Open Access Article
Open Access ArticleSimulated spatial radiation impacts learning and memory ability with alterations of neuromorphology and gut microbiota in mice†
Chen Song abc,
Xin Gaoabc,
Wei Song
abc,
Xin Gaoabc,
Wei Song de,
Deyong Zengabc,
Shan Shanabc,
Yishu Yinabc,
Yongzhi Liaf,
Denis Baranenkog and
Weihong Lu*abc
de,
Deyong Zengabc,
Shan Shanabc,
Yishu Yinabc,
Yongzhi Liaf,
Denis Baranenkog and
Weihong Lu*abc
aInstitute of Extreme Environment Nutrition and Protection, Harbin Institute of Technology, Harbin, China. E-mail: lwh@hit.edu.cn
bNational Local Joint Laboratory of Extreme Environmental Nutritional Molecule Synthesis Transformation and Separation, Harbin, China
cSchool of Chemistry and Chemical Engineering, Harbin Institute of Technology, Harbin, China
dCollege of Food Science and Technology, Northwest University, Xi'an, 710069, China
eLaboratory of Nutritional and Healthy Food-Individuation Manufacturing Engineering, Xi'an, 710069, Shanxi, China
fChina Astronaut Research and Training Centre, Beijing, China
gBiotechnologies of the Third Millennium, ITMO University, Saint-Petersburg, Russia
First published on 23rd April 2020
Abstract
Complex space environments, including microgravity and radiation, affect the body's central nervous system, endocrine system, circulatory system, and reproductive system. Radiation-induced aberration in the neuronal integrity and cognitive functions are particularly well known. Moreover, ionizing radiation is a likely contributor to alterations in the microbiome. However, there is a lacuna between radiation-induced memory impairment and gut microbiota. The present study was aimed at investigating the effects of simulated space-type radiation on learning and memory ability and gut microbiota in mice. Adult mice were irradiated by 60Co-γ rays at 4 Gy to simulate spatial radiation; behavioral experiments, pathological experiments, and transmission electron microscopy all showed that radiation impaired learning and memory ability and hippocampal neurons in mice, which was similar to the cognitive impairment in neurodegenerative diseases. In addition, we observed that radiation destroyed the colonic structure of mice, decreased the expression of tight junction proteins, and increased inflammation levels, which might lead to dysregulation of the intestinal microbiota. We found a correlation between the brain and colon in the changes in neurotransmitters associated with learning and memory. The 16S rRNA results showed that the bacteria associated with these neurotransmitters were also changed at the genus level and were significantly correlated. These results indicate that radiation-induced memory and cognitive impairment can be linked to gut microbiota through neurotransmitters.
1. Introduction
In the context of ongoing human Mars exploration missions and deep space programs, participants will be exposed to large amounts of ionizing radiation in space, and the health risks of these exposures need to be assessed. Exposure of living cells and tissues to ionizing radiation, forms of radiation that can remove electrons from the atoms in these cells or tissues, may result in molecular damage, which can eventually lead to early or late injury. Radiation has been shown to have deleterious effects such as increasing the risk of heart disease, mutations, and accelerated bone loss.1,2 In particular, space radiation can produce neurological and behavioral effects. Ionizing radiation widely exists in our daily lives and has been hazardous to human health due to overexposure from natural and artificial sources, such as diagnostic and therapeutic medical usage.3 Patients undergoing long-term radiotherapy usually experience cognitive impairment due to the brain being sensitive to radiation, which is often manifested as deficits in the hippocampal-dependent functions of learning and memory, including spatial information processing.4–6 Park et al. reported that γ-irradiation as a long-term effect can trigger biological responses resulting in the inhibition of hippocampal neurogenesis.7 Kumar et al. found that the hippocampus is one of the sensitive regions affected by whole-body exposure to gamma rays, leading to profound immediate alterations in cognitive functions.8 Because of the uncertainty in extrapolating dose thresholds from rodents to humans, we should find a proper dose that significantly impacts the CNS. In our previous study, we found that 60Co-γ ray irradiation at 4 Gy induced brain injury.9,10 Therefore, in this study, we chose 60Co-γ ray irradiation at 4 Gy to simulate spatial radiation-induced neurological damage. Cognitive impairment is an early manifestation of neurodegenerative diseases; however, it remains to be further studied whether radiation-induced impairment is associated with the onset of neurodegenerative diseases.The human gastrointestinal tract is colonized by trillions of microorganisms. Recently, there has been emerging interest in how the microbiome may predispose an individual to radiation injury and how the microbiome affect the individual's overall response to radiation.11,12 Researchers have shown that exposure to heavy ions (56Fe) causes oxidative stress and dysregulated prostanoid biosynthesis in the mouse intestinal metabolome.13 Moreover, gut microbiota play a critical role in brain development. It has been demonstrated that the gut–brain axis, a bidirectional communication system, integrates the host gut and brain activities.14 On the one hand, the brain influences gastrointestinal functions as well as immune functions.15 On the other hand, emotional factors such as stress or depression influence chronic gastrointestinal illnesses via the gut–brain axis.15 The gut microbiota have been found to communicate with the brain through various mechanisms. For example, intestinal microbes directly produce neurotransmitters that regulate learning and memory as well as cognitive function.16–18 Intestinal microbial metabolites stimulate endocrine cells to produce neuropeptides and gastrointestinal hormones, which play an important role in the visceral sensory and cognitive functions of the central nervous system.19,20 Besides, gut microbiota or their metabolites directly stimulate the intestinal immune system and produce interferons that interfere with the brain's immune response.21,22 Recent studies have shown that compared to SPF mice, germ-free mice exhibit more exploratory and risk-taking behaviours as well as more locomotion, which can be normalized by bacterial colonization.23,24 Several research efforts have demonstrated that healthy gut microbiota may shift some pathological circumstances such as depression and neurodegenerative diseases that can affect the intestinal system.25–27 The relationship between radiation, neurodegenerative diseases and intestinal microbiota remains unclear.
Given that radiation exposure impairs memory and cognitive performance, we hypothesize that this effect is due to gut dysbiosis, alterations in gut microbiota–brain communication via microbial metabolites, and/or the dysregulation of neuronal signaling systems in the brain. To this end, we examined the effects of simulated space-type radiation (60Co-γ ray irradiation) on learning and memory, spatial discrimination, hippocampal morphology and ultrastructure, gut microbial community, colon pathology and the expression of the tight junction proteins, and neurotransmitters in brain and colon tissues. Through this systematic investigation, we found that simulated space-type radiation-induced memory and cognitive impairment can be linked to gut microbiota through neurotransmitters, which can be considered as a valid approach to understanding gut microbiota–brain relationships, and provide a basis for new drug research aimed at improving gut dysbiosis-associated brain dysfunction.
2. Materials and methods
2.1 Animals
Adult Kunming mice (males: n = 10, females: n = 10) weighing 22 ± 2 g were obtained from the Animal Experimental Center of the 2nd Affiliated Hospital of Harbin Medical University (Harbin, Heilongjiang, China). The certification number was SCXK-(HEI)2019-001. All of the animal experimental procedures followed the National Institutes of Health guidelines for the care and use of laboratory animals and were evaluated and approved by the local ethics committee of the 2nd Affiliated Hospital of Harbin Medical University. All animals were maintained in an environmentally controlled breeding room with a regular 12 h light cycle at 22 ± 2 °C. After a normal diet for one week, the animals were divided into 2 groups (n = 10 of each group, half male and half female) including a radiation group and a control group. The mice in the radiation group were exposed to 60Co-γ ray irradiation at 4 Gy (0.9 Gy min−1) (Corn Research Institute of Heilongjiang Academy of Agricultural Sciences). The control group was exposed to a normal environment without any irradiation treatment.2.2 Behavioral test
2.3 Hematoxylin and eosin staining
Hippocampus and colon tissue were fixed in 4% poly formaldehyde for 4–6 hours. After fixation, the tissues were dehydrated and embedded in paraffin. Tissues were cut at a thickness of 4 μm on a microtome and stained with Hematoxylin & Eosin (HE). The morphology was observed under the light microscope. Cell morphology changes in the hippocampal DG region of mice were evaluated under a light microscope (200 magnification), and the cell number of this area was calculated by the following formula: cell counts per mm hippocampal DG area = normal DG cell counts/DG length. Cells with morphological features of pyknosis or nuclear fragmentation will be excluded.The histological damage to the colon was evaluated by light microscopy and the extent of damage was scored as follows:28 (0) normal histological appearance; (1) histological damage limited to the surface epithelium with mild inflammatory cell infiltration; (2) focal ulcer changes and tissue destruction limited to the mucosa, abnormal intestinal glandular structure, and mild inflammatory cell infiltration; (3) focal and transmural inflammation and ulceration with mild to moderate infiltration of inflammatory cells; (4) large transmural ulcers and inflammation, moderate infiltration of inflammatory cells; (5) large ulcer and inflammation, lesions from mucosal immersion to the serous membrane, severe infiltration of inflammatory cells.
2.4 Transmission electron microscopy
After being deeply anaesthetized, the hippocampus and colon were removed and fixed in 2.5% glutaraldehyde in cold (4 °C) PBS. The fixed tissues were then cut into 1 mm3 and fixed again in 2.5% glutaraldehyde. The samples were subsequently fixed with 1% OSO4 in PBS for 2 h, dehydrated and embedded in resin. Ultrathin sections were cut, stained with lead citrate and observed with a transmission electron microscope (H-7650; Hitachi; Tokyo, Japan).2.5 Elisa
The contents of γ-aminobutyric acid (GABA), serotonin (5-HT), norepinephrine (NE) and acetylcholine (ACH) in the hippocampus and colon were measured using commercially available ELISA kits (Shanghai Lengton Bioscience Co., LTD, Shanghai, China). The concentrations of serum IL-6, TNF-α and IL-10 were determined using ELISA kits (Quanzhou Kenuodi Bioscience Co., LTD, Quanzhou, China).2.6 RNA preparation and real-time qPCR analysis
Total RNA was isolated from the hippocampus and colon tissues by using the TaKaRa MiniBEST Universal RNA Extraction Kit (TaKaRa; Dalian, China) according to the manufacturer's instruction. RNA was reverse transcribed to cDNA using PrimeScript™ RT reagent kit (Perfect Real Time) (TaKaRa; Dalian, China) according to the manufacturer's instruction. qPCR assays were performed in each 25 μL reaction containing cDNA corresponding to 100 ng of total RNA and gene-specific primers (Table 1) supplied by Ruibiotech (Ruibiotech Ltd., Beijing, China) and using TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) (TaKaRa; DaLian, China). Reactions were performed by a CFX96 Real-time PCR Detection System (Bio-Rad, America) with initial denaturation at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 60 °C for 30 s. All samples were analyzed in duplicate and the results are expressed as relative values after normalization to β-actin mRNA.| Genes | Primer sequences (5′–3′) |
|---|---|
| β-actinF | ATCACTATTGGCAACGAGCGGTTC |
| β-actinR | CAGCACTGTGTTGGCATAGAGGTC |
| TNF-αF | GTCCGGGCAGGTCTACTTTG |
| TNF-αR | GGGGCTCTGAGGAGTAGACA |
| IL1-βF | TGCCACCTTTTGACAGTGATG |
| IL1-βR | ATGTGCTGCTGCGAGATTTG |
| IL-6 | TGATGGATGCTACCAAACTGGA |
| IL-6 | TGTGACTCCAGCTTATCTCTTGG |
| OccludinF | GTCCTCCTGGCTCAGTTGAA |
| OccludinR | AGAGTACGCTGGCTGAGAGA |
| Claudin-3F | ACTGCGTACAAGACGAGACG |
| Claudin-3R | TCCCTGATGATGGTGTTGGC |
| ZO-1F | TCTTGCAAAGTATCCCTTCTGT |
| ZO-1R | GAAATCGTGCTGATGTGCCA |
2.7 Analysis of gut microbiota composition
The colonic contents were collected and stored at −80 °C for further detection. Total genomic DNA was extracted using a DNA Extraction Kit following the manufacturer's instructions. The quality and quantity of DNA were verified with NanoDrop and agarose gel. Extracted DNA was diluted to a concentration of 1 ng μl−1 and stored at −20 °C until further processing. The diluted DNA was used as a template for PCR amplification of bacterial 16S rRNA genes with barcoded primers and Takara Ex Taq (Takara). For bacterial diversity analysis, V3–V4 variable regions of 16S rRNA genes were amplified with universal primers 343F 5′-TACGGRAGGCAGCAG-3′ and 798R 5′-AGGGTATCTAATCCT-3′. Amplicon quality was visualized using gel electrophoresis, purified with AMPure XP beads (Agencourt), and amplified for another round of PCR. After being purified again with the AMPure XP beads, the final amplicon was quantified using a Qubit dsDNA assay kit. Equal amounts of purified amplicon were pooled for subsequent sequencing.Paired-end reads were then preprocessed using Trimmomatic software to detect and cut off ambiguous bases (N). It also cut off low-quality sequences with average quality scores below 20 using the sliding window trimming approach. After trimming, the paired-end reads were assembled using FLASH software. Parameters of the assembly were as follows: 10 bp of minimal overlapping, 200 bp of maximum overlapping and 20% of the maximum mismatch rate. Sequences were subjected to further denoising as follows: reads with ambiguous, homologous sequences or sequences below 200 bp were abandoned. Reads with 75% of bases above Q20 were retained. Then, reads with chimera were detected and removed.
Clean reads were subjected to primer sequence removal and clustering to generate operational taxonomic units (OTUs) using the Vsearch software with a 97% similarity cutoff. The representative read of each OTU was selected using the QIIME package. All representative reads were annotated and blasted against the Greengens database using the RDP classifier (confidence threshold was 70%). All representative reads were annotated and blasted against the Unite database (ITSs rDNA) using blast.
2.8 Statistical analysis
All data are presented as mean ± standard deviation (S.D). Statistical comparisons were made using the statistical software package SPSS 19.0. Significant differences between the groups were tested by analysis of the independent sample t-tests with levels of significance of P < 0.05 and P < 0.01.3. Results
3.1 The effects of radiation on the behavioral performance in behavior tests
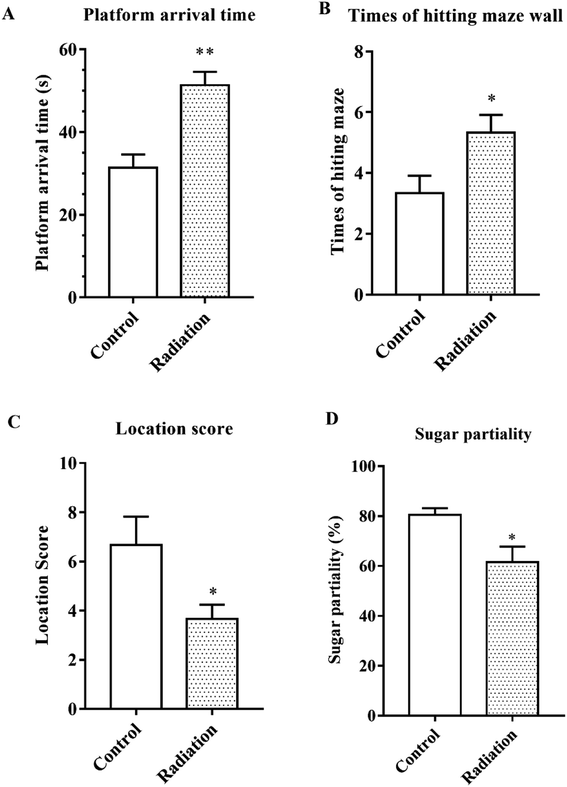
After 60Co-γ irradiation, all the mice were trained for 5 days in the water maze and then tested on the fifth day. As shown in Fig. 1A, the platform arrival time of the radiation group was significantly longer as compared to the control group (P < 0.01). The number of times that the maze was hit and the location score respectively reflected the spatial discrimination and adaptability to stress. Compared to the control group, the number of times the maze was hit increased and the location score decreased in the radiation group (Fig. 1B and C). The sucrose preference test also demonstrated that the radiation caused anhedonia in the mice (Fig. 1D). Therefore, the behavior test indicated that radiation decreased learning and memory in mice and affected nervous system function.3.2 Radiation destroyed the neuronal cell morphology and ultrastructures in the hippocampus
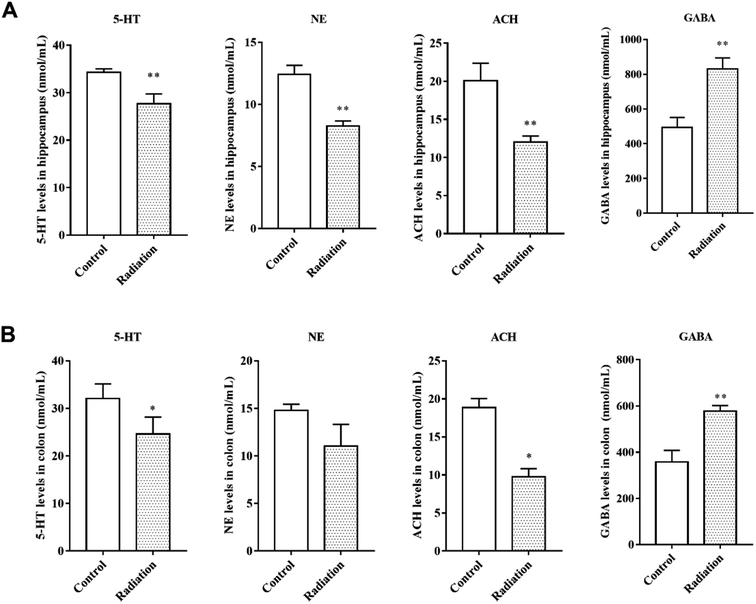
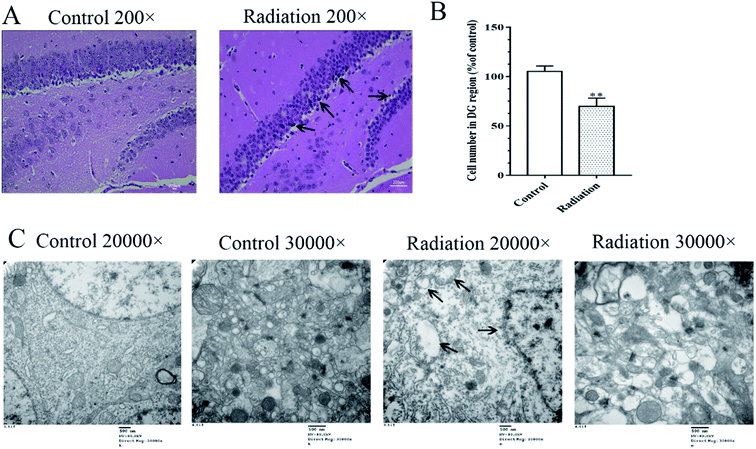
Considering that changes in the cell morphology and the number of the hippocampal neurons are closely associated with learning and memory functions, the histological features of the mice were determined by H&E staining. As shown in Fig. 2A, we observed a large number of neuron cells in the hippocampus of the mice in the control group, which were arranged neatly. However, in the radiation group, the number of neurons decreased and many pyknotic cells could be observed in the hippocampi of the mice. Meanwhile, the ultrastructural features of the hippocampi were detected using transmission electron microscopy. The cytoplasm of the neurons in the control group was abundant and replete with mitochondria, Golgi bodies, and endoplasmic reticuli. The morphology of the nucleus in the control group was normal and the nuclear membrane was clear and complete (Fig. 2C). In the radiation group, the nuclear membrane was unclear and mitochondrial swelling, reduced cristae and vacuolar degeneration were also observed (Fig. 2C). No significant changes were observed in the synaptic structures in the two groups (Fig. 2C).3.3 The effects of radiation on the neurotransmitters in the hippocampus and colon
Neurotransmitters are not only closely related to learning and memory but are also affected by the gut microbiota. Therefore, we measured the levels of neurotransmitters in the brain and colon. After radiation, the levels of 5-HT and NE were significantly decreased in the hippocampus (P < 0.01, Fig. 3A) and colon (P < 0.05, Fig. 3B). We found the same trend in the levels of acetylcholine, which is a cholinergic neurotransmitter (Fig. 3A and B). However, compared to the control group, the GABA levels in both the hippocampus and colon of the radiation group mice increased significantly (P < 0.01, Fig. 3A and B). From these results, we not only observed a consistent trend in the neurotransmitter content in the hippocampus and colon, but we also found a significant correlation between the two groups of data (Table 2).| Object | Pearson R | P value |
|---|---|---|
| a **Significant at P < 0.01. | ||
| ACH | 0.981** | P < 0.01 |
| GABA | 0.896** | P < 0.01 |
| 5-HT | 0.856** | P < 0.01 |
| NE | 0.789 | P = 0.069 |
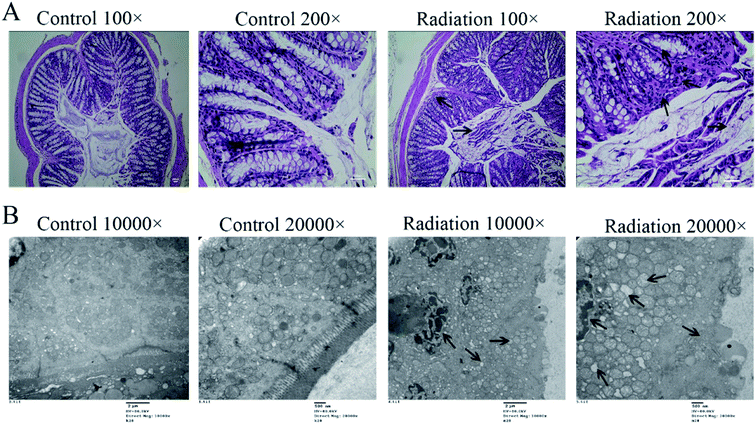
3.4 Radiation destroyed the morphology and ultrastructure of the colon
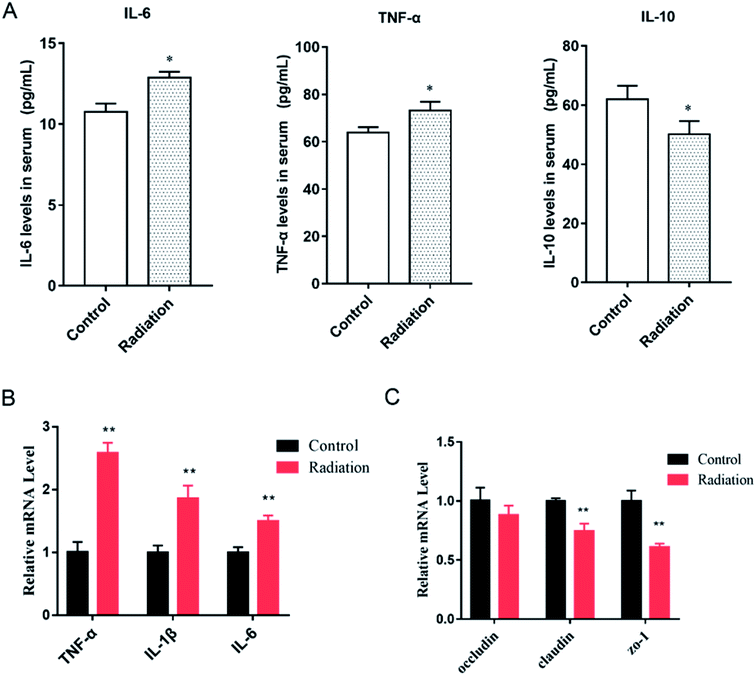
In order to determine the effects of radiation on the colon, the histological features of the colon were determined by H&E staining and the ultrastructures of the colon were determined by transmission electron microscopy. As shown in Fig. 4A (Control), the structure of the colon is normal with the epithelium intact and the glands arranged neatly. In the radiation group (Fig. 4A-Radiation), there was a large number of detached pyknotic epithelial cells in the colonic lumen. The glandular arrangement was slightly disordered, and the lamina propria showed inflammatory hyperplasia. Moreover, the nucleus was deeply stained. The damage to the intestinal epithelium caused by radiation was mainly due to the loss of proliferative capacity of the stem cells at the bottom of the crypt, which led to the loss of villi, mucosal damage, ulcer formation, perforation, bleeding and necrosis. In acute radiation intestinal injury, the intestinal lamina propria was infiltrated by plasma cells and granulocytes with intestinal mucosa atrophy. Moreover, research has demonstrated that radiation can directly cause the recombination or breakage of tight junction proteins, increase the cell gap and cell permeability, and eventually cause the intestinal epithelial barrier to be destroyed.29 From the TEM (Fig. 4B-Control), we found that the tight junctions between the colonic epithelium of the mice showed a dense band with intact structure and clear desmosomes. The microvilli on the surface of the epithelial cells were smooth and well-aligned, and the mitochondria were intact. Conversely, in the radiation group, the mitochondria were swollen and the nucleus showed pyknosis. Furthermore, the microvilli were reduced and even disappeared, and parts of the tight junctions were broken (Fig. 4B-Radiation). In short, the radiation changed the structure of the colon.3.5 The cytokine levels and tight junction proteins in the colon
As we observed, the morphology of colon changed in the radiation group, and the mRNA expression of three tight junction proteins in the colons of the mice were detected. We found that the expressions of claudin and ZO-1 in the radiation group significantly decreased as compared to the control group (Fig. 4C, P < 0.01). This was consistent with the morphological results. At the same time, after radiation, the expressions of inflammatory cytokines (TNF-α, IL-1β, IL-6) were significantly increased (Fig. 5B, P < 0.01). This demonstrated that radiation induced the over-expression of inflammation in the colon, which could lead to the disorder of gut microbiota. From the cytokine levels in the serum, we found that radiation caused a significant increase in the levels of inflammation (TNF-α, IL-6) in the serum, which might be related to the imbalance in the gut microbiota and neuroinflammation (P < 0.05, Fig. 5A). Furthermore, radiation inhibited the anti-inflammatory mediator IL-10 (Fig. 5A).3.6 The effect of radiation on the gut microbiota composition
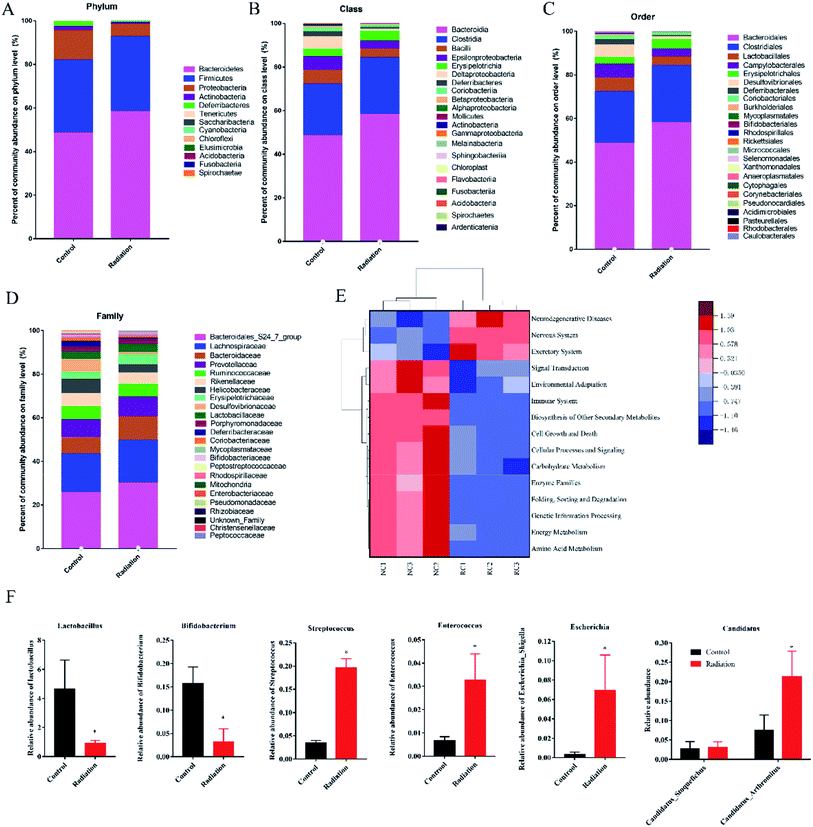
PICRUSt functional predictive analysis is based on 16S sequencing data annotated in the Greengenes database. The PICRUSt software is used to analyze the functional composition of known microorganism genes, to calculate the functional differences between different samples and groups. The heatmap showed significantly different pathways between the radiation group (RC) and the control group (NC). Compared to the control group, 3 pathways (neurodegenerative disease, nervous system, excretory system) were obviously enriched in the radiation group, while the other twelve pathways (signal transduction, environmental adaptation, immune system, biosynthesis of other secondary metabolites, cell growth and death, cellular processes and signaling, carbohydrate metabolism, enzyme families, folding, sorting and degradation, genetic information processing, energy metabolism, amino acid metabolism) were less abundant (Fig. 6E, P < 0.01). This indicated that 60Co-γ irradiation may lead to diseases related to the nervous system.
| Object | Pearson R | P value |
|---|---|---|
| a **Significant at P < 0.01; *significant at P < 0.05. | ||
| Lactobacillus vs. ACH | 0.948** | P < 0.01 |
| Lactobacillus vs. GABA | −0.919** | P < 0.01 |
| Streptococcus vs. GABA | 0.943** | P < 0.01 |
| Streptococcus vs. 5-HT | −0.934** | P < 0.01 |
| Escherichia_Shigella vs. 5-HT | 0.857* | P < 0.05 |
| Enterococcus vs. 5-HT | −0.857* | P < 0.05 |
| Object | Pearson R | P value |
|---|---|---|
| a **Significant at P < 0.01. | ||
| Lactobacillus vs. ACH | 0.895** | P < 0.01 |
| Bifidobacterium vs. GABA | −0.952** | P < 0.01 |
| Streptococcus vs. GABA | 0.958** | P < 0.01 |
| Candidatus_Arthromitus vs. 5-HT | −0.918** | P < 0.01 |
| Group | Shannon | Simpson |
|---|---|---|
| Control group | 6.77 ± 0.19 | 0.97 ± 0.01 |
| Radiation group | 6.79 ± 0.35 | 0.98 ± 0.01 |
4. Discussion
Radiation induces brain neuropathological changes that could induce cognitive impairment in humans and animals. Extensive animal experimental data support the findings. C57BL/J6 mice exposed to the X-rays at 2, 5, 10 Gy showed hippocampal-dependent memory deficits.30 Son et al. demonstrated that mice displayed significant depression-like behaviors after γ-ray irradiation at 10 Gy.31 To our knowledge, water maze can be used as a tool to study neurological diseases, learning and memory in mice. In this article, platform arrival time, the number of hits to the maze and the location score respectively reflect the spatial memory, discrimination and adaptability of the mice to stress. The results demonstrated radiation-induced neurobehavioral impairments by 60Co-γ irradiation at 4 Gy (Fig. 1). In addition, rodents, including mice, are very sensitive to sweets, which are effective rewards. Therefore, sugar partiality was measured to analyze the CNS sensitivity of the mice. It was observed that mice exposed to a dose of 4 Gy showed weak spatial CNS sensitivity (Fig. 1B). Interestingly, similar deficits in spatial memory are an initial sign of a progressive cognitive decline, observed in Alzheimer's disease-related models;32,33 whether this will progress to become far more serious remains to be proven. In mammals, the hippocampus is the key brain region responsible for encoding and consolidating information, and transforming short-term memory into long term memory.34,35 In particular, the CA1 and DG regions of the hippocampus are critical for spatial learning and memory.36 Several studies have revealed that radiation exposures reduce hippocampal neurogenesis.30,37–39 In the present study, we observed not only the decreased neurons but also the mitochondrial and nuclear defects by TEM (Fig. 2). Mitochondria support diverse cellular functions such as the formation of reactive oxygen species, ATP generation and apoptosis.40 Mitochondrial dysfunction is a common feature in neurodegenerative disease, including Alzheimer's and Parkinson's disease.41–43 Therefore, the observed ultrastructural defects in the hippocampus may be related to the deficits in learning and memory induced by 60Co-γ irradiation. Other rays at different doses have also been observed to cause similar hippocampal deficits in mice and rats.44,45 Our study has found that the 60Co-γ ray irradiation impacted learning and memory ability with alterations in the neuromorphology, and we also observed the dysbiosis of gut microbiota in irradiated mice (Fig. 6A).The gut microbiota consist of more than one trillion microorganisms, including bacteria, viruses, fungi and protozoans. The dominant bacteria comprise four main phyla: Bacteroidetes, Firmicutes, Actinobacteria and Proteobacteria.46 The intestinal epithelium, together with the mucus layer, represents a barrier interposed between the luminal contents and the underlying immune, neuronal and muscular compartments that directly interact with enteric bacteria.47 The epithelial barrier integrity was maintained by several subsets of epithelial cells, which are tightly bound together by intercellular junctional complexes (e.g., tight junction proteins such as occludin, zonulin-1 and claudins, gap junctions, adherent junctions, and desmosomes). Besides the intestinal epithelium, gut microbiota interact directly with the enteric immune system, contributing to the maintenance of immune tolerance and shaping the immune responses during inflammation. The morphological results showed that 60Co-γ irradiation destroyed the histological features of the colon (Fig. 4A) and the tight junctions between the colonic epithelium of the mice in the radiation group were broken (Fig. 4B). From the results of qPCR, we also observed that the mRNA expression of tight junction proteins significantly decreased in the colons of irradiated mice (Fig. 5C) and the inflammatory cytokines increased (Fig. 5B). However, the impairments of the colon may have a major impact on the composition and diversity of the gut microbiota. 60Co-γ rays induced disorder in the gut microbiota of exposed mice as compared to the control mice. The relative abundance of some bacteria significantly changed at different levels (Fig. 6A). Notably, the ratio of Firmicutes/Bacteroidetes, which was reported in neurodegenerative disease increased from 0.58 (radiation group) to 0.69 (control group). Moreover, the diversity of the gut microbiota changed (Table 1). After exposure to radiation, neuroinflammatory responses have been well demonstrated to be involved in brain injury, radiation somnolence syndrome, cognitive impairment and aging.48–50 The current findings reveal that the levels of proinflammation increased (Fig. 5A). The gut microbiota form a complex ecosystem and closely interact with the host immune system at the intestinal mucosal surface.51 The effect of ionizing radiation on the gut microbiota is, therefore, a combined outcome of the stress responses from the gut microbes and host cells, and the triangular interactions between the gut microbiota, host, and radiation.
Radiation causes neurophysiological and cognitive impairments as well as intestinal damage and microbiota disturbances. Microbial colonization of the intestine has a significant impact on neurophysiology and behavior.52–54 Experiment confirmed that germ-free (GF) mice exhibited substantial alterations in behaviors and neuropathologies that are relevant to neurodevelopmental, psychiatric and neurodegenerative disorders.54 In our study, the results of PICRUSt functional predictive analysis indicated that the pathway of neurodegenerative disease and the nervous system was significantly enriched in the radiation group (Fig. 6E). There are many potential communication pathways between the gut microbiota and the brain, from complex innervation and highly adjustable neuronal pathways to elusive and difficult-to-measure small molecule information delivery systems.15,16 Microbial endocrinology is becoming an important concept in advancing our knowledge of the microbiota-gut–brain axis. It is well known that some neurotransmitters are related to the nervous system and learning and memory abilities. For example, norepinephrine is historically known for its role in arousal and alertness in the waking state as well as in sensory signal detection, while more recent work has found it is also involved in behavior and cognition, like memory, learning, and attention.55 Serotonin is involved in regulating numerous physiological processes, including gastrointestinal secretion and peristalsis, respiration, vasoconstriction, behavior, and neurological function.56,57 Acetylcholine acts as a neurotransmitter in the central nervous system and is involved in hippocampus-mediated memory function. Meanwhile, GABA is the most widely distributed inhibitory neurotransmitter in the central nervous system. Substantial research supports the link between altered GABAergic neurotransmission and many CNS disorders, including behavioral disorders, pain, and sleep.58,59 We also examined the neurotransmitter levels in order to investigate the effects of radiation on the nervous system (Fig. 3). The levels of these neurotransmitters have significantly changed due to the stimulation of radiation. It has been further proved that radiation damages the nervous system of mice. A great deal of evidence has demonstrated that it is the gut microbiota that influence brain development, neurogenesis, and interact with the enteric and central nervous systems (ENS and CNS, respectively) via communication along the “gut–brain-axis”.60,61 Several different pathways of communication have been identified along the “gut–brain-axis” including those driven by the immune system, by the vagus nerve, by modulation of neuroactive compounds, or by the microbiota. The microbiota have been shown to synthesize and respond to several key neurochemicals including dopamine, norepinephrine, serotonin, or gamma-aminobutyric acid, which are involved in host mood, behavior, and cognition. Many of these host- and microbial-derived neuroactive molecules are also important signaling molecules in host–microbiota interactions at the intestinal interface.16 Several bacteria have also been shown to produce dopamine and norepinephrine, such as Escherichia coli, Escherichia coli (K-12) and Bacillus subtilis.62 Wikoff et al. found that the levels of serotonin were significantly reduced in the blood and colon of germ-free mice as compared to control mice, which can be restored via recolonization with microbiota.63 Based on Matsumoto's research, germ-free animals have substantially reduced luminal and serum levels of GABA, which confirms that the microbiota seem to influence circulating GABA levels.64 At the same time, several bacteria have been reported to produce GABA, including members of the Bifidobacterium and Lactobacillus genera.65,66 Likewise, bacteria express receptors capable of sensing extracellular GABA.67 It is therefore likely that host production of GABA can influence the microbiota. How GABA produced by the microbiota may be involved in human health and disease remains to be determined. In our study, we not only observed the changes in ACH, NE, GABA and 5-HT in the colon and brain of irradiated mice (Fig. 3), but also found the changes in gut microbiota in the genus associated with neurotransmitters (Fig. 6, Tables 3 and 4). This suggests that radiation may affect the behavior and gut microbiota of mice through the “gut–brain axis”.
Indeed, accumulating evidence has shown a link between the gut microbiome and AD.68,69 The cognitive impairments we saw here are only the very beginning of cognitive deficits. However, whether the radiation-induced intestinal bacterial disorder associated with neurodegenerative disease and gut microbes can influence brain function via the gut–brain axis needs to be further studied.
5. Conclusions
We have shown that exposure to 60Co-γ rays at 4 Gy impairs learning and memory ability and even destroys neuronal cell morphology as well as ultrastructures in the hippocampus, specifically disrupting the bacterial community in the colon. Moreover, gut dysbiosis is related to changes in the expression of tight junction proteins and inflammation in the colon as well as the radiation-induced impairment in the histological features and ultrastructures. It also causes distinct alterations in neurotransmitter (ACH, NE, 5-HT, GABA) levels in the brain and colon. Profound alterations in the neurotransmitters of the colon and brain may be relevant to the communication between the gut and the brain. Several of the behavioral and biochemical alterations brought about by radiation are similar to those found in mice with neurodegenerative disease although differences have also been noted. Our findings add to the understanding of the microbiota-gut–brain axis and highlight the potential and limitations of radiation-induced gut dysbiosis as a model system to explore possible mechanisms in the interaction between gut microbiota and the brain.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This research was supported by the National Key Research and Development Program of China: 2017YFC1601900.References
- S. A. Hamilton, M. J. Pecaut, D. S. Gridley, N. D. Travis, E. R. Bandstra, J. S. Willey, G. A. Nelson and T. A. Bateman, J. Appl. Physiol., 2006, 101, 789–793 CrossRef CAS PubMed.
- S. W. Yusuf, S. Sami and I. N. Daher, Cardiol. Res. Pract., 2011, 2011, 317659 Search PubMed.
- N. J. Chao, Exp. Hematol., 2007, 35, 24–27 CrossRef CAS PubMed.
- O. K. Abayomi, Acta Oncol., 1996, 35, 659–663 CrossRef CAS PubMed.
- J. R. Crossen, D. Garwood, E. Glatstein and E. A. Neuwelt, J. Clin. Oncol., 1994, 12, 627 CrossRef CAS.
- O. Surma-Aho, M. Niemel, J. Vilkki, M. Kouri, A. Brander, O. Salonen, A. Paetau, M. Kallio, J. Pyykkonen and J. Jaaskelainen, Neurology, 2001, 56, 1285–1290 CrossRef CAS.
- H. R. Park, U. Jung and S. K. Jo, Radiat. Res., 2007, 168, 446–452 CrossRef CAS PubMed.
- M. Kumar, S. Haridas, R. Trivedi, S. Khushu and K. Manda, Exp. Neurol., 2013, 248, 360–368 CrossRef PubMed.
- Y. Zhou, C. Cheng, D. Baranenko, J. Wang, Y. Li and W. Lu, Int. J. Mol. Sci., 2018, 19, 159 CrossRef PubMed.
- A. Y. Zhou, B. W. Song, C. Y. Fu, D. D. Baranenko, E. J. Wang, F. Y. Li and G. W. Lu, Biomed. Pharmacother., 2018, 99, 781–790 CrossRef PubMed.
- V. Lam, J. E. Moulder, N. H. Salzman, E. A. Dubinsky, G. L. Andersen and J. E. Baker, Radiat. Res., 2012, 177, 573–583 CAS.
- M. Goudarzi, T. D. Mak, J. P. Jacobs, B. H. Moon, S. J. Strawn, J. Braun, D. J. Brenner, A. J. F. Jr and H. H. Li, Radiat. Res., 2016, 186, 219 CAS.
- A. K. Cheema, S. Suman, P. Kaur, R. Singh, A. J. Fornace, Jr. and K. Datta, PLoS One, 2014, 9, e87079 CrossRef PubMed.
- V. C. Lombardi, K. L. De Meirleir, K. Subramanian, S. M. Nourani, R. K. Dagda, S. L. Delaney and A. Palotas, J. Nutr. Biochem., 2018, 61, 1–16 CrossRef CAS PubMed.
- E. A. Mayer, Nat. Rev. Neurosci., 2011, 12, 453–466 CrossRef CAS PubMed.
- M. Lyte, Gut Microbes, 2014, 5, 381–389 CrossRef PubMed.
- K. Pokusaeva, C. Johnson, B. Luk, G. Uribe, Y. Fu, N. Oezguen, R. Matsunami, M. Lugo, A. Major and Y. Mori-Akiyama, Neurogastroenterol. Motil., 2017, 29, e12904 CrossRef PubMed.
- M. H. Beaver and B. Wostmann, Br. J. Pharmacol. Chemother., 1962, 19, 385–393 CrossRef CAS PubMed.
- P. D. Cani, A. Everard and T. Duparc, Curr. Opin. Pharmacol., 2013, 13, 935–940 CrossRef CAS PubMed.
- M. Lyte, J. Endocrinol., 1993, 137, 343–345 CAS.
- B. A. Duerkop, S. Vaishnava and L. V. Hooper, Immunity, 2009, 31, 368–376 CrossRef CAS PubMed.
- K. Takeda and S. Akira, J. Dermatol. Sci., 2004, 34, 73–82 CrossRef CAS PubMed.
- R. Diaz Heijtz, S. Wang, F. Anuar, Y. Qian, B. Bjorkholm, A. Samuelsson, M. L. Hibberd, H. Forssberg and S. Pettersson, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 3047–3052 CrossRef.
- K. M. Neufeld, N. Kang, J. Bienenstock and J. A. Foster, Neurogastroenterol. Motil., 2011, 23, 255–264 CrossRef CAS PubMed.
- S. Wang, X. F. Huang, P. Zhang, K. A. Newell, H. Wang, K. Zheng and Y. Yu, Sci. Rep., 2017, 7, 12203 CrossRef PubMed.
- C. Brandscheid, F. Schuck, S. Reinhardt, K. H. Schäfer, C. U. Pietrzik, M. Grimm, T. Hartmann, A. Schwiertz and K. Endres, J. Alzheimer's Dis., 2016, 56, 775 Search PubMed.
- A. Parashar and M. Udayabanu, Park. Relat. Disord., 2017, 38, 1 CrossRef PubMed.
- H. H. Luk, J. K. Ko, H. S. Fung and C. H. Cho, Eur. J. Pharmacol., 2002, 443, 197–204 CrossRef CAS PubMed.
- S. Shim, H.-S. Jang, H.-W. Myung, J. K. Myung, J.-K. Kang, M.-J. Kim, S. B. Lee, W.-S. Jang, S.-J. Lee and Y.-W. Jin, Toxicol. Appl. Pharmacol., 2017, 329, 40–47 CrossRef CAS PubMed.
- S. Mizumatsu, M. L. Monje, D. R. Morhardt, R. Rola, T. D. Palmer and J. R. Fike, Cancer Res., 2003, 63, 4021–4027 CAS.
- J. S. Kim, J. C. Kim, T. Shin, Y. Son, M. Yang, J. Kim, S. H. Kim, H. Wang, S. K. Jo and U. Jung, Exp. Neurol., 2014, 254, 134–144 CrossRef PubMed.
- J. Raber, D. Wong, M. Buttini, M. Orth, S. Bellosta, R. E. Pitas, R. W. Mahley and L. Mucke, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 10914–10919 CrossRef CAS PubMed.
- R. Jacob, B. Gerold, L. F. Anthony, B. Manuel and M. Lennart, J. Neurosci., 2002, 22, 5204–5209 CrossRef.
- R. P. Kesner, I. Lee and P. Gilbert, Rev. Neurosci., 2004, 15, 333–352 Search PubMed.
- T. Bartsch, R. Schönfeld, F. J. Müller, K. Alfke, B. Leplow, J. Aldenhoff, G. Deuschl and J. M. Koch, Science, 2010, 328, 1412–1415 CrossRef CAS PubMed.
- N. Kazu, T. J. Mchugh, M. A. Wilson and T. Susumu, Nat. Rev. Neurosci., 2004, 5, 361–372 CrossRef PubMed.
- P. Achanta, M. Fuss and J. L. Martinez, Behav. Neurosci., 2009, 123, 1036 Search PubMed.
- T. Kremer, R. Jagasia, A. Herrmann, H. Matile, E. Borroni, F. Francis, H. G. Kuhn and C. Czech, PLoS One, 2013, 8, e59269 CrossRef CAS PubMed.
- l. S. Bã, E. Hajtmanovã, B. Filova, V. Borbelyova and J. Lehotskã, Klin. Onkol., 2015, 28, 191–199 CrossRef PubMed.
- Z. Erpapazoglou, F. Mouton-Liger and O. Corti, Neurochem. Int., 2017, 109, 171–183 CrossRef CAS PubMed.
- J. R. Cannon, V. Tapias, H. M. Na, A. S. Honick, R. E. Drolet and J. T. Greenamyre, Neurobiol. Dis., 2009, 34, 279–290 CrossRef CAS PubMed.
- L. Blazquez-Llorca, S. Valero-Freitag, E. F. Rodrigues, Ã. Merchán-Pérez, J. R. Rodrí-Guez, M. M. Dorostkar, J. Defelipe and J. Herms, Acta Neuropathol. Commun., 2017, 5, 14 CrossRef PubMed.
- S. Gowrishankar, P. Yuan, Y. Wu, M. Schrag, S. Paradise, J. Grutzendler, C. P. De and S. M. Ferguson, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E3699 CrossRef CAS PubMed.
- J. F. Ji, S. J. Ji, R. Sun, K. Li, Y. Zhang, L. Y. Zhang and Y. Tian, Biochem. Biophys. Res. Commun., 2014, 443, 646–651 CrossRef CAS PubMed.
- M. J. Mcginn, D. Sun and R. J. Colello, J. Neurosci. Methods, 2008, 170, 9–15 CrossRef PubMed.
- M. G. Rooks and W. S. Garrett, Nat. Rev. Immunol., 2016, 16, 341 CrossRef CAS PubMed.
- S. C. Bischoff, G. Barbara, W. Buurman, T. Ockhuizen, J.-D. Schulzke, M. Serino, H. Tilg, A. Watson and J. M. Wells, BMC Gastroenterol., 2014, 14, 189 CrossRef PubMed.
- P. Ballesteros Zebadua, CNS Neurol. Disord.: Drug Targets, 2012, 11 Search PubMed.
- B. J. Kelley, J. Lifshitz and J. T. Povlishock, J. Neuropathol. Exp. Neurol., 2007, 66, 989–1001 CrossRef CAS.
- X. Z. Cao, H. Ma, J. K. Wang, F. Liu, B. Y. Wu, A. Y. Tian, L. L. Wang and W. F. Tan, Prog. Neuro-Psychopharmacol. Biol. Psychiatry, 2010, 34, 1426–1432 CrossRef CAS.
- L. V. Hooper, D. R. Littman and A. J. Macpherson, Science, 2012, 336, 1268–1273 CrossRef CAS PubMed.
- S. M. Collins, S. Michael and B. Premysl, Nat. Rev. Microbiol., 2012, 10, 735 CrossRef CAS PubMed.
- J. F. Cryan and T. G. Dinan, Nat. Rev. Neurosci., 2012, 13, 701–712 CrossRef CAS PubMed.
- T. R. Sampson and S. K. Mazmanian, Cell Host Microbe, 2015, 17, 565–576 CrossRef CAS PubMed.
- O. Borodovitsyna, M. Flamini and D. Chandler, Neural Plast., 2017, 2017, 6031478 Search PubMed.
- M. Berger, J. A. Gray and B. L. Roth, Annu. Rev. Med., 2009, 60, 355–366 CrossRef CAS PubMed.
- M. D. Gershon and J. Tack, Gastroenterology, 2007, 132, 397–414 CrossRef CAS PubMed.
- C. G. Ting Wong, T. Bottiglieri and O. C. Snead III, Ann. Neurol., 2003, 54, S3–S12 CrossRef PubMed.
- N. P. Hyland and J. F. Cryan, Front. Pharmacol., 2010, 1, 124 Search PubMed.
- D. H. Rochellys, W. Shugui, A. Farhana, Q. Yu, B. R. Britta, S. Annika, M. L. Hibberd, F. Hans and P. Sven, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 3047–3052 CrossRef PubMed.
- E. S. Ogbonnaya, G. Clarke, F. Shanahan, T. G. Dinan, J. F. Cryan and O. F. O'Leary, Biol. Psychiatry, 2015, 78, e7–e9 CrossRef PubMed.
- P. Strandwitz, Brain Res., 2018, 1693, 128 CrossRef CAS PubMed.
- W. R. Wikoff, A. T. Anfora, J. Liu, P. G. Schultz, S. A. Lesley, E. C. Peters, G. Siuzdak and S. A. Kay, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 3698–3703 CrossRef CAS PubMed.
- M. Matsumoto, R. Kibe, T. Ooga, Y. Aiba, E. Sawaki, Y. Koga and Y. Benno, Front. Syst. Neurosci., 2013, 7, 9 Search PubMed.
- K. Pokusaeva, C. Johnson, B. Luk, G. Uribe, Y. Fu, N. Oezguen, R. K. Matsunami, M. Lugo, A. Major and Y. Mori-Akiyama, Neurogastroenterol. Motil., 2017, 29, e12904 CrossRef PubMed.
- S. Siragusa, A. M. De, C. R. Di, C. G. Rizzello, R. Coda and M. Gobbetti, Appl. Environ. Microbiol., 2007, 73, 7283–7290 CrossRef CAS.
- G. D. Guthrie and C. S. Nicholson-Guthrie, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 7378–7381 CrossRef CAS.
- L. I. Tong-Ju, Y. Zhao, G. M. Dong, D. X. Jia and M. A. Bai-Ping, J. Int. Pharm. Res., 2016, 58, 1 Search PubMed.
- Z. Q. Zhuang, L. L. Shen, W. W. Li, X. Fu, F. Zeng, L. Gui, Y. Lü, M. Cai, C. Zhu and Y. L. Tan, J. Alzheimer's Dis., 2018, 63, 1–10 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra01017k |
| This journal is © The Royal Society of Chemistry 2020 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× and 30
000× and 30