition diagram of each layer (Fig. S1†). Then we selected the representative third layer, the ninth layer and the seventeenth layer for detailed discussion (Fig. 4A). Because the mesophyll cells of Nicotiana benthamiana are densely packed, we use white or orange to outline the borders of two adjacent cells for easy distinction, and the numbers represent different scanning layers. In order to further determine the location of AuNPs-siRNANPR1, AuNPs-FAM-siRNANPR1 was infiltrated into Arabidopsis thaliana leaves (4 weeks-old Col-0 plants). After 1–2 hours of infiltration, plant protoplast was extracted from the leaves and the fluorescence in cells was observed by confocal microscope (Fig. 4B). The movies clearly showed that AuNPs-FAM-siRNANPR1 was located in the plants (ESI Movies 1 and 2†). We also provide an image of protoplasts of Arabidopsis leaf epidermal cells 1 hour after injection by AuNPs-FAM-siRNANPR1 (Fig. S2†). Since there is no chloroplast in epidermal cells, we can exclude the possibility that green fluorescence is the light of chloroplast. This shows that these small spherical bright spots must be AuNPs-FAM-siRNANPR1. Previous research shows AuNPs-siRNA can efficiently transfect animal cells,37,40,41 our experiments show for the first time that the AuNPs-siRNA gene-silencing compounds can be infiltrated into plant cells.
9, diluted three times with the buffer of a buffer of 10 mM MgCl2 and 10 mM MES, incubated at 25 °C under 60 rpm shaking for half an hour, and then diluted five times. In order to investigate whether the silencing complex can enter plant cells, 5 weeks-old leaves of Nicotiana benthamiana were selected for infiltration of AuNPs-FAM-siRNANPR1. AuNPs-FAM-siRNANPR1 infiltration was performed by pressure infiltration with a 1 mL syringe through the abaxial leaf surface. The criterion for successful infiltration was that the whole leaf was wet without obvious mechanical damage. Considering the quenching of fluorescence signal, we only observed the fluorescence of 1 h injection. In a relatively short period of time, only part of the AuNPs-FAM-siRNANPR1 can enter the cell through endocytosis. So fluorescent signals can be detected in the cytoplasm and extracellular matrix. In order to determine the position of AuNPs-FAM-siRNANPR1 in the cell and eliminate the possibility that the FAM fluorescence of the upper layer overlaps with that of the lower layer, the mesophyll cells were scanned 30 layers from top to bottom, and the images of each layer were decomposed. AuNPs-FAM-siRNANPR1 are small spherical bright spots. FAM fluorescence exists in different positions of different layers, which can effectively explain that AuNPs-FAM-siRNANPR1 are in the inner part of Nicotiana benthamiana mesophyll cells. At the same time, we provide the decomposition diagram of each layer (Fig. S1†). Then we selected the representative third layer, the ninth layer and the seventeenth layer for detailed discussion (Fig. 4A). Because the mesophyll cells of Nicotiana benthamiana are densely packed, we use white or orange to outline the borders of two adjacent cells for easy distinction, and the numbers represent different scanning layers. In order to further determine the location of AuNPs-siRNANPR1, AuNPs-FAM-siRNANPR1 was infiltrated into Arabidopsis thaliana leaves (4 weeks-old Col-0 plants). After 1–2 hours of infiltration, plant protoplast was extracted from the leaves and the fluorescence in cells was observed by confocal microscope (Fig. 4B). The movies clearly showed that AuNPs-FAM-siRNANPR1 was located in the plants (ESI Movies 1 and 2†). We also provide an image of protoplasts of Arabidopsis leaf epidermal cells 1 hour after injection by AuNPs-FAM-siRNANPR1 (Fig. S2†). Since there is no chloroplast in epidermal cells, we can exclude the possibility that green fluorescence is the light of chloroplast. This shows that these small spherical bright spots must be AuNPs-FAM-siRNANPR1. Previous research shows AuNPs-siRNA can efficiently transfect animal cells,37,40,41 our experiments show for the first time that the AuNPs-siRNA gene-silencing compounds can be infiltrated into plant cells.
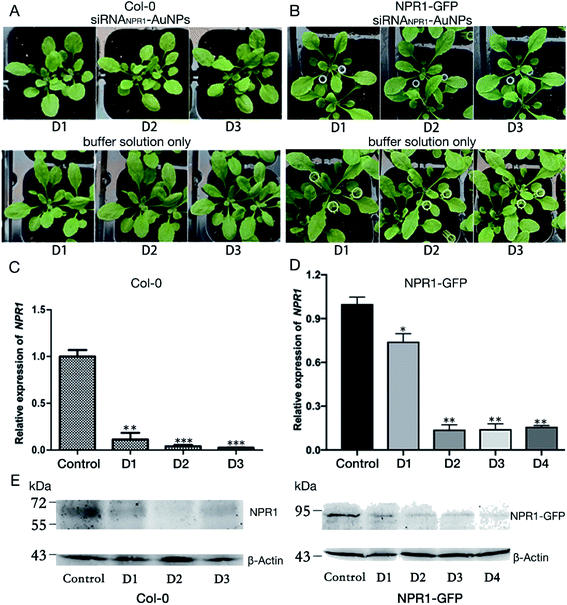
AuNPs-siRNANPR1 silence NPR1 gene in Arabidopsis
Optimization on different combination of chemical compounds and their concentration may yield effective binding to the surface of the nanoparticles and thus save test time and cost. Multi-functional AuNPs have been shown to be very stable without acute toxicity and not to impair cell viability,40 and thus are used in the plant gene silence. We chose Col-0 and NPR1-GFP as the experimental plant for test of duration of gene silencing, and used AuNPs-siRNANPR1 to silence the NPR1 gene for bacteriostatic and ion leakage experiments.We conducted a series of experiments to explore whether AuNPs-siRNANPR1 can successfully silence the NPR1 gene of plants. The AuNPs-siRNANPR1 were infiltrated into leaves of the Col-0 and NPR1-GFP (overexpressing NPR1 gene in npr1-2 background) Arabidopsis. The infiltration buffer was used to dilute the silencing complexes of AuNPs-siRNANPR1 and as the blank control. There was no significant difference in phenotypes between the leaves infiltrated with silencing complex or plain buffer, indicating that the AuNPs-siRNANPR1 do not affect the normal growth of both Col-0 and NPR1-GFP plants (Fig. 5A and B). To find out whether the compound of AuNPs and siRNANPR1 impacts the health of plants, we observed and recorded the phenotype of Arabidopsis after infiltration of the compound. The results showed that the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 ratio of 20 μM siRNANPR1 and 1 mM AuNPs is appropriate and has no impact on the normal growth of plants.
9 ratio of 20 μM siRNANPR1 and 1 mM AuNPs is appropriate and has no impact on the normal growth of plants.
The qPCR analysis shows that the NPR1 gene is effectively silenced after 1 or 2 days of infiltration of AuNPs-siRNANPR1 into Arabidopsis Col-0 and NPR1-GFP plants. The effect of gene silencing is about 80% (Fig. 5C and D). The NPR1 protein in plant is significantly decreased after 1 and 2 days of infiltration of AuNPs-siRNANPR1 to the Col-0 and NPR1-GFP plants (Fig. 5E). These results suggested that we have successfully silenced NPR1 gene.
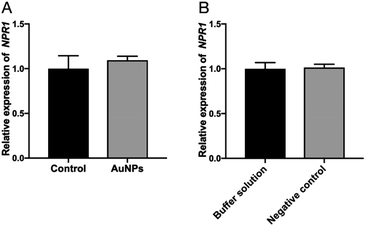
The infiltration of AuNPs and unrelated siRNA did not affect the expression of NPR1 gene
In order to detect whether the AuNPs affect NPR1 gene, the AuNPs were infiltrated into leaves of the NPR1-GFP Arabidopsis. Infiltration buffer was used to dilute the AuNPs and also as the blank control. Then, we combined siRNAn.c. with AuNPs and infiltrated it into NPR1-GFP (the other operations were the same as before). Three days after infiltration, the leaves were collected for qPCR experiment. These experiments suggesting that AuNPs does not affect the expression of NPR1 gene (Fig. 6A), simultaneously, unrelated siRNA does not silence NPR1 gene (Fig. 6B), and that siRNANPR1 induced silence of part of the NPR1 gene (Fig. 5C and D).Bacterial stress and ion leakage experiments confirmed AuNPs-siRNANPR1 silencing NPR1
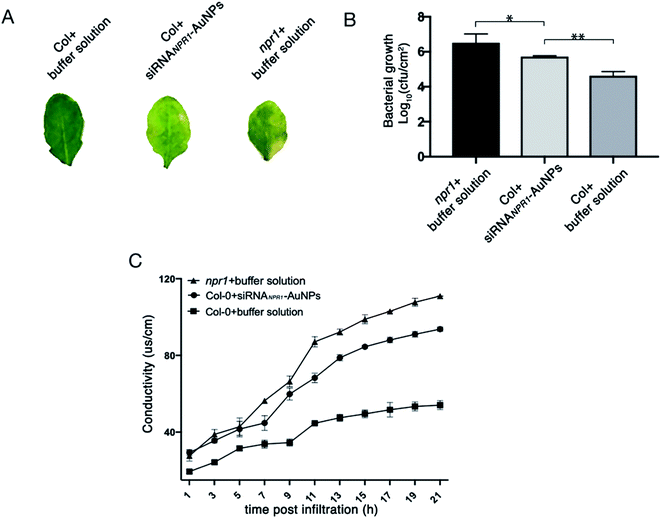
NPR1 is involved in the regulation of cell death and disease resistance in response to non-pathogenic pathogens. After 3 days of infiltrating AuNPs-siRNANPR1 into Arabidopsis Col-0 plants (the infiltration buffer was infiltrated in npr1 and Col-0), Pst DC3000 (AvrRps4) with OD600 = 0.001 was infiltrated. Three days later, the infiltrated leaves were collected and subjected to a bacteriostatic test. The avirulent Pst DC3000 (AvrRps4) infects the leaves of Arabidopsis thaliana, leading to spread of chlorosis symptoms.15 The results showed that the leaves of NPR1 silenced plants with infiltration of Pst DC3000 (AvrRps4) turned to yellow and so do the leaves of npr1 mutant, but the leaves of Col-0 plant remain green (Fig. 7A). The collected leaves were ground, dilute to six concentrations then they were cultured on King's B solid medium resistant to kanamycin and rifampicin. Under Pst DC3000 (AvrRps4) stress, the NPR1 partly silenced plants showed bacterial growth (the number of colony of the plants) more than that of the Col-0 plants, but less than that of the npr1 mutant (Fig. 7B, S3 and Table S1†). Three days after AuNPs-siRNANPR1 were infiltrated into Arabidopsis thaliana Col-0 plants (buffer was infiltrated in npr1 and Col-0), Pst DC3000 (AvrRps4) OD600 = 0.02 was infiltrated and the conductivity was measured one hour later. The NPR1 partly silenced plants showed cell death more than the Col-0 plants, but less than the npr1 mutant (Fig. 7C). These results suggested that AuNPs-siRNANPR1 partially silenced NPR1 gene, thus indicating our gene silencing technique is effective.In short, we show, for the first time, AuNPs-siRNANPR1 can be infiltrated into the leaf cells and partially silence the NPR1 gene in Arabidopsis thaliana. In comparison with the current methods for plant gene silencing, which require pollutant Agrobacterium tumefaciens and are time consuming, our AuNPs-siRNANPR1 for silencing plant genes have advantages of short time, low toxicity, and low dosage. This convenient and rapid method for gene silencing in plants provides a new way to study the gene function of plants.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the National Natural Science Foundation of China (Grants 31570256), and a grant from the science and technology project of Guangzhou (Grant No. 201805010002). We would like to thank Professor Hengming Ke, UNC-Chapel Hill School of Medicine of USA for proofreading of the manuscript.References
- P. Wang, F. J. Zhao and P. M. Kopittke, Engineering Crops without Genome Integration Using Nanotechnology, Trends Plant Sci., 2019, 24, 574–577 CrossRef CAS PubMed.
- M. Oishi, J. Nakaogami, T. Ishii and Y. Nagasaki, Smart PEGylated gold nanoparticles for the cytoplasmic delivery of siRNA to induce enhanced gene silencing, Chem. Lett., 2006, 35, 1046–1047 CrossRef CAS.
- G. Han, C. C. You, B. J. Kim, R. S. Turingan, N. S. Forbes, C. T. Martin and V. M. Rotello, Light-regulated release of DNA and its delivery to nuclei by means of photolabile gold nanoparticles, Angew. Chem., Int. Ed. Engl., 2006, 45, 3165–3169 CrossRef CAS PubMed.
- K. Saha, S. S. Agasti, C. Kim, X. N. Li and V. M. Rotello, Gold Nanoparticles in Chemical and Biological Sensing, Chem. Rev., 2012, 112, 2739–2779 CrossRef CAS PubMed.
- F. Y. Cheng, C. H. Su, Y. S. Yang, C. S. Yeh, C. Y. Tsai, C. L. Wu, M. T. Wu and D. B. Shieh, Characterization of aqueous dispersions of Fe(3)O(4) nanoparticles and their biomedical applications, Biomaterials, 2005, 26, 729–738 CrossRef CAS PubMed.
- G. Jia, H. Wang, L. Yan, X. Wang, R. Pei, T. Yan, Y. Zhao and X. Guo, Cytotoxicity of carbon nanomaterials: single-wall nanotube, multi-wall nanotube, and fullerene, Environ. Sci. Technol., 2005, 39, 1378–1383 CrossRef CAS PubMed.
- N. L. Rosi, D. A. Giljohann, C. S. Thaxton, A. K. Lytton-Jean, M. S. Han and C. A. Mirkin, Oligonucleotide-modified gold nanoparticles for intracellular gene regulation, Science, 2006, 312, 1027–1030 CrossRef CAS PubMed.
- S. K. Lee, M. S. Han, S. Asokan and C. H. Tung, Effective gene silencing by multilayered siRNA-coated gold nanoparticles, Small, 2011, 7, 364–370 CrossRef CAS PubMed.
- R. Schwab, J. F. Palatnik, M. Riester, C. Schommer, M. Schmid and D. Weigel, Specific effects of microRNAs on the plant transcriptome, Dev. Cell, 2005, 8, 517–527 CrossRef CAS PubMed.
- K. Kim, H. B. Lee, J. W. Lee, H. K. Park and K. S. Shin, Self-assembly of poly(ethylenimine)-capped Au nanoparticles at a toluene-water interface for efficient surface-enhanced Raman scattering, Langmuir, 2008, 24, 7178–7183 CrossRef CAS PubMed.
- V. Cebrian, F. Martin-Saavedra, C. Yague, M. Arruebo, J. Santamaria and N. Vilaboa, Size-dependent transfection efficiency of PEI-coated gold nanoparticles, Acta Biomater., 2011, 7, 3645–3655 CrossRef CAS PubMed.
- W. J. Song, J. Z. Du, T. M. Sun, P. Z. Zhang and J. Wang, Gold Nanoparticles Capped with Polyethyleneimine for Enhanced siRNA Delivery, Small, 2010, 6, 239–246 CrossRef CAS PubMed.
- A. Aljabali, Y. Akkam, M. Al Zoubi, K. Al-Batayneh, B. Al-Trad, O. A. Alrob, A. Alkilany, M. Benamara and D. Evans, Synthesis of Gold Nanoparticles Using Leaf Extract of Ziziphus zizyphus and their Antimicrobial Activity, Nanomaterials, 2018, 8(3), 174 CrossRef PubMed.
- Y. H. Bi, W. L. Chen, W. N. Zhang, Q. Zhou, L. J. Yun and D. Xing, Production of reactive oxygen species, impairment of photosynthetic function and dynamic changes in mitochondria are early events in cadmium-induced cell death in Arabidopsis thaliana, Biol. Cell., 2009, 101, 629–643 CrossRef CAS PubMed.
- J. J. Dong and W. L. Chen, The Role of Autophagy in Chloroplast Degradation and Chlorophagy in Immune Defenses during Pst DC3000 (AvrRps4) Infection, PLoS One, 2013, 8, e73091 CrossRef CAS PubMed.
- Y. Jiao, M. Srba, J. C. Wang and W. L. Chen, Correlation of Autophagosome Formation with Degradation and Endocytosis Arabidopsis Regulator of G-Protein Signaling (RGS1) through ATG8a, Int. J. Mol. Sci., 2019, 20(17), 4190 CrossRef PubMed.
- Q. Q. Yan, J. C. Wang, Z. Q. Fu and W. L. Chen, Endocytosis of AtRGS1 Is Regulated by the Autophagy Pathway after D-Glucose Stimulation, Front. Plant Sci., 2017, 8 Search PubMed.
- J. Chen, R. Mohan, Y. Zhang, M. Li, H. Chen, I. A. Palmer, M. Chang, G. Qi, S. H. Spoel, T. Mengiste, D. Wang, F. Liu and Z. Q. Fu, NPR1 Promotes Its Own and Target Gene Expression in Plant Defense by Recruiting CDK8, Plant Physiol., 2019, 181, 289–304 CrossRef CAS PubMed.
- D. Mackey, Y. Belkhadir, J. M. Alonso, J. R. Ecker and J. L. Dangl, Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance, Cell, 2003, 112, 379–389 CrossRef CAS PubMed.
- J. Caplan, M. Padmanabhan and S. P. Dinesh-Kumar, Mant NB-LRR immune receptors: from recognition to transcriptional reprogramming, Cell Host Microbe, 2008, 3, 126–135 CrossRef CAS PubMed.
- X. D. Wang, Y. Y. Gao, Q. Q. Yan and W. L. Chen, Salicylic acid promotes autophagy via NPR3 and NPR4 in Arabidopsis senescence and innate immune response, Acta Physiol. Plant., 2016, 38, 241 CrossRef.
- Y. Y. Gao, X. D. Wang, C. Ma and W. L. Chen, EDS1-mediated activation of autophagy regulates Pst DC3000 (AvrRps4)-induced programmed cell death in Arabidopsis, Acta Physiol. Plant., 2016, 38, 150 CrossRef.
- S. Ossowski, R. Schwab and D. Weigel, Gene silencing in plants using artificial microRNAs and other small RNAs, Plant J., 2008, 53, 674–690 CrossRef CAS.
- R. Schwab, S. Ossowski, M. Riester, N. Warthmann and D. Weigel, Highly specific gene silencing by artificial microRNAs in Arabidopsis, Plant Cell, 2006, 18, 1121–1133 CrossRef CAS.
- L. Gao, K. Wu, Z. Liu, X. Yao, S. Yuan, W. Tao, L. Yi, G. Yu, Z. Hou, D. Fan, Y. Tian, J. Liu, Z. J. Chen and J. Liu, Chromatin Accessibility Landscape in Human Early Embryos and Its Association with Evolution, Cell, 2018, 173, 248–259 e15 CrossRef CAS PubMed.
- L. Huang, J. Jin, P. Deighan, E. Kiner, L. McReynolds and J. Lieberman, Efficient and specific gene knockdown by small interfering RNAs produced in bacteria, Nat. Biotechnol., 2013, 31, 350–356 CrossRef CAS.
- H. Qian, X. Deng, Z. W. Huang, J. Wei, C. H. Ding, R. X. Feng, X. Zeng, Y. X. Chen, J. Ding, L. Qiu, Z. L. Hu, X. Zhang, H. Y. Wang, J. P. Zhang and W. F. Xie, An HNF1alpha-regulated feedback circuit modulates hepatic fibrogenesis via the crosstalk between hepatocytes and hepatic stellate cells, Cell Res., 2015, 25, 930–945 CrossRef CAS PubMed.
- M. Kinkema, W. Fan and X. Dong, Nuclear localization of NPR1 is required for activation of PR gene expression, Plant Cell, 2000, 12, 2339–2350 CrossRef CAS.
- C. P. Jen, Y. H. Chen, C. S. Fan, C. S. Yeh, Y. C. Lin, D. B. Shieh, C. L. Wu, D. H. Chen and C. H. Chou, A nonviral transfection approach in vitro: the design of a gold nanoparticle vector joint with microelectromechanical systems, Langmuir, 2004, 20, 1369–1374 CrossRef CAS PubMed.
- S. Rana, A. Bajaj, R. Mout and V. M. Rotello, Monolayer coated gold nanoparticles for delivery applications, Adv. Drug Delivery Rev., 2012, 64, 200–216 CrossRef CAS PubMed.
- J. S. Lee, J. J. Green, K. T. Love, J. Sunshine, R. Langer and D. G. Anderson, Gold, poly(beta-amino ester) nanoparticles for small interfering RNA delivery, Nano Lett., 2009, 9, 2402–2406 CrossRef CAS PubMed.
- H. A. Meng, M. Liong, T. A. Xia, Z. X. Li, Z. X. Ji, J. I. Zink and A. E. Nel, Engineered Design of Mesoporous Silica Nanoparticles to Deliver Doxorubicin and P-Glycoprotein siRNA to Overcome Drug Resistance in a Cancer Cell Line, ACS Nano, 2010, 4, 4539–4550 CrossRef CAS PubMed.
- R. Thiramanas and R. Laocharoensuk, Competitive binding of polyethyleneimine-coated gold nanoparticles to enzymes and bacteria: a key mechanism for low-level colorimetric detection of gram-positive and gram-negative bacteria, Microchim. Acta, 2016, 183, 389–396 CrossRef CAS.
- E. E. Connor, J. Mwamuka, A. Gole, C. J. Murphy and M. D. Wyatt, Gold nanoparticles are taken up by human cells but do not cause acute cytotoxicity, Small, 2005, 1, 325–327 CrossRef CAS PubMed.
- M. C. Daniel and D. Astruc, Gold nanoparticles: assembly, supramolecular chemistry, quantum-size-related properties, and applications toward biology, catalysis, and nanotechnology, Chem. Rev., 2004, 104, 293–346 CrossRef CAS PubMed.
- J. C. Love, L. A. Estroff, J. K. Kriebel, R. G. Nuzzo and G. M. Whitesides, Self-assembled monolayers of thiolates on metals as a form of nanotechnology, Chem. Rev., 2005, 105, 1103–1169 CrossRef CAS PubMed.
- E. H. Jeong, G. Jung, C. A. Hong and H. Lee, Gold nanoparticle (AuNP)-based drug delivery and molecular imaging for biomedical applications, Arch. Pharmacal Res., 2014, 37, 53–59 CrossRef CAS PubMed.
- G. Han, C. T. Martin and V. M. Rotello, Stability of gold nanoparticle-bound DNA toward biological, physical, and chemical agents, Chem. Biol. Drug Des., 2006, 67, 78–82 CrossRef CAS PubMed.
- M. Thomas and A. M. Klibanov, Conjugation to gold nanoparticles enhances polyethylenimine's transfer of plasmid DNA into mammalian cells, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 9138–9143 CrossRef CAS PubMed.
- J. Conde, A. Ambrosone, V. Sanz, Y. Hernandez, V. Marchesano, F. Tian, H. Child, C. C. Berry, M. R. Ibarra, P. V. Baptista, C. Tortiglione and J. M. de la Fuente, Design of multifunctional gold nanoparticles for in vitro and in vivo gene silencing, ACS Nano, 2012, 6, 8316–8324 CrossRef CAS PubMed.
- C. M. Goodman, K. K. Sandhu, J. M. Simard, S. W. Smith and V. M. Rotello, Gold nanoparticle-mediated transfection of mammalian cells, Abstr. Pap. Am. Chem. Soc., 2002, 224, U131 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra02156c |
| ‡ Co-first authors. |
| This journal is © The Royal Society of Chemistry 2020 |