Open Access Article
Open Access ArticleHistidine-conjugated DNA as a biomolecular depot for metal ions†
Soyoung Park *a,
Haruka Matsuia,
Koyuki Fukumotoa,
Ji Hye Yuma and
Hiroshi Sugiyama
*a,
Haruka Matsuia,
Koyuki Fukumotoa,
Ji Hye Yuma and
Hiroshi Sugiyama *ab
*ab
aDepartment of Chemistry, Graduate School of Science, Kyoto University, Kitashirakawa-oiwakecho, Sakyo-ku, Kyoto 606-8502, Japan. E-mail: hs@kuchem.kyoto-u.ac.jp; oleesy@kuchem.kyoto-u.ac.jp
bInstitute for Integrated Cell-Material Sciences (iCeMS), Kyoto University, Yoshida-ushinomiyacho, Sakyo-ku, Kyoto 606-8501, Japan
First published on 6th March 2020
Abstract
Histidine is a versatile amino acid residue that plays a critical role in the active sites of many metalloenzymes. DNA is an attractive biomolecular scaffold owing to its chemical and thermal stability and easy accessibility. Herein, we report histidine-conjugated DNA oligonucleotides, which were synthesized by combining DNA alphabets and natural metal-binding amino acids, as novel biohybrid materials and demonstrate their use as molecular depots for various metal ions. Moreover, histidine-conjugated DNA oligonucleotides could be successfully used in asymmetric catalysis (up to 90% conversion and 95% ee) as DNA metalloenzymes and in 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) oxidation reactions as horseradish-peroxidase (HRP)-mimicking DNAzymes with suitable metal cofactors. Nature-inspired histidine-DNA hybrids will become an attractive strategy to construct fine-tuned coordination environments as an alternative to bioremediation and the development of multimetal enzymes.
Introduction
Nucleic acids, especially DNA, are useful biomolecules to develop molecular engineering materials for various applications in chemical biology and biomedicine. They have many advantages including straightforward automated synthesis processes, unique complementarity, and high selectivity and affinity for target biomolecules such as proteins.1–4 DNA has been actively explored as a versatile building block in nanotechnology owing to its superior, programmable self-assembling abilities and the high rigidity of its secondary structures.5–7 In addition, DNA is highly amenable to structural modification and introduction of functional groups. The applications of DNA can be further enhanced by the introduction of various functional groups such as fluorescent dyes,8–10 alkylating agents,11–13 and metal-binding ligands.14–16 Among them, site-specific functionalization of DNA with metal ions has received attention because of the wide range of its applications, including molecular magnets,17–19 electron transport chains,20–22 and metalloenzymes.23–26 Several metal-mediated natural or artificial nucleobase pairs have been explored and various metal-binding ligands, such as bipyridine (bpy) and terpyridine, have been used in the development of metal-modified DNA.27–29In our previous study, we established the covalent anchoring strategy for direct incorporation of intrastrand metal-binding ligands such as bpy into the DNA phosphate backbone.24,30–32 We generated a series of Cu (bpy)-containing DNA hybrid catalysts and achieved asymmetric intramolecular Friedel–Crafts alkylation28 and enantioselective hydration reactions.33 We also interspersed a bpy ligand into the DNA strands using an acyclic D-threoninol backbone, which is a versatile moiety for the modification of oligonucleotides with carboxylic acid derivatives.33 In this study, we focused on histidine (His), an amino acid residue that plays a critical role in the active sites of many metalloenzymes. Because the imidazole ring of histidyl peptide residues provides an excellent coordination environment for various metal ions, imidazole-modified oligonucleotides have been investigated. Müller and colleagues synthesized a DNA oligonucleotide containing three consecutive imidazole ligands and reported the nuclear magnetic resonance solution structure of metal-modified DNA with imidazole–Ag(I)-imidazole base pairs.34–36 Recently, Clever and colleagues reported on imidazole-modified G-quadruplex DNA.37,38 They incorporated an imidazole ligand into DNA using a glycidol linker and demonstrated a potential Cu(II)-sensory application involving the peroxidase-mimicking DNAzyme activity of this complex. Surprisingly, to our knowledge, the direct incorporation of His residues into DNA oligonucleotides has not yet been reported. Herein, we report the synthesis and characterization of His-conjugated DNA oligonucleotides with a distinctive metal-binding ability. Furthermore, His-conjugated DNA oligonucleotides could be successfully used in asymmetric catalysis as well as 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) oxidations in the presence of suitable metal cofactors.
Results and discussion
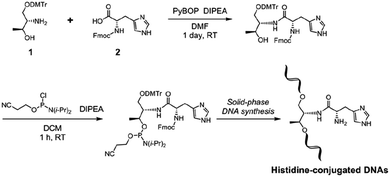
A synthetic route for His-conjugated oligodeoxynucleotides is shown in Scheme 1.Acyclic D-threoninol is a versatile backbone for the modification of DNA, and acyclic threoninol nucleic acids (aTNA) have been well investigated by Asanuma and colleagues.39–41 We have also established a DNA modification approach using the threoninol moiety to incorporate a bpy ligand into the DNA oligonucleotide strands for the synthesis of bpy-containing DNA hybrid catalysts.33 For incorporation of His into oligonucleotides, it is important to select an appropriate amino-protecting group for the His residue that is able to tolerate not only phosphitylation conditions but also the solid-phase DNA synthesis process. In this study, we used fluorenylmethoxycarbonyl (Fmoc)-protected His. The Fmoc group is compatible with the chemical reagents used during phosphitylation and solid-phase oligonucleotide synthesis. In addition, removal of the Fmoc group could be accomplished during standard cleavage and deprotection conditions using an aqueous solution of methylamine and ammonium hydroxide. Regarding the imidazole group of His, an unprotected imidazole group could be incorporated into DNA oligonucleotides. Based on previous studies,24,28,33 DMTr-protected D-threoninol (1) could be conjugated with Fmoc-protected His (Fmoc–His–OH, 2) in the presence of PyBOP and DIPEA. After phosphitylation, the His-functionalized nucleoside surrogate could be incorporated into DNA oligonucleotides by automated solid-phase synthesis (Table S1†). For control experiments, phenylalanine-conjugated derivatives were also prepared (Fig. S1–S4†).
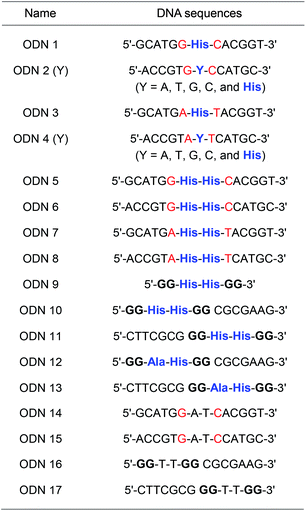
A His residue was incorporated at the center of a 13-mer oligonucleotide (ODN 1: 5′–GCATGGXCACGGT–3′, where X = His) via automated solid-phase synthesis. Complementary strands containing native nucleobases or His in the center (ODN 2(Y): 5′-ACCGTGYCCATGC-3′, where Y = A, T, G, C, and His) were also prepared (Table 1 and Table S2†). The thermal stability of His-containing DNA duplexes was investigated using a DNA duplex containing a His–nucleobase (A, T, G, or C) pair and a duplex containing a His–His pair in the absence and presence of Cu(II) ions. DNA duplexes containing a His–nucleobase pair showed lower melting temperatures (Tm), around 40 °C, than duplexes containing a native A–T or G–C pair (53 °C and 63 °C, respectively). The observed decline in Tm indicated that the threoninol-tethered His residue did not recognize or interact with natural nucleobases. In addition, the presence of Cu(II) ions did not increase the thermal stability of the DNA duplexes containing a His–nucleobase pair. Interestingly, DNA duplexes containing a threoninol-tethered His–His pair showed higher Tm values than the duplexes containing a His–nucleobase pair. Moreover, after addition of 1 equivalent (eq.) of Cu(II) ions, the Tm value of DNA duplexes containing a His–His pair increased, up to 52 °C. For neighboring bases, the DNA duplexes containing ODN 3/ODN 4 (–A–His–T–) generally showed lower Tm values than the DNA duplexes containing ODN 1/ODN 2 (–G–His–C–) regardless of complementary nucleobases (Fig. S5 and S6†). And the thermal stabilization effect caused by the addition of 1 equivalent Cu(II) ion on the His–His pair was observed, however, Tm value decreased by the equivalent of Cu(II) ions increased (Fig. S9†). The results from thermal denaturation experiments with His–His pair-containing DNA duplexes in the presence of Cu(II) ions encouraged us to investigate the metal-binding ability of His-modified DNA duplexes.
We performed Tm measurements with duplexes containing a His–His pair in the presence of various metal ions. Besides Cu(II) ion, the addition of 1 eq of Zn(II) and Ni(II) ions significantly increased Tm values of duplexes to 48.7 °C and 50.3 °C, respectively (Fig. S9–S11†). However, the addition of Fe(II) did not increase the Tm of the duplexes (Fig. S8†). Circular dichroism (CD) measurements showed that the addition of 1 eq. of metal ions did not change the global structures of duplexes as a characteristic feature of typical right-handed B-DNA (Fig. S21–S26†).
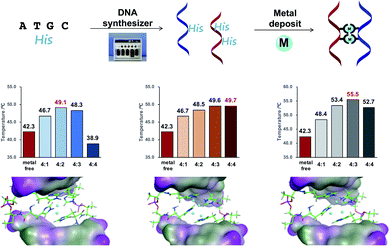
Subsequently, as shown in Fig. 1A, we prepared DNA oligonucleotides with two consecutive His residues incorporated at the center of the DNA strands and investigated their metal-binding ability using thermal denaturation experiments according to the equivalence ratio of metal ion and DNA (Fig. 1B). DNA duplexes ODN 5/ODN 6 have a metal-binding site that consists of four His residues. We changed the molar ratio of metal ions to DNA duplex from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 4
1 to 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and found that Tm varied depending on the equivalence ratio as well as the type of metal ions (Fig. S13–S16†). For instance, the highest Tm was observed with 2 eq. of Cu(II) ions, 3 or 4 eq. of Zn(II) ions, and 3 eq. of Ni(II) ions. This indicated distinct thermal stabilization effects according to the metal ions used. Regardless of the molar ratio of metal ions to DNA duplex, Ni(II) ions led to the highest Tm value in all cases. No thermal stabilization compared with metal-free conditions was observed by the addition of iron ions (Fig. S13†).
1 and found that Tm varied depending on the equivalence ratio as well as the type of metal ions (Fig. S13–S16†). For instance, the highest Tm was observed with 2 eq. of Cu(II) ions, 3 or 4 eq. of Zn(II) ions, and 3 eq. of Ni(II) ions. This indicated distinct thermal stabilization effects according to the metal ions used. Regardless of the molar ratio of metal ions to DNA duplex, Ni(II) ions led to the highest Tm value in all cases. No thermal stabilization compared with metal-free conditions was observed by the addition of iron ions (Fig. S13†).
To verify the thermal stabilization induced by metal-His binding, the His residue was replaced with phenylalanine residues, which are incapable of coordinating to the metal ions. In control experiments with DNA duplexes containing phenylalanine pairs, the thermal stability of the DNA duplexes changed only marginally in the presence of all examined metal ions, indicating that the observed thermal stabilization effects are caused by the metal-binding His residue. To rule out the thermal stabilization effect caused by the interaction between metal ions and the anionic phosphate backbone, we performed thermal denaturation experiments with native DNA duplexes (ODN 14/ODN 15) in the presence of various metal ions. No increase in Tm was observed even if up to 4 eq. of metal ions (copper, zinc, and nickel) were added to the DNA (Fig. S7†). These results support the hypothesis that the increase in Tm of the duplexes is attributed to the binding of metal ions to His residues and clearly indicate the potential of the His–DNA hybrid as a molecular reservoir for metal ions.
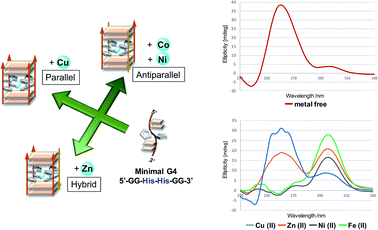
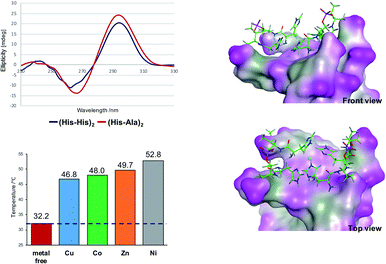
Besides the canonical Watson–Crick duplex, there are various noncanonical secondary structures, such as left-handed Z-DNA, triplex, and quadruplexes, under specific conditions.42–45 Among them, G-quadruplex has been actively explored as a versatile scaffold for the construction of nanostructures because of its structural diversity, which is derived from combinations of parallel and antiparallel strands, different loop sequences, and shapes.46–48 In this context, we incorporated His residues into a G-rich oligonucleotide, 5′-GG–His–His–GG-3′ (ODN 9), to construct a minimal G-quadruplex structure and investigated its conformational properties and metal-binding ability.49 In the presence of K+ ions, the CD spectra of ODN 9 indicated that the His-functionalized oligonucleotide forms a G-quadruplex structure with parallel conformation (Fig. 2B). We then also investigated the conformational changes induced by various other metal ions (Fig. 2A). On the addition of Cu(II) ions, ODN 9 showed a parallel conformation similar to that under metal-free conditions. In contrast, the addition of Co(II) ions induced antiparallel conformations. An antiparallel conformation was also observed in the presence of Ni(II) ions. On the addition of Zn(II) ions, the CD spectra of ODN 9 indicated hybrid or mixed conformations. Interestingly, the His-modified minimal G-quadruplex-forming sequence ODN 9 showed various topologies induced by metal complexations. This result indicated that the His-modified DNA nucleotides could be versatile building blocks for creating metal-triggered switchable nanostructures as well as biomolecular depots for metal ions.
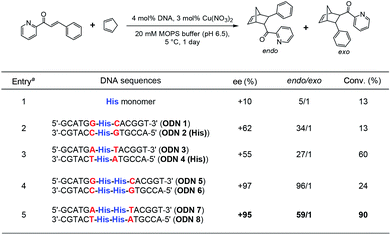
Next, we investigated the utility of DNA duplexes involving threoninol-tethered His residues. DNA-based hybrid catalysts are one of the promising alternatives to develop water-compatible catalysts. Due to thermal and chemical stability, conformational flexibility, and automated solid-phase synthesis procedure, DNA has been actively explored as a biomolecular scaffold for catalysis.50–52 In this study, the well-established asymmetric Diels–Alder reaction between aza-chalcone and cyclopentadiene was selected as a model reaction to investigate the performance of His-conjugated DNA as an asymmetric catalyst. While a control experiment using Cu(II) ions and His-conjugated D-threoninol monomer resulted in low endo/exo selectivity and enantioselectivity (Table 2, entry 1), in the presence of Cu(II) ions, the DNA duplex containing a D-threoninol-tethered His–His pair showed significantly increased enantioselectivity as well as endo/exo selectivity, indicating the potential of the DNA scaffold to induce high enantioselectivity (Table 2, entries 2 and 3). Under optimized conditions, Cu(II)/DNA duplex with four His residues resulted in the corresponding product with high conversion (90%), high endo/exo selectivity, and excellent enantioselectivity (+95% ee, Table 2, entry 5).
| a Experiments were carried out using 3.3 mM aza-chalcone, 80 mM cyclopentadiene, 0.13 mM DNA, and 0.1 mM Cu(NO3)2 at 5 °C in 20 mM MOPS buffer (pH 6.5) for 1 day. The conversion and enantioselectivities were determined by chiral HPLC analysis. |
|---|
 |
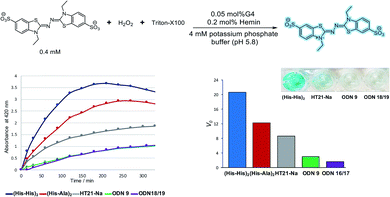
Moreover, we explored the usability of the His-modified G-quadruplex DNA in catalysis. It has been reported that the hemin/G-quadruplex DNA complex promotes the H2O2-mediated oxidation of ABTS to form colorimetric products from ABTS2−.53,54 The ABTS oxidation assay is a useful chromophore-based tool for quantifying oxidative catalytic ability. We evaluated the catalytic performance of the His-modified G-quadruplex DNA with hemin complex based on readout the corresponding radical species in ABTS oxidation reaction.
In this experiment, we designed the His-modified DNA quadruplex–duplex hybrid to focus on the effect of His residue on the well-organized strand topology of DNA. Based on the compatibility of quadruplex and duplex DNAs and their modulated topologies reported by Phan and colleagues,55–57 two stacks of G-tetrad core with four His residues were elongated with duplex-forming sequences (ODN 10/ODN 11). DNA quadruplex–duplex hybrids with two histidine and two alanine residues were also prepared in the same manner (ODN 12/ODN 13). In the presence of K+ ions, ODN 10/ODN 11 and ODN 12/ODN 13 formed antiparallel conformations, as confirmed by CD spectroscopy (Fig. 3A, S32 and S33†). The DNA quadruplex–duplex hybrid with four modified His residues showed a remarkable thermal stabilization effect in the presence of various metal ions, indicating a high storage capacity for metal ions (Fig. 3B, S17 and S18†). Unlike minimal G-quadruplex DNA (5′-GG–His–His–GG-3′, ODN 9), for the His-modified DNA quadruplex–duplex hybrids, we observed consistent antiparallel conformations in the presence of potassium and various other metal ions.
The His-modified DNA quadruplex–duplex hybrids with hemin were examined in the ABTS oxidation assay, and the initial rates (Vo) were determined to quantify the catalytic performance (Fig. 4, S37 and S38†). We found that hemin/quadruplex–duplex hybrids containing His residues catalyzed the ABTS oxidation reaction. Generally, in ABTS oxidation reactions, parallel conformations of G-quadruplex tend to have higher activities than antiparallel conformations.58,59 Surprisingly, our His-modified DNA quadruplex–duplex hybrids with hemin efficiently promoted ABTS oxidation reaction despite their antiparallel conformations. The effectiveness of the His modification was evident compared with unmodified DNA quadruplex–duplex hybrid (ODN 16/ODN 17) in the control experiment. In the native horseradish peroxidase enzymes, a His residue binds to the hemin iron atom and promotes the oxidation reaction.60–62 Our results indicated that the catalytic ability of unconventional antiparallel-based DNAzymes could be enhanced by a direct incorporation of His residue into the DNA scaffold.
Conclusions
In this study, we incorporated His, a representative metal-binding amino acid residue, into DNA oligonucleotides via an acyclic D-threoninol backbone. His-modified DNA duplexes showed remarkable metal-binding ability depending on the type of metal ion and the secondary structures of DNA. Furthermore, His-modified DNA oligonucleotides could be successfully used in DNA-based asymmetric catalysis as well as ABTS oxidation reactions. This will find important applications as an attractive approach to construct fine-tuned coordination environment for the biomimetic system of multimetal enzymes and the alternative of bioremediation. It is noteworthy that the present approach is as applicable to any other amino acid as to His. The present study is significant as it provides new insights into the incorporation of amino acid residues into nucleic acid scaffolds. Furthermore, we have also synthesized phenylalanine, alanine, and lysine-modified DNA oligonucleotides for conducting experiments comparable with those in this study (Fig. S2–S4, S19, S20, S34 and S35†). Research related to the development of new hybrid biomolecular systems by combining amino acids and nucleic acids is already under way.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We express sincere thanks for a Grant-in-Aid Priority Research (16H06356 for H. S) from Japan Society for the Promotion of Science (JSPS). We also thank KAKENHI program (Grant-in-Aid for scientific research C, 18K05315) for support to S. P. We like to thank Karin Nishimura (Graduate School of Engineering, Kyoto University) for technical assistance to measure mass spectra of synthetic compounds.Notes and references
- P. H. von Hippel and O. G. Berg, Proc. Natl. Acad. Sci. U. S. A., 1986, 83, 1608 CrossRef CAS PubMed.
- S. L. Beaucage and R. P. Iyer, Tetrahedron, 1992, 48, 2223 CrossRef CAS.
- N. C. Seeman, Nature, 2003, 421, 427 CrossRef PubMed.
- L. Pray, Nature Education, 2008, 1, 100 Search PubMed.
- P. W. Rothemund, Nature, 2006, 440, 297 CrossRef CAS PubMed.
- J. Bath and A. J. Turberfield, Nat. Nanotechnol., 2007, 2, 275 CrossRef CAS PubMed.
- J. L. Mergny and D. Sen, Chem. Rev., 2019, 119, 6290 CAS.
- H. S. Rye, S. Yue, D. E. Wemmer, M. A. Quesada, R. P. Haugland, R. A. Mathies and A. N. Glazer, Nucleic Acids Res., 1992, 20, 2803 CrossRef CAS PubMed.
- J. Ju, C. Ruan, C. W. Fuller, A. N. Glazer and R. A. Mathies, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 4347 CrossRef CAS PubMed.
- C. T. Wittwer, M. G. Herrmann, A. A. Moss and R. P. Rasmussen, Biotechniques, 1997, 22, 130 CrossRef CAS PubMed.
- S. T. Crooke, FASEB J., 1993, 7, 533 CrossRef CAS PubMed.
- S. T. Crooke and B. Lebleu, Antisense research and applications, 1993, CRC press Search PubMed.
- L. H. Hurley, Nat. Rev. Cancer, 2002, 2, 188 CrossRef CAS PubMed.
- D. R. A. J. L. B. L. Magda, R. A. Miller, J. L. Sessler and B. L. Iverson, J. Am. Chem. Soc., 1994, 116, 7439 CrossRef CAS.
- N. S. Oltra and G. Roelfes, Chem. Commun., 2008, 45, 6039 RSC.
- P. Fournier, R. Fiammengo and A. Jäschke, Angew. Chem., Int. Ed., 2009, 48, 4426 CrossRef CAS PubMed.
- K. Tanaka, A. Tengeiji, T. Kato, N. Toyama and M. Shionoya, Science, 2003, 299, 1212 CrossRef CAS PubMed.
- E. Meggers, P. L. Holland, W. B. Tolman, F. E. Romesberg and P. G. Schultz, J. Am. Chem. Soc., 2000, 122, 10714 CrossRef CAS.
- H. Weizman and Y. Tor, J. Am. Chem. Soc., 2001, 123, 3375 CrossRef CAS.
- C. J. Murphy, M. R. Arkin, Y. Jenkins, N. D. Ghatlia, S. H. Bossmann, N. J. Turro and J. K. Barton, Science, 1993, 262, 1025 CrossRef CAS.
- T. J. Meade and J. F. Kayyem, Angew. Chem., Int. Ed., 1995, 34, 352 CrossRef CAS.
- O. I. Wilner and I. Willner, Chem. Rev., 2012, 112, 2528 CrossRef CAS.
- J. Bos and G. Roelfes, Curr. Opin. Chem. Biol., 2014, 19, 135 CrossRef CAS PubMed.
- S. Park, I. Okamura, S. Sakashita, J. H. Yum, C. Acharya, L. Gao and H. Sugiyama, ACS Catal., 2015, 5, 4708 CrossRef CAS.
- S. Dey, C. L. Rühl and A. Jäschke, Chem.–Eur. J., 2017, 23, 12162 CrossRef CAS PubMed.
- J. Mansot, S. Aubert, N. Duchemin, J. J. Vasseur, S. Arseniyadis and M. Smietana, Chem. Sci., 2019, 10, 2875 RSC.
- H. A. Wagenknecht, Angew. Chem., Int. Ed., 2003, 42, 3204 CrossRef CAS PubMed.
- S. Park, L. Zheng, S. Kumakiri, S. Sakashita, H. Otomo, K. Ikehata and H. Sugiyama, ACS Catal., 2014, 4, 4070 CrossRef CAS.
- E. Golub, H. B. Albada, W. C. Liao, Y. Biniuri and I. Willner, J. Am. Chem. Soc., 2015, 138, 164 CrossRef.
- S. Park and H. Sugiyama, Angew. Chem., Int. Ed., 2010, 49, 3870 CrossRef CAS.
- S. Park and H. Sugiyama, Molecules, 2012, 17, 12792 CrossRef CAS PubMed.
- J. H. Yum, S. Park and H. Sugiyama, Org. Biomol. Chem., 2019, 17, 9547 RSC.
- J. H. Yum, S. Park, R. Hiraga, I. Okamura, S. Notsu, S. Park and H. Sugiyama, Org. Biomol. Chem., 2019, 17, 2548 RSC.
- S. Johannsen, N. Megger, D. Böhme, R. K. Sigel and J. Müller, Nat. Chem., 2010, 2, 229 CrossRef CAS PubMed.
- P. Scharf and J. Müller, ChemPlusChem, 2013, 78, 20 CrossRef CAS.
- K. Schweizer, J. Kösters and J. Müller, J. Biol. Inorg Chem., 2015, 20, 895 CrossRef CAS PubMed.
- P. M. Punt and G. H. Clever, Chem. Sci., 2019, 10, 2513 RSC.
- P. M. Punt and G. H. Clever, Chem.–Eur. J., 2019, 25, 13987 CrossRef CAS PubMed.
- H. Kashida, K. Sekiguchi, X. Liang and H. Asanuma, J. Am. Chem. Soc., 2010, 132, 6223 CrossRef CAS.
- H. Kashida, K. Murayama, T. Toda and H. Asanuma, Angew. Chem., Int. Ed., 2011, 50, 1285 CrossRef CAS PubMed.
- K. Murayama, Y. Tanaka, T. Toda, H. Kashida and H. Asanuma, Chem.–Eur. J., 2013, 19, 14151 CrossRef CAS PubMed.
- G. Gupta, M. Bansal and V. Sasisekharan, Proc. Natl. Acad. Sci. U. S. A., 1980, 77, 6486 CrossRef CAS.
- R. D. Wells, D. A. Collier, J. C. Hanvey, M. Shimizu and F. Wohlrab, FASEB J., 1988, 2, 2939 CrossRef CAS.
- A. T. Phan, V. Kuryavyi and D. J. Patel, Curr. Opin. Struct. Biol., 2006, 16, 288 CrossRef CAS.
- J. Choi and T. Majima, Chem. Soc. Rev., 2011, 40, 5893 RSC.
- S. Burge, G. N. Parkinson, P. Hazel, A. K. Todd and S. Neidle, Nucleic Acids Res., 2006, 34, 5402 CrossRef CAS PubMed.
- A. T. Phan, V. Kuryavyi, K. N. Luu and D. J. Patel, Quadruplex Nucleic Acids, 2006, 7, 81 Search PubMed.
- J. L. Huppert, FEBS J., 2010, 277, 3452 CrossRef CAS.
- J. Thevarpadam, I. Bessi, O. Binas, D. P. Gonçalves, C. Slavov, H. R. Jonker, C. Richter, J. Wachtveitl, H. Schwalbe and A. Heckel, Angew. Chem., Int. Ed., 2016, 55, 2738 CrossRef CAS PubMed.
- D. A. Baum and S. K. Silverman, Cell. Mol. Life Sci., 2008, 65, 2156 CrossRef CAS PubMed.
- A. J. Boersma, R. P. Megens, B. L. Feringa and G. Roelfes, Chem. Soc. Rev., 2010, 39, 2083 RSC.
- N. Duchemin, I. Heath-Apostolopoulos, M. Smietana and S. Arseniyadis, Org. Biomol. Chem., 2017, 15, 7072 RSC.
- P. Travascio, Y. Li and D. Sen, Chem. Biol., 1998, 5, 505 CrossRef CAS PubMed.
- P. Travascio, A. J. Bennet, D. Y. Wang and D. Sen, Chem. Biol., 1999, 6, 779 CrossRef CAS PubMed.
- K. W. Lim, Z. J. Khong and A. T. Phan, Biochemistry, 2013, 53, 247 CrossRef PubMed.
- K. W. Lim and A. T. Phan, Angew. Chem., Int. Ed., 2013, 52, 8566 CrossRef CAS PubMed.
- K. W. Lim, T. Q. N. Nguyen and A. T. Phan, J. Am. Chem. Soc., 2014, 136, 17969 CrossRef CAS PubMed.
- X. Cheng, X. Liu, T. Bing, Z. Cao and D. Shangguan, Biochemistry, 2009, 48, 7817 CrossRef CAS PubMed.
- D. M. Kong, W. Yang, J. Wu, C. X. Li and H. X. Shen, Analyst, 2010, 135, 321 RSC.
- T. L. Poulos and J. Kraut, J. Biol. Chem., 1980, 255, 8199 CAS.
- R. A. Marcus and N. Sutin, Biochim. Biophys. Acta, Rev. Bioenerg., 1985, 811, 265 CrossRef CAS.
- J. H. Dawson, Science, 1988, 240, 433 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details of the material, compound synthesis, catalytic reactions, and molecular modeling information. See DOI: 10.1039/d0ra01267j |
| This journal is © The Royal Society of Chemistry 2020 |