Open Access Article
Open Access ArticleDocking and in vitro molecular biology studies of p-anisidine-appended 1-hydroxy-2-acetonapthanone Schiff base lanthanum(III) complexes†
V. Sathiyanarayananab,
P. Varun Prasathc,
P. Chandra Sekharde,
K. Ravichandran c,
D. Easwaramoorthy*a,
Faruq Mohammad
c,
D. Easwaramoorthy*a,
Faruq Mohammad f,
Hamad A. Al-Lohedanf,
Won Chun Oh
f,
Hamad A. Al-Lohedanf,
Won Chun Oh g and
Suresh Sagadevan
g and
Suresh Sagadevan *h
*h
aDepartment of Chemistry, B. S. Abdur Rahman Crescent Institute of Science and Technology, Vandalur, Chennai 600048, Tamilnadu, India. E-mail: easwar@bsauniv.ac.in
bDepartment of Analytical Chemistry, Vanta Bioscience Limited, SIPCOT Industrial Complex Gummidipundi, Tamilnadu – 601201, India
cDepartment of Analytical Chemistry, Madras University, Guindy, Chennai 600 025, Tamil Nadu, India
dDepartment of Genetic Toxicology, Palamur Biosciences Pvt Ltd, Mahabubnagar 509002, Telangana, India
eSchool of Life Sciences, Manipal University, Manipal, 576104, Karnataka, India
fDepartment of Chemistry, College of Science, King Saud University, P. O. Box 2455, Riyadh, 11451, Kingdom of Saudi Arabia
gDepartment of Advanced Materials Science and Engineering, Hanseo University, Seosan-si, Chungnam 356-706, Korea
hNanotechnology & Catalysis Research Centre, University of Malaya, Kuala Lumpur 50603, Malaysia. E-mail: drsureshnao@gmail.com
First published on 24th April 2020
Abstract
A new series of lanthanum(III) complexes was synthesized using a p-anisidine-appended 1-hydroxy-2-acetonapthanone (3) Schiff base and characterized via spectroscopic methods. The ligand was synthesized via sonication and the crystalline product was characterized using X-ray crystallography. The genotoxicity of the compound was assessed primarily by the bacterial reverse mutation (Ames) test and the in vitro mammalian chromosome aberration test; in both cases, the samarium complex 5 was found to be non-mutagenic. The anti-tumor activity of complexes 4, 5, and 6 was assayed against HeLa tumor cells and screened using the MTT assay. The IC50 value of complex 5 was found to be 34 ± 1.2 μg mL−1 and this compound exhibited superior activity towards the cells compared to 4 and 6. These results were further confirmed by Hoechst 33258 staining and AO/EI dual staining, which indicated that the cells underwent an apoptosis mechanism in a dose-dependent manner. The apoptosis was further confirmed by the formation of ladders in the DNA fragmentation assay, and the western blot analysis of complex 5 suggested that the cells underwent the caspase-3-dependent pathway with PARP cleavage. Furthermore, the docking studies of complex 5 with HSA showed that it was situated in a hydrophilic cavity held by the electrostatic attraction of four hydrogen-bonding interactions. PDB ID:1BNA binds with complex 5 via strong π–π stacking interactions, which facilitate binding with the major grooves of DNA strands. The above-mentioned results illustrate that for complex 5, mitochondrion-mediated apoptosis occurs via caspase-3 activation. Complex 5 binds with DNA via intercalation because of S-phase cell cycle arrest in the HeLa cells.
1 Introduction
Therapeutic drugs developed based on the use of organometallic chemistry and its principles have been successfully applied for many years to fight malignant diseases such as cancer.1 Cisplatin was the first potential organometallic drug used for the treatment of various cancers, which was discovered by an Italian researcher in the 19th century but only reported in the 1960s.2,3 However, despite its potential therapeutic activity, its side-effects such as nephrotoxicity, neurotoxicity, and inherited or acquired resistance limit its utility in the treatment of cancer.4 In recent decades, chemists have synthesized therapeutically selective metal-based drugs by exploiting the properties of metal ions such as different coordination numbers, geometry, variable redox states, thermodynamic and kinetic characteristics, and intrinsic properties. Besides, targeted drug delivery has been a growing area with a significant impact on the field of cancer therapeutics. Most chemotherapeutic drugs such as cisplatin, carboplatin, and oxaliplatin5 target the nucleus, while the photodynamic therapy drug Photofrin® targets the mitochondria.6 Thus, different targeting of cancer cells requires more careful drug design and therefore, the data on the specific three-dimensional structure of targets have been utilized to improve the drug efficacy at the targeted sites. Accordingly, to visualize these activities on computer-aided systems without actual experimentation, molecular docking approaches have been attracting increasing attention in recent years to enhance targeted drug delivery and the simultaneous protection of non-targeted sites.The chemistry of Schiff base complexes has been the focus in organometallic chemistry and these complexes have potential applications in clinical and pharmaceutical chemistry due to their excellent biological activities, including anti-tumor activity.7 Schiff bases are electron donors and form multidentate ligands, resulting in an increase in binding constants upon complexation with d- and f-block metal ions.8 The introduction of salicylic acid as an auxiliary ligand in a Schiff base Cu(II) complex9 resulted in stronger anti-cancer activity than that of the parent compound, thereby suggesting the synergistic effects of Schiff base complexes, and non-steroidal anti-inflammatory drugs (NSAIDs) may be involved in the cell-killing process. Similarly, some new nanoscale water-soluble complexes of Ni and Zn have been reported, in which the Zn(II)-derived complexes exhibited higher cytotoxicity against the human hepatoma (HepG2) and HeLa cell lines than that of Ni(II) complexes.10 Also, some novel tumor-targeting photo-activated anti-cancer agents, ternary oxovanadium(IV) complexes of (acridinyl)dipyridophenazine and vitamin B6 Schiff base ligands, have been reported and found to be non-toxic in the dark.11 These complexes show photo-induced apoptotic cell death via singlet oxygen 1O2 generation and specific localization to the endoplasmic reticulum (ER). The apoptotic cell death possibly results from the generation of 1O2, which triggers the ER stress response (ERSR).11 An iron(III) complex of a Schiff base ligand derived from vitamin B6 and thiosemicarbazide12 induced apoptosis by the generation of ROS (reactive oxygen species) upon irradiation with visible light (400–700 nm). The biological preference of enantiomeric fluoro-substituted benzothiazole Schiff base-valine Cu(II) and Zn(II) complexes was evaluated via DNA binding studies. When tested in human cervical cancer cells, the L-enantiomer of the Cu(II) complex was found to be highly selective and the biologically preferred chiral form of the drug exhibited DNA binding propensity and cleavage activity.13
Many recent reports have described the biological applications of lanthanide-coordinated complexes and great attention has been paid recently to their excellent potency as anti-tumor agents. A mitochondria-targeted selective photocytotoxic ferrocenyl neodymium(III) complex of terpyridine and coumarin was reported by Akthar et al., which showed remarkable phototoxicity towards HeLa cells.12 MRI contrast reagents such as crystalline Gd(III) and Gd(III) chelates grafted on macromolecular nanoparticles14 have been reported as selective and sensitive functionalities for the diagnosis of cancer and occurrence of metastasis. Subsequently, trinuclear (RuII–Gd2III) and tetranuclear (RuII–Gd3III) d–f heterometallic complexes were reported to act as contrast-enhancing agents for MRI and optical probes for fluorescence imaging.15 Recently, the anti-cancer activities of trinuclear d–f heterometallic complexes such as (RuII–Gd2III) have also been reported.16
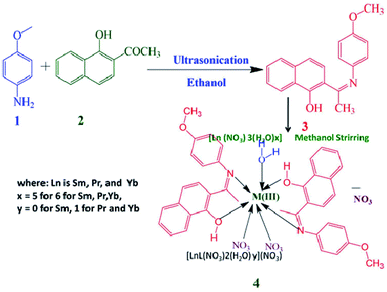
Schiff base ligands are a class of organic compounds containing an imine functional group, which are formed via a simple synthetic strategy, the condensation of primary amines with carbonyl compounds. They are flexible and can coordinate many lanthanide systems12 due to their excellent binding capacity towards rare-earth ions, and they can also sensitize the properties of lanthanide ions.16 Furthermore, Schiff bases are excellent chemo-sensors for the selective recognition and sensing of trace metal ions in biological and environmental settings.17 Jean-Pierre et al. reported the synthesis of bimetallic Cu and Gd complexes with 2-hydroxy-3-methoxy benzaldehyde.18 The significant photosensitive character of lanthanides has been used in various fields of life sciences such as luminescent probes in biomedical assays, electroluminescent devices and UV dosimeters,19 fluorescent lighting and luminescence sensors by taking advantage of their long excited-state lifetime and large Stoke shifts.20 Accordingly, the present work is focused on the synthesis and characterization of a bidentate Schiff base ligand (labeled as 3) and its three lanthanides(III) complexes, praseodymium (Pr), samarium (Sm), and ytterbium (Yb).21,22 The reason why these lanthanide(III) complexes were selected is due their superior oxidoreduction ability and magnetic property even in the presence of some intracellular reduction agents such as ascorbate and thiols, in addition to their favorable luminescent nature provided by fast charge transfer, and 4f ↔ 5d and f ↔ f transitions.22–24 Subsequently, this series of compounds was characterized, and then we examined their in vitro anti-cancer activity, toxicity, and DNA cleavage mechanism, together with molecular docking studies of their binding with DNA.
2 Results and discussion
Synthesis and characterization
The Schiff base ligand 3 was synthesized via the condensation reaction between 1-hydroxy-2-acetonapthone and p-anisidine in methanol. The lanthanide complexes of Pr (labelled as 4), Sm (labelled as 5), and Yb (labelled as 6) were prepared as shown in Scheme 1. The yields of the prepared complexes were found to be moderate to high (75% to 80%). The elemental analysis and conductivity of the ligand and complexes are presented in Table 1. Based on the elemental analysis, the complexes have the general formula [Ln(3)2(NO3)2·xH2O]·NO3, where x = 0 for samarium and 1 for praseodymium and ytterbium.| Compound | Molecular weight | Observed m/z value [M + H] | Molar conductance | Elemental analysis (%) | ||
|---|---|---|---|---|---|---|
| C | N | H | ||||
| C19H17NO2 (3) | 291.13 | 292.25 | — | 78.269 (78.33) | 4.758 (4.81) | 5.872 (5.88) |
| PrC38H36N5O14 (4), [Pr(3)2(NO3)2H2O]·NO3 | 927.624 | 928.14 | 109 | 49.109 (49.20) | 7.452 (7.35) | 3.824 (3.91) |
| SmC38H34N5O13 (5), [Sm(3)2(NO3)2]·NO3 | 919.061 | 920.15 | 118 | 49.628 (49.66) | 7.521 (7.62) | 3.760 (3.73) |
| YbC38H36N5O14 (6), [Yb(3)2(NO3)2H2O]·NO3 | 959.756 | 960.15 | 114 | 47.489 (47.55) | 7.019 (7.30) | 3.592 (3.78) |
NMR spectra of the ligand
The 1H NMR spectrum of ligand 3 recorded in CDCl3 is shown in the ESI, Fig. S1.† In the spectrum, the peak at δ 2.473 ppm is attributed to the chemical shift of the acetyl protons (CH3) of the naphthyl ring, and the methoxy proton signals of the p-anisidine ring are observed at δ 3.844 ppm. These two signals confirm the condensation of an amine and a ketone, leading to the formation of the imine functionality. The aromatic protons of the naphthyl and toluidine moieties are observed at δ 8.522–7.450 and 6.944–7.065 ppm, respectively. The peak at δ 12.168 belongs to the hydroxyl protons in the molecule. Similarly, Fig. S2 (ESI†) presents the 13C NMR spectrum of ligand 3, where the signals observed in the range of δ 124.96–169.5 ppm are attributed to the chemical shift of the naphthyl carbons. The peak at δ 170.56 ppm is due to the carbon of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and the peak appearing at δ 16.54 ppm is due to the methyl carbon attached to C
N and the peak appearing at δ 16.54 ppm is due to the methyl carbon attached to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N. The peaks appearing at δ (C12, C13, C14, C15, C16, and C17) 124.74, 110.68, and 157.87 ppm are due to the carbon atoms in the p-anisidine ring system. The peak at δ 55.55 ppm is due to C18 on the methoxy group present in the p-anisidine ring system.
N. The peaks appearing at δ (C12, C13, C14, C15, C16, and C17) 124.74, 110.68, and 157.87 ppm are due to the carbon atoms in the p-anisidine ring system. The peak at δ 55.55 ppm is due to C18 on the methoxy group present in the p-anisidine ring system.
ESI-MS for ligand and complexes
An ESI-MS experiment was performed in the positive ionization mode for ligand 3 and complexes 4, 5, and 6. The ESI-MS spectrum of ligand 3 (ESI, Fig. S3†) displays a molecular ion peak at m/z 292.25, which is attributed to the protonated form of 3, [M − H]+. The mass spectra of lanthanum complexes 4, 5, and 6 are shown in Fig. S4, S5, and S6,† respectively, where the molecular ion peak was not observed for all the complexes in the spectra. The Pr(III) complex exhibited a peak at m/z 928.14, corresponding to the product of ([Pr(3)2(NO3)2(H2O)]NO3) + H+. Similarly, the ([Yb(3)2(NO3)2(H2O)]NO3) +H+ fragment was observed at m/z 960.15 for the Yb(III) complex. A different scenario was observed for the Sm(III) complex, where the coordination of two water molecules was observed at m/z 920.15, corresponding to the ([Sm(3)2(NO3)2(H2O)2]NO3) + H+ fragment ion. Since most of the lanthanides possess several stable and radioactive isotopes, their mass spectrum is more complicated than that of transition metal complexes. Thus, the LC-MS/MS patterns were studied to record the isotopic pattern of the prepared complexes, and from the results, a cluster of peaks with appropriate isotopic abundance was observed (m/z 928.14–931.16) for [Pr(3)2(NO3)2H2O]·NO3 (Fig. S4†). For the Sm complex, the pattern due to [Sm(3)2(NO3)2]·NO3 was observed in the region of m/z 913.14–925.15 (Fig. S5†). Similarly, for [Yb(3)2(NO3)2H2O]·NO3, an isotopic pattern was observed in the region of m/z 957.16–964.14 (Fig. S6†). Thus, these results support the elemental analysis results shown in Table 1.X-ray diffraction studies
Table 2 summarizes the crystal and refinement data for ligand 3, where the compound crystallized in the monoclinic space group P21/c and the unit cell parameters are: a = 7.931(1) Å, b = 17.938(1) Å, c = 10.999(1) Å, α = 90°, β = 103.853(2)°, γ = 90°, and V = 1519.3(2) Å3. The ORTEP plot of this compound with displacement thermal ellipsoids drawn at 30% probability level is shown in Fig. 1. In ligand 3, the atoms in the naphthalene moiety lie on a plane and the O1, C11, C19, and N1 atoms lie in the same plane as the naphthalene moiety, as evident from the torsion angle values of −179.6(2)° (C8–C9–C10–O1), 177.09(2)° (C7–C8–C9–C11), −177.11(2)° (C10–C9–C11–C19) and 179.39(2)° (C8–C9–C11–N1), respectively. The atoms in the anisole moiety are arranged in a planar fashion. The naphthalene ring is oriented equatorially to the planar anisole moiety, which can be detected from the dihedral angle value of 78.8(1)°. The intramolecular interaction (O–H⋯N) observed in this molecule allows the crystal packing, as described in Table 2.| Empirical formula | C19H17NO2 | |||
|---|---|---|---|---|
| Formula weight | 291.34 | |||
| Temperature | 293(2) K | |||
| Wavelength | 0.71073 Å | |||
| Crystal system, space group | Monoclinic, P21/c | |||
| Unit cell dimensions | a = 7.9308(6) Å, α = 90°, | |||
| b = 17.9382(12) Å, β = 103.853(2)°, | ||||
| c = 10.9994(7) Å, γ = 90° | ||||
| Volume | 1519.31(18) Å3 | |||
| Z, calculated density | 4, 1.274 mg m−3 | |||
| Absorption coefficient | 0.083 mm−1 | |||
| F(000) | 616 | |||
| Crystal size | 0.19 × 0.15 × 0.12 mm | |||
| Theta range for data collection | 2.22° to 27.94° | |||
| Limiting indices | −10 ≤ h ≤ 10, −23 ≤ k ≤ 23, −14 ≤ l ≤ 14 | |||
| Reflections collected/unique | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 493/3650 [Rint = 0.0283] 493/3650 [Rint = 0.0283] |
|||
| Completeness to theta = 27.94 | 99.80% | |||
| Refinement method | Full-matrix least-squares on F2 | |||
| Data/restraints/parameters | 3650/0/200 | |||
| Goodness-of-fit on F2 | 1.095 | |||
| Final R indices [I > 2σ(I)] | R1 = 0.0634, wR2 = 0.1494 | |||
| R indices (all data) | R1 = 0.1061, wR2 = 0.1791 | |||
| Extinction coefficient | 0.0071(14) | |||
| Largest diffraction peak and hole | 0.235 and −0.222 e Å−3 | |||
| Hydrogen bond interaction of ligand 3 | ||||
| D–H⋯A | d(D–H) | d(H⋯A) | d(D⋯A) | <(DHA) |
| O1–H1⋯N1 | 0.82 | 1.77 | 2.505(3) | 148 |
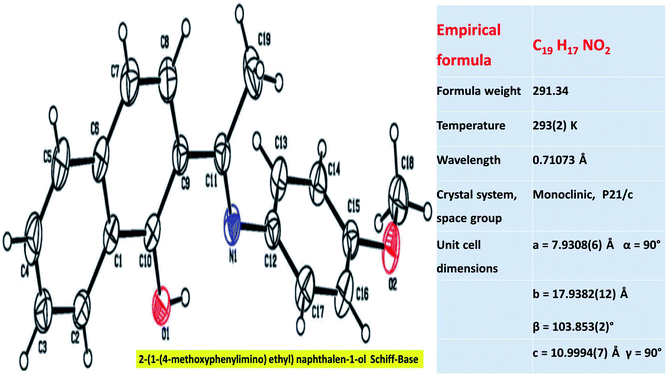 | ||
| Fig. 1 ORTEP plot of ligand 3 with the displacement thermal ellipsoids drawn at the 30% probability level. | ||
Molar conductivity
The ionic nature and type of electrolyte for the ligand and complexes were inferred from the room temperature molar conductivity measurements (Λm) performed in DMF solvent (Table 1). The conductivity values for complexes 4, 5, and 6 were calculated to be 109 S m2 mol−1, 118 S m2 mol−1 and 114 S m2 mol−1, respectively. From these values, it was inferred that these complexes are in the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, confirming that the free nitrate ions remain outside the coordination sphere in all the complexes.25 These results are in good agreement with those obtained from the FTIR studies discussed below.
1, confirming that the free nitrate ions remain outside the coordination sphere in all the complexes.25 These results are in good agreement with those obtained from the FTIR studies discussed below.
FTIR spectra
The representative FTIR spectra for 3, 4, 5, and 6 are shown in Fig. S7 (ESI†), where the shifts in the vibration frequency of ligand 3 upon complexion were observed for all the rare earth complexes. The free ligand 3 exhibited a strong stretching vibrational absorption band ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) in the region of 1630 cm−1. After complexation, this stretching frequency shifted to 1612 cm−1, similar to that reported in the literature.20 This illustrates that there was an increase in the double bond character of the imine due to the chelation of the imine group containing nitrogen in ligand 3 with the Pr(III) metal atom.26 This was further established by the presence of the medium band in the region of 498 cm−1, indicating the Pr(III) metal bonded with the nitrogen ν(Pr–N). The presence of a broad band at around 3264 cm−1 signifies the aromatic hydroxyl stretching due to the existence of intramolecular hydrogen bonding between the phenolic hydrogen and imine nitrogen atoms [O–H⋯N].27 The participation of OH groups in the coordination without the displacement of a proton from the phenolic OH28 group was ascertained by the presence of the strong band in the region of 3453 cm−1. The new non-ligand medium intensity band that appeared at around 542 cm−1 is assigned to the stretching frequency of Pr–O.29 The peaks observed at around 1489 cm−1 (ν1), 1024 cm−1 (ν2), 817 cm−1 (ν3), and 1294 cm−1 (ν4) confirm the participation of a nitrate ion present inside the coordination sphere.30 From the above data, it can be concluded that ligand 3 coordinates the praseodymium metal atom in a bidentate manner via the nitrogen of the amine group and the oxygen of the hydroxyl group.31 Complexes 5 and 6 show coordination behaviors similar to that of complex 4.
N) in the region of 1630 cm−1. After complexation, this stretching frequency shifted to 1612 cm−1, similar to that reported in the literature.20 This illustrates that there was an increase in the double bond character of the imine due to the chelation of the imine group containing nitrogen in ligand 3 with the Pr(III) metal atom.26 This was further established by the presence of the medium band in the region of 498 cm−1, indicating the Pr(III) metal bonded with the nitrogen ν(Pr–N). The presence of a broad band at around 3264 cm−1 signifies the aromatic hydroxyl stretching due to the existence of intramolecular hydrogen bonding between the phenolic hydrogen and imine nitrogen atoms [O–H⋯N].27 The participation of OH groups in the coordination without the displacement of a proton from the phenolic OH28 group was ascertained by the presence of the strong band in the region of 3453 cm−1. The new non-ligand medium intensity band that appeared at around 542 cm−1 is assigned to the stretching frequency of Pr–O.29 The peaks observed at around 1489 cm−1 (ν1), 1024 cm−1 (ν2), 817 cm−1 (ν3), and 1294 cm−1 (ν4) confirm the participation of a nitrate ion present inside the coordination sphere.30 From the above data, it can be concluded that ligand 3 coordinates the praseodymium metal atom in a bidentate manner via the nitrogen of the amine group and the oxygen of the hydroxyl group.31 Complexes 5 and 6 show coordination behaviors similar to that of complex 4.
Electronic spectroscopy
Fig. S8† presents the electronic spectra of Schiff base ligand 3 and its complexes, which were recorded in methanol. The UV-Vis absorption maximum values (λmax), molar absorptivity (ε) and absorption band assignments are listed in Table 3. The absorption of ligand 3 is characterized by four main absorption bands at 212, 235, 271, and 355 nm. The two bands at lower wavelengths correspond to the n → π* transition due to the excitation of the lone pair of electrons of the p orbital of nitrogen in C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N with the π bonds of the two phenyl rings.28 The two bands at higher wavelengths are due to the π → π* transitions32 of the aromatic ring and the π* → π* transition of the C
N with the π bonds of the two phenyl rings.28 The two bands at higher wavelengths are due to the π → π* transitions32 of the aromatic ring and the π* → π* transition of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N group possibly because the n → π* transition state of the C
N group possibly because the n → π* transition state of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N group is involved in intramolecular charge transfer.28 The UV-Vis spectra for all the Ln(III) complexes 4, 5, and 6 are quite similar in the nature of their bands, but differ significantly in comparison with that of ligand 3, illustrating the effect of the coordination of ligand 3 with the Ln(III) ions. Due to the formation of coordination bonds, the two bands that appeared at a higher energy for the complexes were redshifted for the free ligand 3. Further, the two bands that appeared at a lower energy for the free ligand overlapped and blue shifted. This shift may be attributed to the ligand-to-metal charge transfer (LCMT), i.e. a pair of electrons from the ligand N-atoms transferring to the Ln(III) ions (N → Ln), a phenomenon consistent with the report by Keshavan et al.33 The comparison of the spectral data for ligand 3 with that of the La(III) complexes 4, 5, and 6 confirms the successful formation of these Ln(III) complexes. No bands due to the f–f transition were observed for all the complexes because of the weak nature of f–f transitions and they are Laporte-forbidden.34
N group is involved in intramolecular charge transfer.28 The UV-Vis spectra for all the Ln(III) complexes 4, 5, and 6 are quite similar in the nature of their bands, but differ significantly in comparison with that of ligand 3, illustrating the effect of the coordination of ligand 3 with the Ln(III) ions. Due to the formation of coordination bonds, the two bands that appeared at a higher energy for the complexes were redshifted for the free ligand 3. Further, the two bands that appeared at a lower energy for the free ligand overlapped and blue shifted. This shift may be attributed to the ligand-to-metal charge transfer (LCMT), i.e. a pair of electrons from the ligand N-atoms transferring to the Ln(III) ions (N → Ln), a phenomenon consistent with the report by Keshavan et al.33 The comparison of the spectral data for ligand 3 with that of the La(III) complexes 4, 5, and 6 confirms the successful formation of these Ln(III) complexes. No bands due to the f–f transition were observed for all the complexes because of the weak nature of f–f transitions and they are Laporte-forbidden.34
| Compound | λmax (nm) | ε × 104 (cm−1 mol−1) | Band assignment |
|---|---|---|---|
| 3 | 212, 235, 271, 355 | 5.42, 4.21, 3.06, 1.05 | π → π*, n → π* |
| 4 | 224, 268, 322, 299, 450 | 8.85, 0.32, 0.04 | π → π*, LMCT, n → π* |
| 5 | 223, 265, 328, 271, 446 | 3.12, 2.11, 1.19, 0.38, 0.02 | π → π*, LMCT, n → π* |
| 6 | 224, 306, 325, 266, 433 | 3.57, 1.59, 0.21, 0.34, 0.05 | π → π*, LMCT, n → π* |
Fluorescence spectroscopy
In the present study, we explored the photophysical behavior of the synthesized compounds. Fig. S9 (traces 3, 4, 5, and 6 in the ESI†) presents the photophysical characteristics of ligand 3 and complexes 4, 5, and 6 in ethanol solution at 25 °C. The emission spectra for the synthesized compounds 3, 4, 5, and 6 were obtained at λex 300 nm. From the comparison of the spectra of 4, 5, and 6 after complexation with ligand 3, a decrease in peak intensity was observed and the spectrum was shifted. This phenomenon of the shift in the fluorescence spectrum may be due to the excited state reactions, molecular rearrangements, energy transfer, ground-state complex formation and quenching due to collisions.35–37 The emission spectrum of Schiff base complex 4 displays three luminescence bands at 525 nm (3P0 → 3H5), 616 nm (5D4 → 7F3) and 689 nm (4G5/2 → 6H9/2). According to the transitions observed above, it is obvious that the energy-transfer mechanism occurs via the excitation of the ligand to the emission of the Pr(III) central metal ion.38 The signals of complexes 4, 5, and 6 related to these 3 transitions are tabulated in Table 4.| Compound | λex (nm) | λem (nm) | Band assignment |
|---|---|---|---|
| 3 | 300 | 353 | π* → π |
| 612 | π* → π | ||
| 687 | π* → π | ||
| 4 | 300 | 525 | 3P0 → 3H5 |
| 616 | 5D4 → 7F3 | ||
| 689 | 4G5/2 → 6H9/2 | ||
| 5 | 300 | 496 | 5D6 → 7F3 |
| 554 | 5D4 → 7F3 | ||
| 690 | 4G5/2 → 7F3 | ||
| 6 | 300 | 501 | π* → π |
| 616 | π* → π | ||
| 683 | π* → π |
MTT assay in vitro anti-tumor activity screening
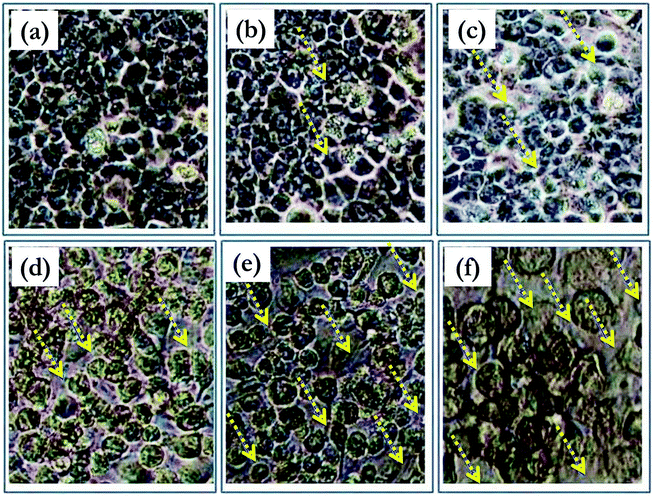
The reduction of the tetrazolium dye MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolim bromide) depends on the NAD(P)H oxidoreductase enzymes, which are present mainly in the cytosol. Rapidly dividing cells exhibit higher rates of MTT reduction, while cells with lower metabolic rates reduce MTT much less. The synthesized ligand 3 and complexes 4, 5, and 6 were tested in vitro against the HeLa cell line39 via the MTT assay. After the ligand and the complexes were incubated with the cells for 48 h, the percentage of viable cells in each case was estimated using the MTT assay. Nanjundan et al.40 reported that the treatment of HeLa cells with an N, S donor Schiff base and its mononuclear Ni(II), Cu(II) and Zn(II) complexes resulted in IC50 values of 39.74 ± 1.79, 38.34 ± 2.23, and 48.32 ± 1.02 μM (respectively). Fig. 2 represents the microscopy images of complex 5 administered to the HeLa cell line at various concentrations. All the tested compounds were inhibitory in a dose-dependent manner in the range of 5–50 μg mL−1 (ESI, Fig. S10†). Significantly, there was a decrease in cell viability with an increase in concentration. For the HeLa cells treated at higher concentrations (50 μg mL−1) of ligand 3 and its complexes 4, 5, and 6, the cell viability values observed were 65.23%, 49.26%, 20.15% and 55.16%, respectively. According to the IC50 values recorded for the tested ligand and its complexes, complex 5 showed the highest cytotoxic potency against HeLa cells (IC50 of 34.0 ± 1.2 μg mL−1), with a potency approximately 2.5 times higher than that of the other complexes for 48 h incubation. Therefore, further investigations were carried out only with complex 5.Flow cytometric analysis
Cell cycle analysis via flow cytometry with the aid of propidium iodide (PI) is extensively used for the evaluation of apoptosis. Fig. 3 depicts the flow cytometric analysis and presents the % distribution of cells during the cell cycle following the treatment of synchronized HeLa cells with complex 5. In the cell cycle, the S phase is the DNA replication phase, the G2 phase is the preparation phase for the M phase and the M phase is when “mitosis” occurs, which is when nuclear and cytoplasmic division occurs.41 The phase distribution histograms (ESI, Fig. S11†) were further quantitatively analyzed for each phase of the cell cycle (G1, S, and G2) and compared with the corresponding phase of the untreated control of HeLa cells. Treatment with 35 μg mL−1 of complex 5 resulted in a 2.1 fold increase in G1 phase cells, 1.5 fold increase in S phase cells, and 17.2 fold decrease in G2 phase cells, as shown in Fig. 3. Indeed, treating the cells with complex 5 resulted in a delay or inhibition of cell cycle progression through the S phase42 with little opportunity for progressing into mitotic division or remaining cancerous.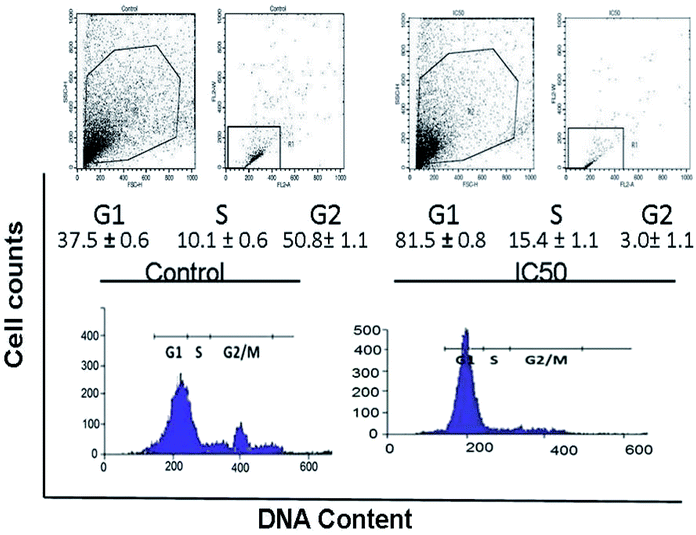 | ||
| Fig. 3 Cell apoptosis induced by complex 5 as examined using PI via flow cytometric analysis. The HeLa cells were treated with an IC50 concentration of 5 (a = control and b = 35 μg mL−1, IC50). | ||
DNA fragmentation studies
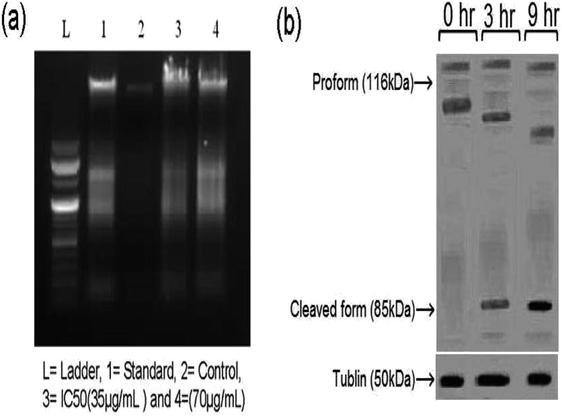
A DNA fragmentation assay was conducted by seeding HeLa cells into a 60 mm sterilized Petri dish and over an incubation period of 24 h at 37 °C in a 5% CO2 atmosphere. The cells were washed with medium and treated with the sample and standard (Doxorubicin), and incubated according to the above-specified conditions. After completion of the incubation period, the HeLa cells were prepared using a G Biosciences, USA apoptotic DNA ladder kit. Total genomic DNA was extracted and resolved on 1% agarose gel. Apoptotic DNA fragmentation was visualized via ethidium bromide (EB) staining under a UV transilluminator and photographed after the DNA ladder assay.43 Nithyakumar et al.15 reported that the treatment of HeLa cells with tetranuclear (RuII–Gd2III) and (RuII−Gd3III) d–f heterometallic complexes of 4-aminopyrine promoted apoptosis based on DNA fragmentation studies. The treatment of HeLa cells with complex 5, a simple mono rare earth Schiff base complex, resulted in the formation of a DNA ladder, which represents the enrichment of apoptosis in these cells. It is noteworthy that the simple Schiff base complex compares well with the d–f heterometallic complexes,15 with equivalent apoptosis in the HeLa cells.Fig. 4(a) shows the DNA fragmentation related to programmed cell death in the treated sample compared with the control-1 and Doxorubicin treated cells both at 24 h. Sample concentrations above 100 μg mL−1 resulted in moderate apoptosis (programmed cell death) compared to concentrations lower than 70 μg mL−1. Based on the comparison (lanes 3 and 4 in Fig. 4(a)), complex 5 forms a ladder, representing the promotion of apoptosis.
Western blotting
Induced apoptosis44 is the vital phenomenon that determines the efficacy of anti-cancer agents, and therefore it was necessary to evaluate the mechanism of the major cell death pathway by which complex 5 exerts its apoptotic effect on HeLa cells. The extent of apoptosis in HeLa cells can be estimated based on the cleavage of poly ADP-ribose polymerase (PARP) since it is a specific indicator of the apoptotic mechanism. PARP is an exact substrate for the active form of caspase-3, one of the “executioners” of the caspase family, including caspase-6 and caspase-7,45 thus the levels of PARP were monitored at different time points (0, 3, and 9 h). The Western blot analysis (Fig. 4(b)) showed that the HeLa cells underwent apoptosis upon treatment with complex 5 at its IC50 value, as demonstrated by the increase in the levels of PARP in its cleaved form. Initially, there was no significant presence of PARP in the cleaved form; however, there was a substantial increase in the PARP levels (85 kDa) after 3 h and also a further decrease in the proform (118 kDa). Furthermore, after 9 h, there was a substantial increase in the PARP levels compared to that at 3 h. The results obtained in the blot analysis are consistent with the previously reported western blot results for HeLa cells.46Hoechst 33258 and AO/EB dual staining
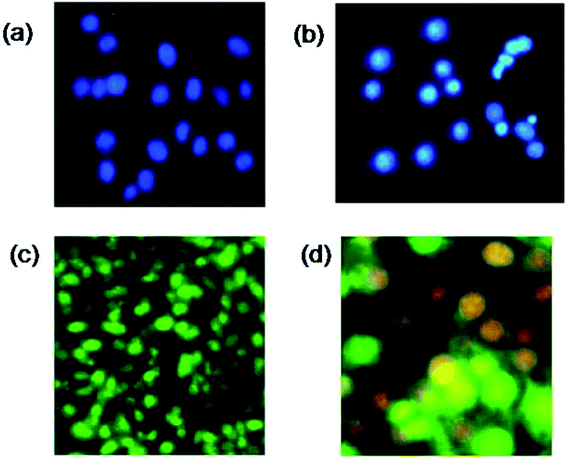
Visible nuclear changes and apoptotic body formation are the characteristic changes due to apoptosis. Accordingly, HeLa cells were treated with complex 5 at its IC50 value and stained with Hoechst 33258 for 24 h. As shown in Fig. 5(a and b), Hoechst 33258 staining47 showed regular contours and uniform round nuclei for the control. In the complex 5 treated sample, apoptotic cells with apoptotic features such as nuclear pyknosis, round shapes, and an enhanced fluorescent signal were observed compared to that in the control. The reduced nuclei and the apoptotic fragments in the tumor cell can be observed in Fig. 5(b). Intrinsic and extrinsic pathways are the two plausible mechanisms by which cell apoptosis can occur. The presence of caspase in apoptotic cells specifies an apoptotic morphology.48 The representative photographic image of HeLa cells treated with a 35 μg mL−1 solution of complex 5 and incubated at 37 °C for 24 h under CO2, followed by staining with EB and AO solutions is shown in Fig. 5. An enhancement in green and orange fluorescence can be observed in Fig. 5(c and d), which indicates that complex 5 has excellent cell membrane permeability, and thus resulted in effective apoptosis in the HeLa cells.Molecular docking study
Molecular modeling of drugs helps to predict the mode of drug binding with the targeted protein/other biomolecule, which is usually done by molecular docking. DNA is a widely used therapeutic target for diseases/disorders involving intracellular interactions.49 The adenine–thymine and guanine–cytosine base pairs in the complementary double helix structure in DNA are bonded by extensive hydrogen bonding. Intercalation50 is the process when a charged planar aromatic molecule comes in between the adjacent strands held by the van der Waals forces and hydrophobic interactions.51,52Docking studies were done using DNA (PDB ID: 1BNA) with complex 5 to determine its type of interaction and binding nature. The study revealed that the complex interacts in a purely electrostatic manner, involving outside-edge stacking interactions with the oxygen atoms of the phosphate backbone. Although no hydrogen bonding was detected for any of the ligands, steric interaction was determined to play a dominant role in the binding of complex 5 to DNA. The planar structure of complex 5 supports strong π–π stacking interactions and it is situated in the best position in between the DNA structure. Intercalation seems to occur in the minor to moderate grooves with the sequence (CGCGAATTCGCG)2. Fig. 6 shows the resultant docking of complex 5 with 1BNA. The docking score value is 5136 and complex 5 resides inside the B-DNA dodecamer in position DA 18-B-O4′ and DC 9-A-O2 with the C of complex 5. Table S2 (ESI†) represents the docking results, binding nature, binding site(s) and mode of interaction for complex 5 binding to 1BNA. Molecular docking studies were also performed to determine the binding site of complex 5 with the high-affinity site of human serum albumin (HAS). Table S3 (ESI†) presents the docking results, binding nature, binding site and mode of interaction. The studies reveal that complex 5 has an excellent binding orientation with a geometrical shape complementary score of 5492, indicating its strong affinity toward the binding site. The essential amino acid residues from the protein are mainly from Ala194, Arg145, Arg197, Asp108, Gln459, Glu425, His146, Lys190, Phe149, Pro147, Trp150, and Ser193. The three homologous domains, I (residues 1–195), II (196–383), and III (384–585) were obtained from the docking studies. Complex 5 binds with HSA and lies in the hydrophilic cavity of I and II, and held by electrostatic attraction and four hydrogen-bonding interactions with HAS (Tyr150 OH to O of complex 5 and Tyr150 OH to N of complex 5). The hydrogen bonding distances from the studies are 3.07, 3.23, 2.94 and 2.68 Å. Fig. 7(c) illustrates that the complex is close to tyrosine 150, designated here as TRY 150, TRY 150, TRY 150 and TRY 150. Fig. 7(c) presents the four hydrogen-bonding interactions generated when complex 5 intercalates with HSA. Nithyakumar et al.16 reported that the intercalation of d–f heterometallic complexes with HSA in the hydrophilic cavity is due to the presence of three hydrogen bonds near Arg197, Ser193, and Ser193.
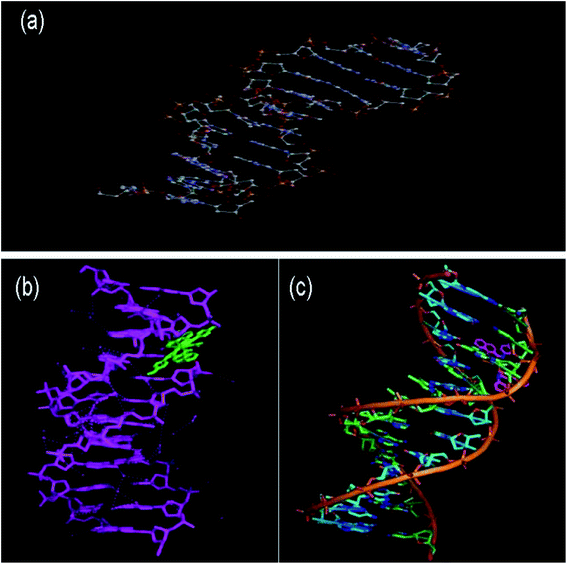 | ||
| Fig. 6 Graphical representation of the intercalation of complex 5 with DNA through π–π stacking: (a) 1B DNA (PDB ID: 1BDNA) alone, (b) wireframe molecular surface and (c) stick models. | ||
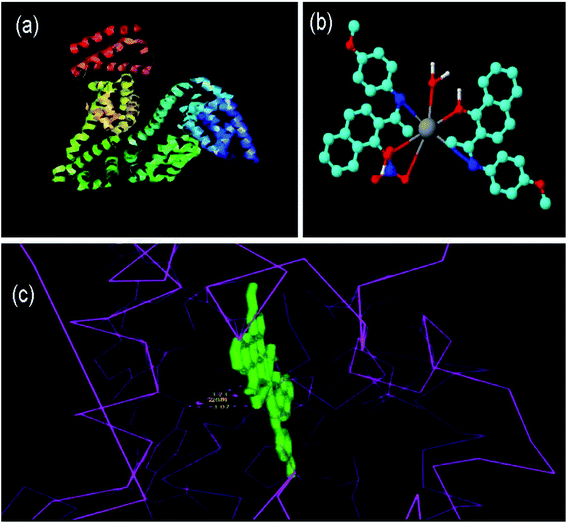 | ||
| Fig. 7 Molecular docked model of 5 with HSA through hydrogen bonding: (a) HSA alone (PDB ID: 1h9z), (b) 3D image of 5 and (c) bonding structures. | ||
Toxicity screening by bacterial cell culture
Any compound that generates free radicals can be lethal to cells, and this cytotoxic potential of can be evaluated in terms of the zone of inhibition (ZoI) and minimum inhibitory concentration (MIC) for the test strain E. coli AB1157. Therefore, this study provides an early indication of the capability of a compound to generate free radicals and its carcinogenic or mutagenic potential. Fig. S14(a–c) (ESI†) show a graphical representation of the bacterial strain-based assay53 for ligand 3 and its complexes 4, 5, and 6. Compound 3 and its complexes 5 and 6 displayed a significant MIC of 0.25 mg mL−1, as shown in Fig. S14(c) (ESI†). Compounds 3, 4, and 6 showed a lower MIC value than stannous chloride (superior value compared to stannous chloride). On the other hand, the MIC value of complex 6 is more than 0.5 mg mL−1 (inferior value compared to stannous chloride). Based on the comparative results obtained for the ligand and its complexes, complex 5 displays the best therapeutic value among the candidates in this study. Thus, the toxicity of complex 5 was further assessed by a bacterial reverse mutation test.Bacterial reverse mutation test
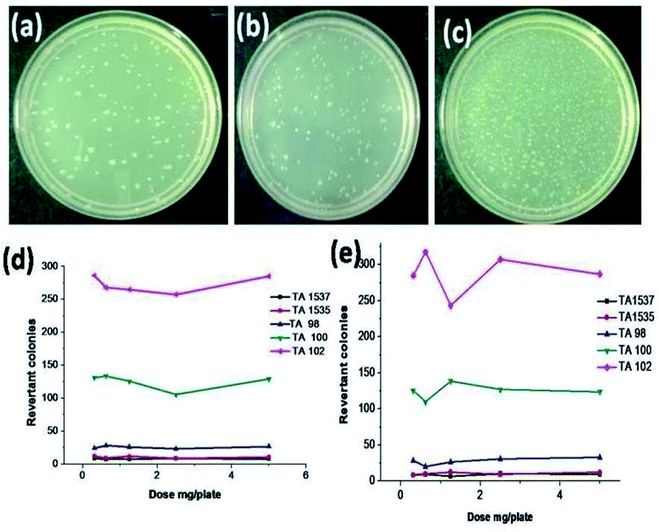
The bacterial reverse mutation54 or Ames test is a widely employed method to test whether chemical compounds can cause mutations in the DNA of test organisms, which in this case, was a bacterial strain. After the incubation period was complete, the colonies were counted, and the results were evaluated by comparison with the negative control. Fig. 8 presents the images of the Salmonella typhimurium test strain TA100. No significant or dose-dependent increase in colonies was observed in TA100 and the results were quite similar for all the test strains of Salmonella typhimurium, both in the presence and absence of a metabolic activation system. Identical results were observed for all the test strains and a representative image of the tested strain TA 100 is shown in Fig. 8. Cytotoxicity or reduction in the bacterial background lawn was not observed in all the test concentrations for all the strains. The dose-dependent curves for all the bacterial strains at the specified test doses are shown in Fig. 8(d and e), and based on the results, it can be concluded that complex 5 is non-mutagenic for concentrations up to 5 mg per plate.In vitro chromosomal aberration test
The micronucleus test55 is recognized as one of the most successful and reliable assays for the toxicological screening of potentially genotoxic compounds. Both chromatid and chromosome aberrations were observed and recorded under a microscope at 100× magnification. Gaps and ploidy were not considered for evaluating the results, while statistical and biological relevance were considered. No significant increase in chromosome aberrant cells was observed at any of the test doses (1.25, 2.50 and 5.00 mg mL−1). The results are tabulated in Table S4 (ESI†), and according to the table, a significant increase in the aberrant cells was observed with the positive control mitomycin C and no aberrations were observed in the negative control. Fig. S15(a and b) (ESI†) show a representative image for the chromosomal aberration of complex 5 administered to cells in the normal metaphase and mitomycin-treated cells with metaphase aberration exchange. The significant increase in aberrant cells in the positive control group demonstrates the suitability and sensitivity of the test procedure followed. Thus, according to the results, it can be concluded that the test chemical Sm3 (5) is not genotoxic at concentrations of 1.25, 2.5 and 5.0 mg mL−1 of culture medium in the chromosomal aberration test57 using the CHO–K1 cell line, both upon short term (4 h) and long term (21 h) treatment under the test conditions.3 Experimental
Materials
Hams F12 K medium, colchicine, and mitomycin-C (50-07-7) were obtained from Sigma. Fetal bovine serum (FBS) and antibiotic solutions (penicillin/streptomycin and amphotericin B) were purchased from Himedia Laboratories Ltd. The lanthanide nitrates, 1-hydroxy-2-aceto napthanone and p-anisidine, phosphate buffered saline (PBS), RPMI-1640 cell culture medium, and trypsin were purchased from Sigma Aldrich. EDTA, glucose, and antibiotics were obtained from Hi-Media Laboratories Ltd. The apoptotic DNA ladder kit was purchased from Roche, Germany. The HeLa cell culture was procured from the National Centre for Cell Sciences (NCCS), Pune, India. Methanol, diethyl ether and dimethylformamide (DMF) were purchased in spectroscopically pure form from Merck. Doubly distilled deionized water was used throughout the experiments.Physical measurements
Fourier transform infrared (FTIR) spectra were collected using KBr pellets on a Jasco spectrophotometer (4000–400 cm−1). A PerkinElmer, Lambda 45 (UV-Vis) spectrophotometer was used to record electronic spectra. 1H and 13C NMR spectra were recorded on a Bruker Avance 400 MHz spectrometer using TMS as an internal standard in CDCl3. Fluorescence spectra were measured on a PerkinElmer LS45 fluorescence spectrophotometer using ethanol as the solvent. Crystallographic data was collected with a Bruker Kappa APEXII area-detector diffractometer, using Mo Kα radiation with a wavelength of 0.71073 Å at 296 K. A colorless block-shaped single crystal of the compound with a size of 0.19 × 0.15 × 0.12 mm was selected, and data was collected. The data set was processed using the SAINT PLUS software.56 The structure was solved by direct methods and refined using the full-matrix least-squares procedure on F2 using the SHELXS and SHELXL programs.57 All hydrogen atoms were positioned geometrically (C–H = 0.93–0.97 Å) and refined using a riding model with Uiso(H) = 1.2Ueq(C). The final difference Fourier maps showed peaks of no chemical significance. The geometrical calculations and molecular diagrams were generated using the PLATON program.58MS (mass spectrometry) and liquid chromatography-mass spectrometry (LC-MS) were performed using a TSQ Quantum Endura mass spectrometer (Thermo Fisher Scientific, USA). All compounds were tested via electron spray ionization (ESI) using an eluent of 0.01 M ammonium formate/acetonitrile (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 v/v). The mass spectra of the selected compounds were obtained from the injection of 20 μg mL−1 of each compound at a flow rate of 0.8 mL min−1 using both negative and positive ion mode for ESI, using a Waters Xtera MS, 150 × 4.6 × 5 μcolumn. The Xcalibur software (rev.2.0 SP2, Thermo Fisher Scientific) was used for acquisition and data processing.
50 v/v). The mass spectra of the selected compounds were obtained from the injection of 20 μg mL−1 of each compound at a flow rate of 0.8 mL min−1 using both negative and positive ion mode for ESI, using a Waters Xtera MS, 150 × 4.6 × 5 μcolumn. The Xcalibur software (rev.2.0 SP2, Thermo Fisher Scientific) was used for acquisition and data processing.
Synthesis of 2-(1-(4-methoxyphenylimino)ethyl)naphthalen-1-ol (3)
In a typical synthesis, p-anisidine (1 mol) was added to a mixture containing 1-hydroxy-2-acetonapthanone (1 mol) dissolved in ethanol. The condensation reaction was carried in the slurry phase and the solution was subjected to ultrasonic irradiation for a particular duration. The progress of the reaction was monitored using TLC (thin layer chromatography) plates and an ethyl acetate/hexane solvent system. After completion of the reaction, the mixture was extracted in ethyl acetate and the extract was filtered through Na2SO4. The content was evaporated to dryness to obtain ligand 3. Ligand 3 was recrystallized using an equal-volume mixture of acetonitrile and ethanol to obtain fine crystals suitable for analysis. The product yield was found to be 95%.1H NMR (400 MHz, CDCl3): δ = 12.168 (s, 1H), 8.522–8.502 (s, 1H), 7.694–7.674 (d, 1H), 7.575–7.576 (m, 1H), 7.489–7.450 (m, 2H), 7.065–7.037 (m, 3H), 6.966–6.944 (m, 2H), 3.844 (s, 1H), 2.473 (s, 3H).
13C NMR (62.5 MHz, CDCl3): δ = 128.48, 124.8, 129.29, 125.15, 135.14, 115.49, 127.20, 114.47, 169.50, 170.56, 124.74, 110.68, 157.87, 55.55, and 16.54.
MS (EI) m/z = 292.25 (M + H).
Synthesis of lanthanide complexes (4–6)
A solution of praseodymium(III) nitrate (1 mol) in distilled methanol was added dropwise to 3 (2 mol) in methanol. The reaction mixture was then stirred for 3 h under reflux conditions. The precipitated complex 4 was then filtered, washed with ether, and dried in a vacuum oven for 8 h. The same procedure was used for the preparation of the samarium (5) and ytterbium (6) complexes.[Pr (3)2 (NO3)2·H2O]·NO3 [M + H+]: LCMS-MS; retention time tR = 5.81 minutes; (EI) m/z = *928.14, *929.16, and *931.6.
[Pr (3)2 (NO3)2·H2O]·NO3 [M + H+]: LCMS-MS; retention time tR = 8.31 minutes; (EI) MS/MS(EI) m/z = *850.14, *851.15, *853.14,*913.14, *914.15, *916.14,*917.14,*918.16, *919.15, *920.15, *921.15, *922.15, *923.14 *924.15 and *925.15.
MS (EI) m/z = 917.36 [Yb (3)2 (NO3)2·H2O] H3O+ adduct: LCMS-MS retention time tR = 5.75 minutes; MS/MS(EI) m/z = *957.16, *985.16, *959.14, *960.15, *961.15, *962.16, *963.14, and *964.14. *Isotopic peaks.
The details for the srm values and the collision energy for the complexes are shown in Table S1 (ESI†).
Cell culture and MTT-induced cytotoxicity
HeLa cells were cultured using RPMI-1640 medium supplemented with FBS (10%), penicillin (100 μg mL−1), and streptomycin (100 μg mL−1). Then there were incubated at 37 °C under 5% CO2 and 95% air. Cells from the culture were further diluted to 2.5 × 103 cells per well with medium, and 100 μL per well were seeded and incubation was continued for 24 h at 37 °C. Then the cells were treated with compounds 3, 4, 5, and 6 (5–50 μg mL−1). The control samples were the cultures in the absence of any test compounds in the medium. The incubation of the test compounds was continued for 48 h, and then a solution of MTT reagent (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide)29 (100 μL, 5 mg mL−1) was added to each well. Further, approximately 100 μL of cell lysate was added after incubation at 4 h at 37 °C. After incubation, the plates were analyzed on a microplate reader at 570 nm (Biorad 680). The percentage growth inhibitory rate of the treated cells was calculated as follows:where A is the average value from three determinations. A logarithmic plot between percentage viability versus concentration and concentration at which 50% of cells were viable, the IC50 value, was obtained.
Flow cytometric analysis
Complex 5 at its IC50 concentration of 50 μg mL−1 was incubated with HeLa cells in an incubator at 37 °C for 24 h. Subsequently, cell harvesting was performed, and the cells were further centrifuged 15 min at 1200 rpm, and approximately 2 mL of 70% (v/v) ethanol in water was added. The incubation period was continued for 12 h at −20 °C after the incubated cells were centrifuged for 15 min and treated with ice-cold PBS buffer. A staining solution of propidium iodide (PI) at a concentration of 10 μg mL−1 and DNase-free RNase (100 μg mL−1) were prepared. An aliquot of 200 μL of PI and 100 μL of DNase-free RNase were mixed and the cells were suspended in the mixture. The solution was analysed using a BD FACS Calibur™ cytometer (Becton Dickinson, Heidelberg, Germany). The cell count of each sample was 1000, and triplicate measurements were recorded using the CellQuest™ Pro software and analysed using the ModFit LT 2.0 software.DNA ladder assay
HeLa cells were cultured in 25 cm2 culture flasks at 37 °C in a humidified atmosphere of 5% CO2 using RPMI-1640 cell culture medium and supplemented with 10% FBS, penicillin (150 IU mL−1), streptomycin (150 μg mL−1) and amphotericin B (10 μg mL−1). The cells were treated with TPVG solution (0.2% trypsin, 0.02% EDTA, and 0.05% glucose in PBS) for dissociation.A known quantity of complex 5 was dissolved in HPLC-grade DMSO and further diluted with the cell culture medium to obtain a concentration of 1 mg mL−1 and the solution was filtered through a sterilized filter. The medium was supplemented with 2% inactivated FBS. A known aliquot of sample was transferred to Eppendorf tubes and centrifuged. The solution was further extracted in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 phenol and chloroform mixture and centrifuged again for 10 min. Then, upon the addition of cold ethanol and 0.1% of sodium acetate solution, the extract was precipitated. Then 50 μL of deionized water-RNase solution (0.4 mL water and 5 μL RNase) and 10 μL of buffer were added to the precipitate and it was kept at 37 °C for 30 min. Initially, 1.2% agarose gel was run at 5 V for 5 min, and thereafter the voltage was further increased to 100 V. The DNA marker (3 μL) was kept on the outer lanes before the start of the run. The results were recorded under a 312 nm UV illuminator.
1 phenol and chloroform mixture and centrifuged again for 10 min. Then, upon the addition of cold ethanol and 0.1% of sodium acetate solution, the extract was precipitated. Then 50 μL of deionized water-RNase solution (0.4 mL water and 5 μL RNase) and 10 μL of buffer were added to the precipitate and it was kept at 37 °C for 30 min. Initially, 1.2% agarose gel was run at 5 V for 5 min, and thereafter the voltage was further increased to 100 V. The DNA marker (3 μL) was kept on the outer lanes before the start of the run. The results were recorded under a 312 nm UV illuminator.
Preparation of cell lysates for western blotting
HeLa cells (1.5 × 106) were seeded in 60 mm culture plates and allowed to adhere overnight in a humidified 95% air, 5% CO2 incubator at 37 °C. The cells were treated with complex 5 (35 μg mL−1) for 48 h. Following the incubation, each plate was washed twice with cold PBS buffer (pH 7.4) and the cells were lysed using cold RIPA buffer (40–100 μL) (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA, and 0.25% sodium deoxycholate) containing a protease inhibitor cocktail (Roche, Indianapolis, IN, USA). The volume of RIPA buffer was adjusted according to the confluence of the cells, pouring it evenly throughout the plate and incubating for 10 min on ice. The cells were scraped from the plate and the cell lysate was transferred using a micropipette in a pre-cooled 1.5 mL centrifuge tube. The tube was placed on ice for 20 min and centrifuged (Eppendorf, 5804R) at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min at 4 °C. The collected supernatant was separated in a new pre-cooled centrifuge tube and the solution was used for quantification. A dilution was prepared using 2–5× loading buffer (Sigma) and the tubes were covered with a lid and boiled for 3–5 min.
000 rpm for 10 min at 4 °C. The collected supernatant was separated in a new pre-cooled centrifuge tube and the solution was used for quantification. A dilution was prepared using 2–5× loading buffer (Sigma) and the tubes were covered with a lid and boiled for 3–5 min.
PARP cleavage and western blot analysis
Proteins for western blot analysis were separated by SDS-PAGE using 8% gel. Equal amounts of protein and complex 5 were placed on the stacking gels. The acrylamide gels loaded with the buffer in empty wells were run at 250 V, 50 mA for 1 h. Further equilibration of the gels was performed using buffer (30 mM tris + 200 mM glycine and pH 8.0–8.8) for 15 min. The semi-dry electroblotting technique was used to transfer the proteins in the gel using nitrocellulose.The proteins in the gels were transferred to the nitrocellulose membrane by semi-dry electroblotting. The acrylamide gel was kept on the nitrocellulose membrane, and eight pre-saturated filter papers were placed on top of the gel. Rolling throughout the filter paper was performed to remove the excess air. By using 15 V at 34 mA for 25 min, the proteins were transferred from the gel to the nitrocellulose. A revolving apparatus (Stovall, Belly Dancer) was utilized throughout the process to ensure the even blot of all reagents. The nitrocellulose membranes were sterilized with 5% non-fat dried milk (Cadburys, Marvel skimmed milk) in tris-buffered saline (TBS) with 0.8% Tween-20 (Sigma) and rinsed for 15 min using 15 mM tris, 200 mM NaCl, at 0.6% Tween-20, at pH 7.1. Rabbit polyclonal antibody to poly(ADP-ribose) polymerase (PARP) (Santa Cruz Biotech) or mouse monoclonal antibody to α-tubulin (Sigma Aldrich) was used. An HRP substrate kit (Millipore, Billerica, MA) and secondary peroxidase-labelled anti-mouse or anti-rabbit immunoglobulin G antibody (Santa Cruz Biotech) were used in accordance with the manufacturers' directions. Bound antibodies were detected using enhanced chemiluminescence (ECL, Bio-Rad).
Cell viability assay by Hoechst-33342 staining
Hoechst staining is useful to determine apoptosis by staining DNA,59 which was developed by Hoechst AG. This protocol involves the use of Hoechst 33342 (2′-(4-ethoxyphenyl)-5-(4-methyl-1-piperazinyl)-2,5′-bi-1H-benzimidazoletrihydrochloridetrihydrate) to label the nuclear DNA of cells grown in cultures. The characteristic features of the Hoechst stain are that it is highly cell-permeable, produces blue fluorescence, and it stains live cells by intercalation in the basic residues of the adenine–thymine (A–T) rich region of DNA in the minor grooves. Thus, it produces blue fluorescence upon excitation in the UV region. HeLa cells were incubated with complex 5 at its IC50 concentration (34 μg mL−1) for 24 h at 37 °C and stained with Hoechst-33342 and examined by fluorescence microscopy.Dual AO/EB fluorescence staining
The cells were treated with the test compound at its IC50 concentration and incubated for 24 h in a CO2 incubator at 37 °C. The cells were removed by trypsinization and collected by centrifugation, including the non-adherent cells. The cell pellet was re-suspended in the medium and cell suspensions (25 μL) were transferred to glass slides. Dual fluorescent staining solution containing (1 μL) 100 μg mL−1 AO and 100 μg mL−1 EB (AO/EB, Sigma) was added to the suspension and then covered with a coverslip. The morphology of the apoptotic cells was examined, and the cells were counted within 20 min using a fluorescent microscope.Molecular docking study
Molecular docking studies were performed using the PatchDock software and the Q-Site finder molecular graphics program was used for docking and interaction calculations for identifying the possible binding sites for the drug molecules. The structure of the complex was obtained as a.mol file, and subsequently converted to the pdb format using the RasMol software. The crystal structures of BDNA (PDB ID: 1BNA) and human serum albumin (PDB ID: 1h9z were obtained from the RCSB protein data bank http://www.rcsb.org/.pdb). Default parameters were used for the docking calculations with the correlation type shape and FFT mode at the 3D level, grid dimension of 6 with a receptor range of 180, ligand range of 180 with a twist range of 360, and distance range of 40.Cytotoxicity screening using a bacterial strain-based assay
The carcinogenicity and mutagenicity studies of the test compounds were assessed using a bacterial strain-based assay27 using E. coli AB1157, which is a wild-type strain proficient in DNA damage repair. Identification of molecules that have implications in oxidative stress at an early stage helps in better understanding the structure-based drug design (SBDD). This bacterial strain was incubated with the compounds or chemical structures of interest for the analysis of any associated lethal effects. Initially, the stock culture of bacteria was revived by inoculation in broth medium and grown at 37 °C for 18 h. LB agar plates were prepared and wells were made in the solidified LB agar plate. Each plate was inoculated with 18 h-old cultures (100 μL, 10−4 CFU) and spread evenly on the plate. After 20 min, the wells were filled with test compounds 3, 4, 5, and 6 at different concentrations. The standard compound plate was also prepared in the same manner. All the plates were incubated at 37 °C for 24 h and the diameter of the zone of inhibition (ZoI) was recorded.Bacterial reverse mutation test using Salmonella typhimurium
The mutagenicity of complex 5 was tested in the bacterial reverse mutation (Ames) test according to the method of Maron and Ames, 1983, followed by the incorporation method.60,61 The study was conducted with Salmonella typhimurium strains TA 1537, TA 1535, TA 98, TA 100 and TA 102, both in the presence and absence of a metabolic activation system (rat liver S9 fraction 10% v/v). Five different test concentrations, 0.312, 0.625, 1.25, 2.5 and 5.0 mg per plate were prepared in DMSO and 100 μL of each test concentration, 100 μL of each Salmonella typhimurium test strain (1–2 × 109/mL) and 0.5 mL of PBS in the absence or S9 mix (10% v/v) in the presence of the metabolic activation system were added to a 2 mL overlay agar tube and poured on Minimal Glucose Agar (MGA) plates. Triplicate plates were prepared for each concentration and the control. Incubation was performed at 37 °C for 48 h and colony counting was done manually. 2-Aminoanthracene was used as a positive mutagen in the presence of the metabolic activation system, and 9 amino acridine, sodium azide, 2-nitrofluorene and mitomycin C were applied in the absence of the metabolic activation system.Chromosomal aberration test
CHO-K1 cells (ATCC No. CCL-61) were cultured in Hams F12 K culture medium supplemented with 10% FBS and antibiotic solution at 37 °C and under 5% CO2 in a 25 cm2 tissue culture flask (BD).57 The cultured CHO–K1 cells (3.0 × 105/mL) were treated with SM 3 (5) at 1.25, 2.5 and 5.0 mg mL−1 in culture medium for periods of 4 and 21 h. Mitomycin C and 0.3 μg mL−1 of culture medium was used as a positive control. After 4 h of treatment, the medium was removed, and the culture flasks were supplemented with 5 mL culture medium and further incubated for 17 h. Then 2 h before the harvesting period of 21 h, 0.4 μg colchicine was added to each culture flask, and incubation was continued. After 21 h, the incubated cells were harvested from the flasks by trypsinization. The cells were further treated with a hypotonic solution (0.075 M KCl solution) for 20 min at 37 °C and fixed in Carnoy's fixative (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 methanol
1 methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetic acid). In each treatment, the cells were pelleted by centrifugation at 1100 rpm for 10 min. Slides were prepared by dropping cell suspensions on cleaned, chilled slides. Staining was performed with 5% Giemsa stain and 300 well-spread metaphases from each concentration were observed under a microscope for chromosome aberrations.59 Statistical analysis (Chi-square trend test) was conducted to check for significant differences between the negative control and all other treatment groups, while Fisher's exact test was conducted for comparing the negative control vs. the positive control.
acetic acid). In each treatment, the cells were pelleted by centrifugation at 1100 rpm for 10 min. Slides were prepared by dropping cell suspensions on cleaned, chilled slides. Staining was performed with 5% Giemsa stain and 300 well-spread metaphases from each concentration were observed under a microscope for chromosome aberrations.59 Statistical analysis (Chi-square trend test) was conducted to check for significant differences between the negative control and all other treatment groups, while Fisher's exact test was conducted for comparing the negative control vs. the positive control.
4 Conclusion
The present study demonstrated the anti-cancer effects of tumor-targeting Schiff base Ln(III) bidentate complexes. Based on the analytical results, ligand 3 coordinates to the central Ln(III) ion by its azomethine nitrogen atom and hydroxyl atoms in the naphthalene ring with a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry. The E. coli AB1157 wild-type assay showed that complex 5 has the highest therapeutic value among the corresponding complexes 4 and 6. The bacterial reverse mutation (Ames) test further revealed that complex 5 is non-mutagenic at a test dose of 2.5 mg per plate, both in the presence and absence of a metabolic activation system (7.5% v/v S9 mix). The cell viability, as assessed by the MTT assay, demonstrated that complex 5 caused apoptosis at an IC50 concentration of 34.2 μg mL−1 compared to 4 and 6 complexes. The flow cytometric analysis demonstrated that cell death occurred at the G1 phase, where there was no replication of cells. Similarly, the western blotting suggested that the anti-cancer effect of complex 5 is likely based on the apoptosis of a caspase 3-specific substrate (PARP cleavage). Further, the in vitro imaging of cultured HeLa cells showed favorable responses, thereby opening up avenues for the application of our synthesized compounds as leads for the development of new anti-cancer drugs.
1 stoichiometry. The E. coli AB1157 wild-type assay showed that complex 5 has the highest therapeutic value among the corresponding complexes 4 and 6. The bacterial reverse mutation (Ames) test further revealed that complex 5 is non-mutagenic at a test dose of 2.5 mg per plate, both in the presence and absence of a metabolic activation system (7.5% v/v S9 mix). The cell viability, as assessed by the MTT assay, demonstrated that complex 5 caused apoptosis at an IC50 concentration of 34.2 μg mL−1 compared to 4 and 6 complexes. The flow cytometric analysis demonstrated that cell death occurred at the G1 phase, where there was no replication of cells. Similarly, the western blotting suggested that the anti-cancer effect of complex 5 is likely based on the apoptosis of a caspase 3-specific substrate (PARP cleavage). Further, the in vitro imaging of cultured HeLa cells showed favorable responses, thereby opening up avenues for the application of our synthesized compounds as leads for the development of new anti-cancer drugs.
Conflicts of interest
All authors declare no conflict of interest.Acknowledgements
We are thankful to SAIF, IIT – Madras, India for recording crystal structure and error rectification. We are grateful to B.S. Abdur Rahman Crescent Institute of Science and Technology for FTIR, UV-Vis, and fluorescence spectroscopy analysis. We thank Pondicherry Centre for Biological Sciences (PCBS) Pondicherry, India, for performing MTT Assay, Hoechst staining, and AO/EB staining, bacteria stain toxicity analysis, we gratefully acknowledge Palamur Biosciences Private Limited, Telangana, India for conducting conductivity measurements, LC-MS/MS analysis, western blot, chromosomal aberration, and Ames test. Interdisciplinary Institute of Indian System of Medicine, SRM Institute of Science and Technology, Kattankulathur for NMR and LCMS analysis. The King Saud University authors acknowledge funding of this work by the Research Supporting Project (RSP-2019/54), King Saud University, Riyadh, Saudi Arabia.References
- (a) E. R. Jamieson and S. J. Lippard, Chem. Rev., 1999, 99, 2467–2498 CrossRef CAS PubMed; (b) P. C. Bruijnincx and P. J. Sadler, Curr. Opin. Chem. Biol., 2008, 12, 197–206 CrossRef CAS PubMed.
- (a) F. Arnesano and G. Natile, Coord. Chem. Rev., 2009, 253, 2070–2081 CrossRef CAS; (b) T. W. Hambley, Dalton Trans., 2007, 43, 4929–4937 RSC; (c) C. Santini, M. Pellei, V. Gandin, M. Porchia, F. Tisato and C. Marzano, Chem. Rev., 2014, 114, 815–862 CrossRef CAS PubMed; (d) M. L. Jain, P. Y. Bruice and T. C. Bruice, Chem. Rev., 2012, 112, 1284 CrossRef CAS PubMed.
- (a) S. Doldi, Michele Peyrone e il suo sale, Chim. Ind., 1995, 77, 989–994 CAS; (b) B. Rosenberg, L. VanCamp, J. E. Trosko and V. H. Mansour, Nature, 1969, 222, 385–386 CrossRef CAS PubMed.
- Y. Jung and S. J. Lippard, Chem. Rev., 2007, 107, 1387–1407 CrossRef CAS PubMed.
- (a) A. M. Florea and D. Büsselberg, Cancers, 2011, 3, 1351–1371 CrossRef CAS PubMed; (b) T. C. Johnstone, J. J. Wilson and S. J. Lippard, Inorg. Chem., 2013, 52, 12234–12249 CrossRef CAS PubMed.
- (a) M. Chakrabarti, N. L. Banik and S. K. Ray, PLoS One, 2013, 8, 55652 CrossRef PubMed; (b) A. B. Ormond and H. S. Freeman, Materials, 2013, 6, 817–840 CrossRef CAS PubMed.
- M. Hong, H. Geng, M. Niu, F. Wang, D. Li, J. Liu and H. Yin, Eur. J. Med. Chem., 2014, 86, 550–561 CrossRef CAS PubMed.
- (a) S. Salehzadeh, S. M. Nouri, H. Keypour and M. Bagherzadeh, Polyhedron, 2005, 24, 1478 CrossRef CAS; (b) Focus on Organometallic Chemistry Research, ed. M. Andruh and F. Tuna, M. A. CatoNova Publishers, Hauppauge, 2005 Search PubMed.
- W.-J. Lian, X.-T. Wang, C.-Z. Xie, H. Tian, X.-Q. Song, H. T. Pan, X. Qiao and J.-Y. Xu, Dalton Trans., 2016, 1–3 Search PubMed.
- Z. Mandegani, Z. Asadi, M. Asadi, H. R. Karbalaei-Heidari and B. Rastegari, Dalton Trans., 2016, 45, 6592–6611 RSC.
- S. Banerjee, A. Dixit, A. A. Karande and A. R. Chakravarty, Dalton Trans., 2016, 45, 783–796 RSC.
- T. Sarkar, S. Banerjee and A. Hussain, RSC Adv., 2015, 5, 29276–29284 RSC.
- R. Alizadeh, I. Yousuf, M. Afzal, S. Srivastav, S. Srikrishna and F. Arjmand, J. Photochem. Photobiol., B, 2015, 143, 61–73 CrossRef CAS PubMed.
- C.-T. Yang, P. Padmanabhan and B. Z. Gulyas, RSC Adv., 2016, 6, 60945–60966 RSC.
- A. Nithyakumar and V. Alexander, New J. Chem., 2015, 1–10 Search PubMed.
- A. Nithyakumar and V. Alexander, Dalton Trans., 2015, 44, 17800–17809 RSC.
- A. M. Ajlouni, Z. A. Taha, K. A. Al-Hassan and A. M. AbuAnzeh, J. Lumin., 2012, 1357–1360 CrossRef CAS.
- J.-P. Costes, F. Dahan and A. Dupuis, Inorg. Chem., 2000, 39, 165–168 CrossRef CAS PubMed.
- (a) J.-C. G. Bunzli, S. Comby, A.-S. Chauvin and C. D. B. Vandevyver, J. Rare Earths, 2007, 25, 257–274 CrossRef; (b) Li Quan, T. Li and J. Wu, J. Phys. Chem. B, 2001, 105, 12293–12296 CrossRef; (c) S. alder, A. Bhattacharjee, A. Roy, S. Chatterjee and P. Roy, RSC Adv., 2016, 6, 39118–39124 RSC; (d) H.-r. Cheng and Y. Qian, RSC Adv., 2015, 5, 82887–82893 RSC.
- (a) B. A. El-Sayed, M. M. Abo Aly, A. A. A. Emara and S. M. E. Khalil, Vib. Spectrosc., 2002, 30, 93–100 CrossRef CAS; (b) E. M. Nour, A. A. Taha and I. S. Alnaimi, Inorg. Chim. Acta, 1988, 141, 139–144 CrossRef CAS.
- K. Andiappan, A. Sanmugam, E. Deivanayagam, K. Karuppasamy, H. S. Kim and D. Vikraman, Sci. Rep., 2018, 8, 3054 CrossRef PubMed.
- B. T. Vhanale, N. J. Deshmukh and A. T. Shinde, Heliyon, 2019, 5, e02774 CrossRef CAS PubMed.
- R. D. Teo, J. Termini and H. B. Gray, J. Med. Chem., 2016, 59(13), 6012–6024 CrossRef CAS PubMed.
- K. P. Neelima, S. Siddiqui, M. Arshad and D. Kumar, Spectrochim. Acta, Part A, 2016, 155, 146–154 CrossRef CAS PubMed.
- W. J. Geary, Coord. Chem. Rev., 1971, 7, 81–120 CrossRef CAS.
- (a) G. Leniec, S. M. Kaczmarek, J. Typek, B. Kołodziej, E. Grech and W. Schilf, J. Phys.: Condens. Matter, 2006, 18, 9871–9880 CrossRef CAS; (b) S. M. Kaczmarek and G. Leniec, J. Non-Cryst. Solids, 2009, 355, 1325–1332 CrossRef CAS.
- A. M. Ajlouni, Z. A. Taha, W. A. Momani, A. K. Hijazi and M. Ebqa'ai, Inorg. Chim. Acta, 2012, 388, 120 CrossRef CAS.
- T. A. Youssef, J. Coord. Chem., 2008, 61, 816 CrossRef CAS.
- L. J. Bellamy, The infrared Spectra of complex Molecules, J. Wiley, New York, 1971 Search PubMed.
- S. M. Jadhav, V. A. Shelke, S. G. Shankarwar, A. S. Munde and T. K. Chandrashekar, J. Saudi Chem. Soc., 2014, 18, 27–34 CrossRef CAS.
- (a) P. Yan, W. Sun, Z. Li, C. Net, T. Gao and Z. Yue, J. Coord. Chem., 2007, 60, 1973 CrossRef CAS; (b) W. Wang, Y. Huang and N. Tang, Spectrochim. Acta, 2007, 66, 1058 CrossRef PubMed.
- (a) L. Guo, S. Wu, F. Zeng and J. Zhao, Eur. Polym. J., 2006, 42, 1670–1675 CrossRef CAS; (b) R. C. Felico, E. T. G. Canalheiro and E. R. Dockal, Polyhedron, 2001, 20, 261 CrossRef.
- B. Keshavan, P. G. Chandrashekara and N. M. J. Made Gowda, J. Mol. Struct., 2000, 553, 193–197 CrossRef CAS.
- C. Gorller-Walrand, K. Binnemans, Handbook on the Physics Chemistry of Rare Earths, North-Holland Publishers, Amsterdam, 1998, vol. 25, p. 101 Search PubMed.
- A. Pui, T. Malutan, L. Tataru, C. Mualtuan, D. Humelnicu and G. Carja, Polyhedron, 2011, 30, 2127 CrossRef CAS.
- N. Sabbatini, M. Guardigli and J. M. Lehn, Coord. Chem. Rev., 1993, 123, 201 CrossRef CAS.
- (a) A. G. Kolchinski, Coord. Chem. Rev., 1998, 174, 207 CrossRef CAS; (b) A. V. Haynes and H. G. Drickamer, J. Chem. Phys., 1982, 76, 114 CrossRef; (c) K. Inoue, Y. Sasaki and T. Itaya, Eur. Polym. J., 1997, 33, 841 CrossRef CAS.
- L. Guo, S. Wu, F. Zeng and J. Zhao, Eur. Polym. J., 2006, 42, 1670 CrossRef CAS.
- (a) A. A. van de Loosdrecht, et al., J. Immunol. Methods, 1994, 174, 311–320 CrossRef CAS PubMed; (b) M. Ferrari, et al., J. Immunol. Methods, 1990, 131, 165–172 CrossRef CAS PubMed; (c) D. Gerlier and N. Thomasset, J. Immunol. Methods, 1986, 94, 57–63 CrossRef CAS PubMed; (d) M. C. Alley, et al., Cancer Res., 1988, 48, 589–601 CAS; (e) T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS PubMed.
- N. Nanjundan, R. Narayanasamy, S. Geib, K. Velmurugan, R. Nandhakumar, M. D. Balakumaran and P. T. Kalaichelvan, Polyhedron, 2016, 110, 203–220 CrossRef CAS.
- (a) A. J. Krishan, Cell Biosci., 1975, 66, 188–193 CAS; (b) C. Riccardi and I. Nicoletti, Nat. Protoc., 2006, 1, 1458–1461 CrossRef CAS PubMed.
- M. Li, L. k. Lin, Y. Gou, F. Yang and H. Liang, Spectrochim. Acta, Part A, 2014, 128, 686–693 CrossRef CAS PubMed.
- J. Zhang and M. Xu, Trends Cell Biol., 2002, 12, 84–89 CrossRef CAS PubMed.
- W. Hu and J. J. Kavanagha, Lancet Oncol., 2003, 4, 721 CrossRef CAS PubMed.
- J. G. Walsh, S. P. Cullen, C. Sheridan, A. U. Luthi, C. Gerner and S. J. Martin, Proc. Natl. Acad. Sci. U. S. A, 2008, 105, 12815 CrossRef CAS PubMed.
- B. Jung, Y. C. Jeong, J. H. Min, J. E. Kim, Y. J. Song, J. K. Park, J. H. Park and J. D. Kim, J. Mater. Chem., 2012, 22, 9385–9394 RSC.
- (a) S. A. Latt, G. Stetten, L. A. Juergens, H. F. Willard and C. D. Scher, J. Histochem. Cytochem., 1975, 23, 493–505 CrossRef CAS PubMed; (b) S. A. Lattand and G. Stetten, J. Histochem. Cytochem., 1976, 24, 24–33 CrossRef PubMed.
- (a) M. R. Gill, J. Garcia-Lara, S. J. Foster, C. Smythe, G. Battaglia and J. A. Thomas, Nat. Chem., 2009, 1, 662–667 CrossRef CAS PubMed; (b) T. Chen, Y. Liu, W.-J. Zheng, J. Liu and Y.-S. Wong, Inorg. Chem., 2010, 49, 6366–6368 CrossRef CAS PubMed; (c) L. Li, Y.-S. Wong, T. Chen, C. Fan and W. Zheng, Dalton Trans., 2012, 41, 1138–1141 RSC.
- E. Quin, J. R. Devlin, D. Cameron, K. M. Hannan, R. B. Pearson and R. D. Hannan, Biochim. Biophys. Acta, Mol. Basis Dis., 2014, 1842, 802–816 CrossRef PubMed.
- (a) V. V. Kostjukov, N. M. Khomytova, A. A. Hernandez Santiago, R. Licona Ibarra, D. B. Davies and M. P. Evstigneev, Int. J. Quantum Chem., 2011, 111, 711–721 CrossRef CAS; (b) A. J. Mukherjee, J. Phys. Chem. Lett., 2011, 2, 3021–3026 CrossRef CAS; (c) S. Zhang, B. Ling, F. Qu and X. Sun, Spectrochim. Acta, Part A, 2012, 97, 521–525 CrossRef CAS PubMed.
- (a) N. C. Seeman, H. Wang, X. Yang, F. Liu, C. Mao, W. Sun, L. Wenzler, Z. Shen, R. Sha, H. Yan, M. H. Wong, P. Sa-Ardyen, B. Liu, H. Qiu, X. Li, J. Qi, S. M. Du, Y. Zhang, J. E. Mueller, T.-J. Fu, Y. Wang and J. Chen, Nanotechnology, 1998, 9, 257–273 CrossRef CAS; (b) S. Arnott, Nature, 1986, 320, 313 CrossRef CAS PubMed.
- P. R. G. De Lemos, O. N. Terra Junior and M. L. Amantea, Asian J. Pharm. Health Sci., 2011, 1(2), 89–90 Search PubMed.
- (a) Organization for Economic Co-operation and Development, Guidelines for Testing of Chemicals No. 471: Bacterial reverse mutation test, OECD, Paris, France, 1997, Adopted: 21st July 1997 Search PubMed; (b) K. Mortelmans and E. Zeiger, Mutat. Res., 2000, 455, 29–60 CrossRef CAS PubMed.
- (a) J. Dorthy Maron and B. N. Ames, Mutat. Res., 1981, 88, 343–350 CrossRef; (b) D. M. Maron and B. N. Ames, Mutat. Res., 1983, 113, 173–215 CrossRef CAS PubMed.
- (a) R. I. Freshney, Culturing of animal cells: A manual of basic technique, Wiley-liss, N.Y., 5th edn, 2005 CrossRef; (b) The Organization for Economic Co-operation and Development (OECD), Guidelines for the Testing of Chemicals, Volume II, adopted by the council on, 28th September, 2014 Search PubMed.
- Bruker, SAINT: Area-Detector Integration Program, Bruker Analytical X-ray Systems, Inc., Madison, Wis, USA, 2007 Search PubMed.
- G. M. Sheldrick, Acta Crystallogr. A, 2008, 64(pt 1), 112–122 CrossRef CAS PubMed.
- A. Spek, J. Appl. Crystallogr., 2003, 36(1), 7–13 CrossRef CAS.
- L. Yan, Y. Wang, X. Xu, C. Zeng, J. Hou, M. Lin, J. Xu, F. Sun, X. Huang, L. Dai, F. Lu and Y. Liu, Chem. Res. Toxicol., 2012, 25(6), 1265–1270 Search PubMed.
- M. M. Hossain, M. F. Aziz, R. Ahmed, M. Hossain, A. Mahmuda, T. Ahmed and M. H. Mazumder, Int. J. Pharm. Sci., 2010, 2, 60–63 Search PubMed.
- A. A. Sakur and H. Fael, Int. J. Pharm. Sci. Rev. Res., 2010, 4, 60–63 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra01936d |
| This journal is © The Royal Society of Chemistry 2020 |