Open Access Article
Open Access ArticleIn silico and in vivo studies of gp120-HIV-derived peptides in complex with G4-PAMAM dendrimers†
Rodríguez-Fonseca Rolando Alberto a,
Bello Martiniano
a,
Bello Martiniano a,
Rojas-Hernández Saúl
a,
Rojas-Hernández Saúl b,
García-Machorro Jazmín
b,
García-Machorro Jazmín c,
Gutiérrez-Sánchez Mara
c,
Gutiérrez-Sánchez Mara b,
Estrada-Pérez Alan Rubéna,
Fragoso-Vázquez Manuel Jonathan
b,
Estrada-Pérez Alan Rubéna,
Fragoso-Vázquez Manuel Jonathan d,
Méndez-Méndez Juan Vicente
d,
Méndez-Méndez Juan Vicente e and
Correa-Basurto José
e and
Correa-Basurto José *a
*a
aLaboratorio de Diseño y Desarrollo de Nuevos Fármacos e Innovación Biotécnológica (Laboratory for the Design and Development of New Drugs and Biotechnological Innovation), Escuela Superior de Medicina, Instituto Politécnico Nacional, Plan de San Luis y Díaz Mirón, Ciudad de México 11340, Mexico. E-mail: corrjose@gmail.com
bLaboratorio de Inmunología Molecular, Escuela Superior de Medicina, Instituto Politécnico Nacional, Plan de San Luis y Díaz Mirón,, Ciudad de México 11340, Mexico
cLaboratorio de Medicina de Conservación, Escuela Superior de Medicina, Instituto Politécnico Nacional, Plan de San Luis y Díaz Mirón, Ciudad de México, 11340, Mexico
dDepartamento de Química Orgánica, Escuela Nacional de Ciencias Biológicas. Instituto Politécnico Nacional, Prolongación Carpio y Plan de Ayala s/n, Ciudad de México 11340, Mexico
eCentro de Nanociencias y Micro y Nanotecnología. Instituto Politécnico Nacional, México City, Mexico. Av. Luis Enrique Erro s/n, Nueva Industrial Vallejo, 07738 Ciudad de México, Mexico
First published on 27th May 2020
Abstract
Novel synthetic vaccines as immunotherapy approaches for HIV are interesting strategies that imply big challenges as they increase the poor immunogenic properties of peptide epitopes and their structural damage from the physiological environment. In this work, we used fourth-generation polyamidoamine dendrimers (G4-PAMAM) to increase the immunoglobulin responses (in vivo) induced by two peptide epitopes (pPGT122: DIIGDIRQAH and pVRC03: DGGANNTSNETFR), both recognized by broadly neutralizing antibodies (bNAb) on gp120-HIV type 1. pPGT122 and pVRC03 were identified on the gp120 surface via recognition by bNAb by using X-ray diffraction-derived structures obtained from the Protein Data Bank. pPGT122 and pVRC03 were coupled to the G4-PAMAM molecule by ligand diffusion using molecular dynamics (LDMDS) simulations and their energetic values were calculated by using the MMGBSA approach. Additionally, docking and MD simulations showed the affinity of pPGT122 and pVRC03 for MHC-I/II. G4-PAMAM–peptide complexes were chemically characterized through MALDI-TOF-MS, LC-ESI-QTOF-MS, atomic force microscopy (AFM) and 1H NMR spectroscopy. Then, the G4-PAMAM–peptide complexes were assayed in vivo by intranasal administration in female BALB/cJ mouse groups, showing that both peptides were immunogenic systemically and in the mucous membrane (in nasal and vaginal washes) via increase in IgG and IgA, respectively. This demonstrated that G4-PAMAM can be used as a nanocarrier for immunogenic peptides.
1. Introduction
Human immunodeficiency virus infection and acquired immune deficiency syndrome (HIV/AIDS) are worldwide health issues that affect 36.7 million people living with HIV-1. In 2016, 1.8 million people newly infected with HIV were diagnosed, and 1.0 million people died from AIDS-related diseases.1 Currently, there are no prophylactic vaccines or immunotherapies that significantly decrease HIV morbidity or prevent AIDS-related diseases.2 Meanwhile, combined antiretroviral therapy is susceptible to viral resistance and is not a cure for HIV/AIDS. Sexual intercourse is the most common HIV transmission route.3 Therefore, mucosal immunity is crucial to block HIV-sexual transmission; thus, it is the best anatomical region to serve as a focus for strategies for HIV-1 prevention and/or treatment.4 The HIV envelope glycoprotein (i.e., gp120) is the major target for the design of prophylactic vaccines against HIV;5 gp120 consists of five variable regions (V1–V5) and five constant regions (C1–C5) connected by an antiparallel sheet.6 It interacts with the CD4 receptor in T-CD4+ lymphocytes, leading to HIV infection.6 To prevent HIV infection by T-CD4+ lymphocytes, antibodies must bind to gp120 to prevent gp120–CD4 complex formation. The structural characterization of broadly neutralizing antibody (bNAb)–gp120 complexes is useful for identifying conserved peptide epitopes in gp120 by using in silico tools. Thus, one can decipher the gp120 regions targeted by antibodies, such as the CD4 binding site, the V3 loop, and the C4/V3 and V2/V1 conserved protein regions.5,7,8 Antibodies against V2 or V3 decreased the risk of HIV infection in an RV144 vaccine trial.9 However, only small cohorts of patients with chronic HIV-1 infection produce bNAbs. Because of this, it is necessary to elicit immune responses to conserved peptide epitopes to prevent HIV-1 transmission.10 Vaccines based on synthetic peptide epitopes from gp120-C4/V3 have been evaluated in animal models using intranasal administration, which demonstrates their capability to induce strong immune responses in vaginal mucous membrane and in sera.4 The use of bNAbs (PGT122 and VRC03) in treatment with passive immunization has been evaluated in HIV-seropositive patients, in which they induced sustained serological remission and neutralization of the virus.11 PGT122 is a bNAb IgG isotype that binds to V3 in gp120, which neutralizes 60–70% of HIV isolates and recognizes glycosylated regions via an allosteric mechanism.7 VRC03 is a bNAb IgG isotype that binds CD4 in gp120. In this sense, it is necessary to identify gp120 peptide epitopes recognized by PGT122 and VRC03 to evaluate their immune responses in mucosal tissues. However, the individual peptide epitopes could be poorly immunogenic, non-absorbable in mucosal tissue, or damaged due to physiological environmental conditions, among other disadvantages. These challenges may be resolved by the use of nanocarriers as adjuvants, such as fourth-generation polyamidoamine dendrimers (G4-PAMAM), which have been proposed to improve cellular uptake in antigen-presenting cells.12–14In this work, two peptide epitopes were identified by the in silico dissociation of crystallographic models of gp120-PGT122 and gp120-VRC03 and then subjected to docking and molecular dynamics simulations with G4-PAMAM and MHC-I/II. These peptides (denoted as pPGT122 and pVRC03) alone or in complex with G4-PAMAM were administered by the intranasal route in a mouse model to evaluate their immune responses in sera as well as in vaginal and nasal washes.
2. Materials and methods
2.1. Computational methods
Prior to the peptide diffusion of G4-PAMAM using LDMDS, the 3D model of the peptides was submitted to 100 ns-long MD simulations to obtain the most populated conformer using the Amber 16 package20 and the ff14SB force field.21 Peptides were embedded in a dodecahedral box containing a TIP3P water model and neutralized using counterions. The water model extended 1 nm between the peptide and the edge of the box. The solvated peptide was equilibrated by performing energetic minimization using the steepest descent through 1000 steps followed by 1 ns equilibration at 300 K, in which the solvent was allowed to relax, whereas the protein was restrained. MD simulations were run with an NPT ensemble at 300 K using the V-rescale algorithm22 and 1 bar pressure using Parrinello–Rahman dynamics. The LINCS algorithm23 was used to constrain all bonds, including hydrogen atoms, and the SETTLE algorithm was used24 to constrain the water molecules. The time step for the simulations was 2 femtoseconds (fs). The van der Waals forces were treated using a cutoff of 1.2 nm. The particle mesh Ewald method25 was used to treat the long-range electrostatic forces, and the coordinates were saved every 0.5 ps. After obtaining the most populated conformation, each peptide was placed around the charge-neutral G4-PAMAM at a ratio of 10 peptides to 1 dendrimer at distances between 7.0 and 15 Å from the dendrimer.
LEaP was used to place the counterions to mimic neutral physiological conditions in a rectangular box of water using a TIP3P water model of 12.0 Å,28 where a single G4-PAMAM molecule was solvated and neutralized. The molecule was minimized and equilibrated by the SHAKE algorithm,29 using 5000 steps of steepest descent minimization, 1000 picoseconds (ps) of heating, 1000 ps of density equilibration with weak restraints on the complex, and 10 ns of constant pressure equilibration at 310 K with hydrogen atoms to allow a 2 femtosecond (fs) time step, and the use of Langevin dynamics for temperature control. The particle mesh Ewald method (PME)30 was used to treat the long-range electrostatic interactions under periodic conditions with a direct space cutoff of 10 Å, and a similar cutoff was employed for the van der Waals interactions. The equilibration was run as 100 ns-long MD simulations without position restraints with periodic boundary conditions (PBC) and an NPT ensemble at 310 K. The time step of the MD simulations was set to 2.0 fs, and the SHAKE algorithm29 was used to constrain the bond lengths at their equilibrium values. Temperature and pressure were maintained using the weak-coupling algorithm31 with values of the coupling constants τT and τP of 1.0 and 0.2 ps, respectively (310 K, 1 atm). The coordinates were retrieved every 20 ps for analysis. AmberTools 16 was used to analyze the time-dependence of the solvent-accessible surface area (SASA), and the radius of gyration (RG) was used to obtain the different G4-PAMAM conformers.
2.2. Experimental procedures
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 μM using 500 μL of deionized water and were incubated at ∼28 °C for 24 h. Subsequently, the mixture was ultrafiltered using a 10 kDa Millipore filter to separate the free peptides (MW < 2 kDa) from G4-PAMAM (mw = 14
1 μM using 500 μL of deionized water and were incubated at ∼28 °C for 24 h. Subsequently, the mixture was ultrafiltered using a 10 kDa Millipore filter to separate the free peptides (MW < 2 kDa) from G4-PAMAM (mw = 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 214 kDa). The solution in the cartridge was used for the sample analysis in 500 μL of deionized water and stored at 4 °C. The mass spectra were obtained by Autoflex MALDI-TOF(/TOF). The sample was prepared with 1 μL of 2′,4′,6′-trihydroxyacetophenone solution and 1 μL of G4-PAMAM or G4-PAMAM–peptide complex. The following parameters were used: reflector voltage 20 kV, laser beam 2, attenuation laser beam focus 40, laser repetition rate 500 Hz, ion source voltage 2 18.15 kV, detector voltage 1802 kV, number of shots 1000, positive voltage POS polarity Pulsed Ion Extraction PIE delay 300 ns, ion source voltage 1 19.5 kV, lens voltage 7 kV, linear detector voltage 2784 kV.
214 kDa). The solution in the cartridge was used for the sample analysis in 500 μL of deionized water and stored at 4 °C. The mass spectra were obtained by Autoflex MALDI-TOF(/TOF). The sample was prepared with 1 μL of 2′,4′,6′-trihydroxyacetophenone solution and 1 μL of G4-PAMAM or G4-PAMAM–peptide complex. The following parameters were used: reflector voltage 20 kV, laser beam 2, attenuation laser beam focus 40, laser repetition rate 500 Hz, ion source voltage 2 18.15 kV, detector voltage 1802 kV, number of shots 1000, positive voltage POS polarity Pulsed Ion Extraction PIE delay 300 ns, ion source voltage 1 19.5 kV, lens voltage 7 kV, linear detector voltage 2784 kV.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution in deionized water. The samples (G4-PAMAM, G4-PAMAM–pPGT122 complex and G4-PAMAM–pVRC03 complex) were distributed on glass slides surface and dried for 3 min at 37 °C in an incubator; the dry sample deposited on the glass slide surface was placed posteriorly in the AFM microscope to perform the measurement. Each glass slide was scanned at three random locations, and images of 3 × 3 μm were obtained. A total of 9 images per sample were obtained. To verify the absence of adsorbed contaminants, the interface was first scanned in the presence of pure solvent.
100 dilution in deionized water. The samples (G4-PAMAM, G4-PAMAM–pPGT122 complex and G4-PAMAM–pVRC03 complex) were distributed on glass slides surface and dried for 3 min at 37 °C in an incubator; the dry sample deposited on the glass slide surface was placed posteriorly in the AFM microscope to perform the measurement. Each glass slide was scanned at three random locations, and images of 3 × 3 μm were obtained. A total of 9 images per sample were obtained. To verify the absence of adsorbed contaminants, the interface was first scanned in the presence of pure solvent.2.3. In vitro biological assays
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution (100 μL) and undiluted nasal washes and vaginal washings (100 μL). The plates were incubated overnight at 4 °C and washed with PBS-Tween (in triplicate) and then secondary antibody was added to the plate per well as follows: 100 μL of peroxidized mouse anti-IgG (Thermo Scientific) or anti-IgA (Zymed Laboratories, San Francisco, CA) was added at a 1
100 dilution (100 μL) and undiluted nasal washes and vaginal washings (100 μL). The plates were incubated overnight at 4 °C and washed with PBS-Tween (in triplicate) and then secondary antibody was added to the plate per well as follows: 100 μL of peroxidized mouse anti-IgG (Thermo Scientific) or anti-IgA (Zymed Laboratories, San Francisco, CA) was added at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6000 and 1
6000 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution, respectively, and incubated for 2 h at 37 °C, and then washed with PBS-Tween (in triplicate). The developing solution was added to the plate per well as follows: 100 μL of 0.5 mg mL−1 o-phenylenediamine in 50 mM phosphate–citrate buffer, pH 5.2, in the presence of H2O2. After 15 min, the reactions were stopped with 50 μL of 2.5 M H2SO4, and the absorbance at 492 nm (A492) was measured in a Multiscan Ascent (Thermo Labsystems) microplate reader. Samples were analyzed in duplicate.
500 dilution, respectively, and incubated for 2 h at 37 °C, and then washed with PBS-Tween (in triplicate). The developing solution was added to the plate per well as follows: 100 μL of 0.5 mg mL−1 o-phenylenediamine in 50 mM phosphate–citrate buffer, pH 5.2, in the presence of H2O2. After 15 min, the reactions were stopped with 50 μL of 2.5 M H2SO4, and the absorbance at 492 nm (A492) was measured in a Multiscan Ascent (Thermo Labsystems) microplate reader. Samples were analyzed in duplicate.2.4. Statistics
The data were statistically analyzed by means of univariate analysis of variance (ANOVA) and Tukey's post hoc test with PRISM Software (GraphPad). A significance level of P < 0.05 was considered to indicate whether there was a significant difference between two groups.3. Results
3.1. Computational methods
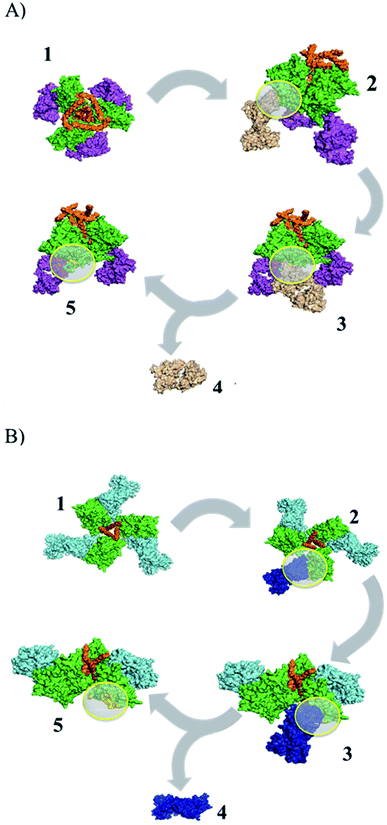 | ||
| Fig. 1 Identification of peptide epitopes in silico using structural dissociation in USCF CHIMERA v1.9. (A) gp120–PGT122 complex, green: gp120, orange: gp41, purple: pPGT122 (PDB: 4NCO). (B) gp120–VRC03 complex, green: gp120, blue: pVRC03 (PDB: 3CC8). Yellow circles show the gp120 surface coupled to pPGT122 or pVRC03. | ||
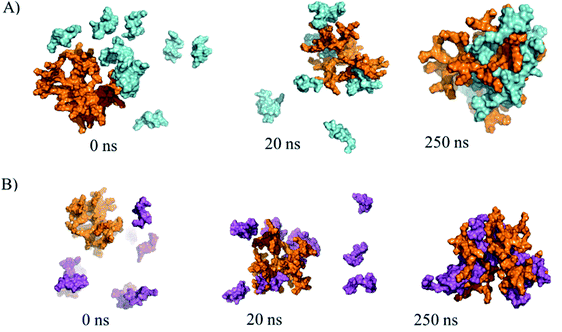
The G4-PAMAM–peptide complexes were obtained with the Amber 16 package20 by employing ligand diffusion in MD simulations36 to reach the most energetically favorable relative binding free energy (ΔGmmgbsa) values, which were obtained by MMGBSA calculations and indicated that the coupling of both peptides can be performed in all conformational states on the surface of G4-PAMAM. Eight pPGT122 or ten pVRC03 molecules were capable of being coupled on G4-PAMAM in the 250 ns-long MD simulations in aqueous environments (Fig. 2A and B). Based on the ΔG values and the number of peptides coupled to G4-PAMAM, pVRC03 exhibited a more favorable affinity for G4-PAMAM than pPGT122 (Table 1). Furthermore, there were differences in the ΔGmmgbsa value between the conformers of the same peptide epitope, depending on the location where they were internally coupled, which was close to the ethylenediamine nucleus or between the branches in a tertiary amine, or in a primary amine group (NH2) on the surface of the dendrimer. In addition, the nonpolar interactions (ΔEnon-polar = ΔEvwd + ΔGnpol,sol) were the main energetic contributors to G4-PAMAM dendrimer–peptide complex stabilization and formation (Table 1).
| System | ΔEvdw | ΔEele | ΔGele,sol | ΔGnpol,sol | ΔEpolar | ΔEnon-polar | ΔGmmgbsa |
|---|---|---|---|---|---|---|---|
| G4-PAMAM–peptide complex | |||||||
| PGT122 | −26.28 (0.22) | −1177.31 (−3.9) | 1189.86 (3.71) | −4.44 (−0.26) | 12.55 | −30.72 | −18.17 (0.20) |
| Conformer 1 | |||||||
| PGT122 | −70.82 (0.33) | −1248.23 (−3.33) | 1261.64 (3.25) | −10.73 (−0.4) | 13.41 | −81.55 | −68.14 (0.41) |
| Conformer 2 | |||||||
| PGT122 | −38.32 (0.22) | −1242.67 (−2.58) | 1254.15 (2.51) | −6.42 (−0.28) | 11.48 | −44.74 | −33.26 (0.27) |
| Conformer 3 | |||||||
| PGT122 | −24.25 (0.30) | −1120.23 (−3.2) | 1130.63 (3.18) | −3.99 (−0.4) | 10.4 | −28.24 | −17.84 (0.24) |
| Conformer 4 | |||||||
| PGT122 | −68.34 (0.28) | −1147.41 (−2.38) | 1178.33 (2.27) | −10.19 (−0.4) | 30.92 | −78.53 | −47.61 (0.41) |
| Conformer 5 | |||||||
| PGT122 | −34.24 (0.17) | −1269.89 (−3.68) | 1273.37 (3.52) | −6.01 (−0.3) | 3.48 | −40.25 | −36.77 (0.33) |
| Conformer 6 | |||||||
| PGT122 | −36.85 (0.29) | −1293.62 (−2.74) | 1302.75 (2.72) | −6.01 (−0.3) | 9.13 | −42.86 | −33.73 (0.30) |
| Conformer 7 | |||||||
| PGT122 | −46.32 (0.22) | −1102.09 (−2.5) | 1129.74 (2.41) | −6.98 (−0.3) | 27.65 | −53.3 | −25.66 (0.29) |
| Conformer 8 | |||||||
| VRC03 | −6.63 (0.10) | −929.01 (−2.19) | 922.67 (2.13) | −2.25 (−0.1) | −6.34 | −8.88 | −15.22 (0.16) |
| Conformer 1 | |||||||
| VRC03 | −79.56 (0.28) | −1238.39 (−2.23) | 1270.78 (2.22) | −11.64 (−0.4) | 32.39 | −91.2 | −58.81 (0.39) |
| Conformer 2 | |||||||
| VRC03 | −59.63 (0.52) | −1044.03 (−3) | 1069.28 (3.10) | −9.28 (−0.6) | 25.25 | −68.91 | −43.66 (0.42) |
| Conformer 3 | |||||||
| VRC03 | −39.03 (0.33) | −1087.17 (−4.27) | 1112.10 (4.18) | −6.15 (−0.6) | 24.93 | −45.18 | −20.25 (0.45) |
| Conformer 4 | |||||||
| VRC03 | −55.98 (0.48) | −1066.67 (−3) | 1088.65 (3.00) | −8.66 (−0.7) | 21.98 | −64.64 | −42.66 (0.45) |
| Conformer 5 | |||||||
| VRC03 | −52.53 (0.20) | −1404.99 (−2.36) | 1423.97 (2.26) | −8.53 (−0.2) | 18.98 | −61.06 | −42.08 (0.28) |
| Conformer 6 | |||||||
| VRC03 | −15.93 (0.40) | −1071.42 (−4.06) | 1077.69 (3.94) | −3.04 (−0.5) | 6.27 | −18.97 | −12.70 (0.30) |
| Conformer 7 | |||||||
| VRC03 | −36.24 (0.31) | −963.24 (−2.7) | 978.36 (2.66) | −5.38 (−0.3) | 15.12 | −41.62 | −26.50 (0.37) |
| Conformer 8 | |||||||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| MHC-II HLA-DRA4 (PDB: 1D5M)–peptide complex | |||||||
| PGT122 | −56.26 (−0.59) | 60.73 (−1.81) | −37.99 (1.61) | −8.73 (−0.6) | 22.74 | −64.99 | −42.25 (0.63) |
| VRC03 | −56.53 (−0.41) | 80.51 (−2.68) | −39.50 (2.11) | −9.48 (−0.5) | 41.01 | −66.01 | −25.00 (0.65) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| MHC-I HLA-B5703 (PDB: 2VBQ)–peptide complex | |||||||
| PGT122 | −57.91 (−0.39) | −133.42 (−4.3) | 157.90 (4.14) | −8.82 (0.6) | 24.48 | −66.73 | −42.25 (0.48) |
| VRC03 | −54.87 (−0.36) | −25.5 (−1.73) | 60.23 (1.50) | −8.13 (0.4) | 34.73 | −63 | −28.27 (0.43) |
3.2. Experimental procedures
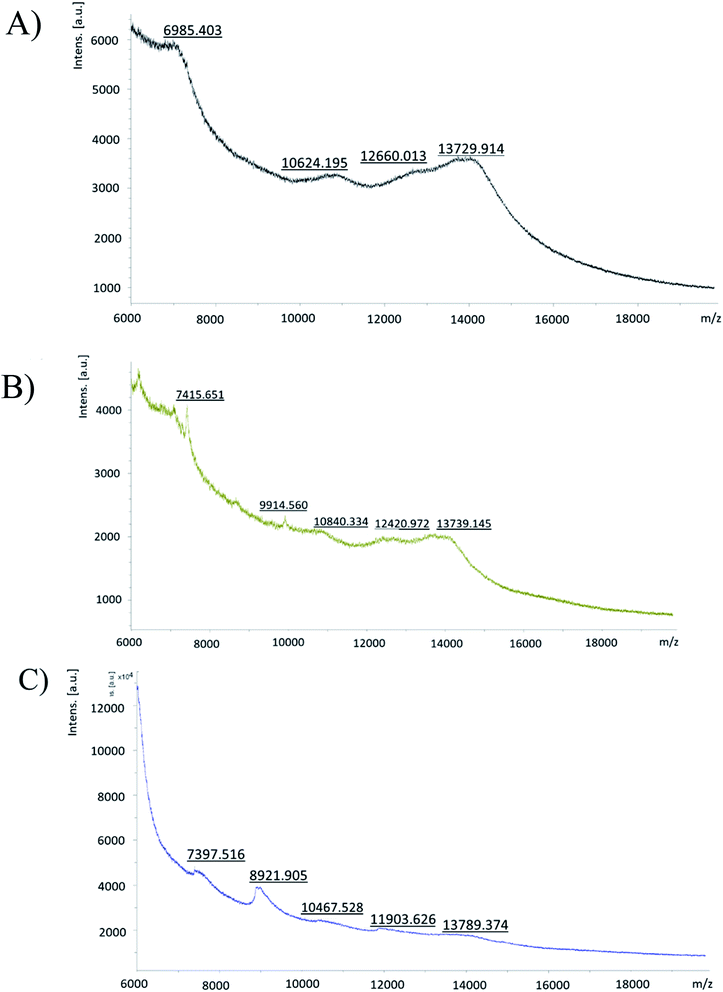
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 m/z, which belonged to a peptide bound to a G4-PAMAM molecule (Fig. 3). The calculation was as follows: peak m/z = 7416; this corresponds to pPGT122. m.w. G4-PAMAM + m.w. dendrimer pPGT122 = 14
000 m/z, which belonged to a peptide bound to a G4-PAMAM molecule (Fig. 3). The calculation was as follows: peak m/z = 7416; this corresponds to pPGT122. m.w. G4-PAMAM + m.w. dendrimer pPGT122 = 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 214 + 7416 = 21
214 + 7416 = 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 630; m.w. G4-PAMAM–peptide complex = 21
630; m.w. G4-PAMAM–peptide complex = 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 630, z = 2; m/z = 10
630, z = 2; m/z = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 815–10
815–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 840. Consequently, the peak at m/z = 10
840. Consequently, the peak at m/z = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 840 corresponds to pPGT122. G4-PAMAM + m.w. dendrimer pVRC03 = 14
840 corresponds to pPGT122. G4-PAMAM + m.w. dendrimer pVRC03 = 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 214 + 1382 = 15
214 + 1382 = 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 596; m.w. G4-PAMAM–peptide complex = 15
596; m.w. G4-PAMAM–peptide complex = 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 596, z = 2; m/z = 7798–7397. Consequently, the peak at m/z = 7397 corresponded to pVRC03.
596, z = 2; m/z = 7798–7397. Consequently, the peak at m/z = 7397 corresponded to pVRC03.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 212.854 g mol−1, according to eqn (1) and (2) with the following parameters: mn = 1422.2934 and mn+1 = 1293.0857; therefore, the load is n = 10. It should be noted that this mass was corroborated by the Large Molecular Feature tool and the deconvolution algorithm (data not shown); both tools were included in the equipment software. For the G4-PAMAM–pPGT122 complex (Fig. 6), mn = 1142, mn+1 = 1080, and n = 19; thus, the corresponding pPGT122 molecules represent seven molecules per single G4-PAMAM molecule. Regarding the G4-PAMAM–pVRC03 complex, mn = 1137, mn+1 = 1089 and n = 23, which thus correspond to nine pVRC03 molecules per G4-PAMAM.
212.854 g mol−1, according to eqn (1) and (2) with the following parameters: mn = 1422.2934 and mn+1 = 1293.0857; therefore, the load is n = 10. It should be noted that this mass was corroborated by the Large Molecular Feature tool and the deconvolution algorithm (data not shown); both tools were included in the equipment software. For the G4-PAMAM–pPGT122 complex (Fig. 6), mn = 1142, mn+1 = 1080, and n = 19; thus, the corresponding pPGT122 molecules represent seven molecules per single G4-PAMAM molecule. Regarding the G4-PAMAM–pVRC03 complex, mn = 1137, mn+1 = 1089 and n = 23, which thus correspond to nine pVRC03 molecules per G4-PAMAM.
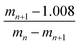 | (1) |
| m = (mnn) − (1.008n) | (2) |
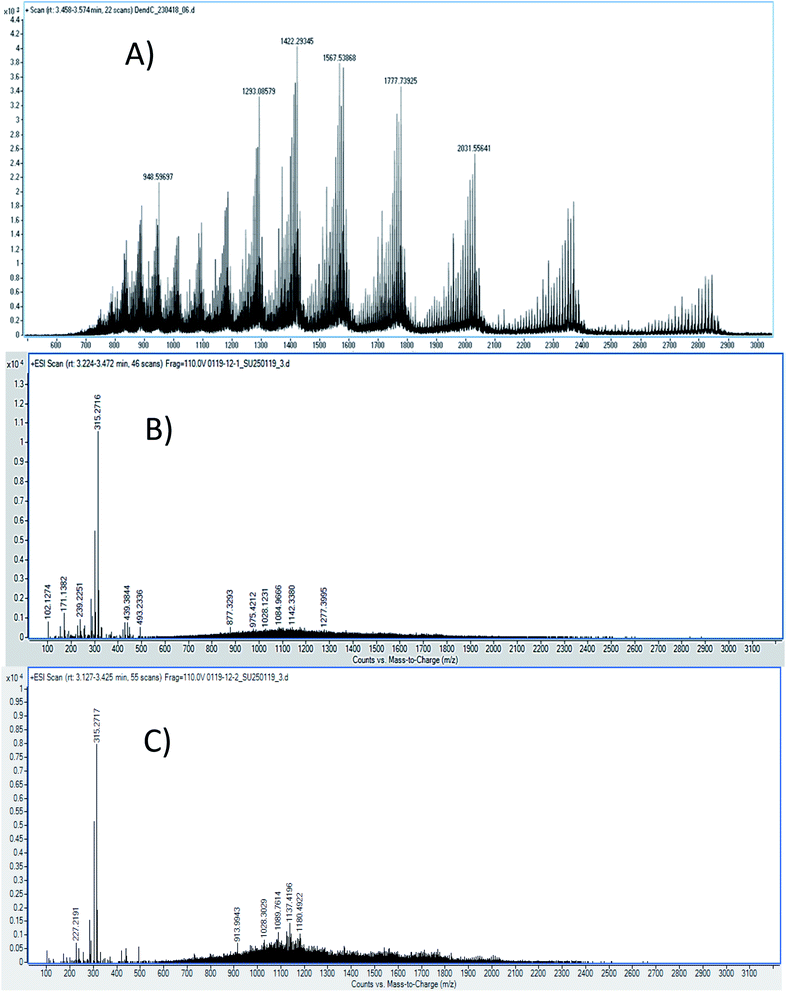 | ||
| Fig. 6 LC-ESI-QTOF-MS spectra of (A) G4-PAMAM, (B) the G4-PAMAM–pPGT122 complex and (C) the G4-PAMAM–pVRC03 complex. | ||
LC-ESI-QTOF-MS allowed the characterization of several G4-PAMAM populations according to their protonation patterns, as has been reported for other PAMAM dendrimer generations.53,54
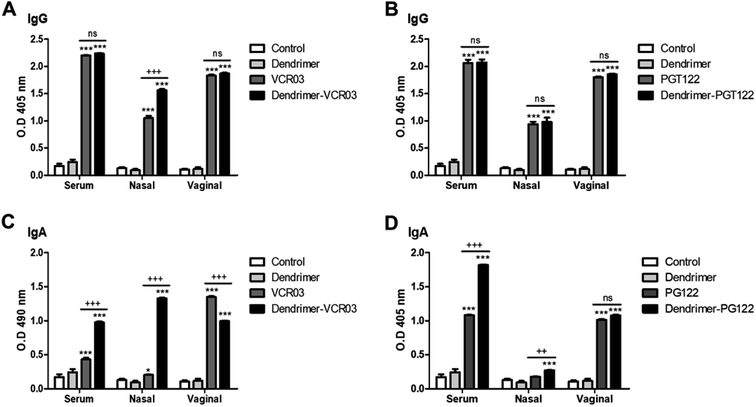
3.2.5.1. IgG and IgA responses. In all the fluids tested, a statistically significant increase was observed in IgG antibodies in the groups administered the peptide alone or G4-PAMAM–peptide complexes, in comparison with that in the control or G4-PAMAM groups (Fig. 7). In addition, the IgG responses were not significantly different for the G4-PAMAM–pPGT122 complex and G4-PAMAM–pVRC03 complex, except for the response to pVRC03 in nasal washes (Fig. 7), in which G4-PAMAM also exhibited adjuvant effects.
Regarding IgA in all of the fluids tested, there was a statistically significant increase in antibodies in the groups administered the peptide alone as compared with that in the controls, except for the nasal washes. In contrast, for the G4-PAMAM–peptide complexes, there were statistically significant differences in comparison with the control or G4-PAMAM groups (Fig. 7). In some cases, G4-PAMAM could be an adjuvant, except for vaginal IgA responses, in which G4-PAMAM-pVRC03 decreased the response. In this work, G4-PAMAM–peptide complexes were administered via the intranasal route to induce humoral immune responses in the genital tract and systemically, as has been reported previously.13,14
4. Discussion
In this work, we sought to identify immunogenic epitopes in gp120 using in silico studies (Fig. 1). However, it is known that peptides are not enough to induce immunogenic responses and that they are subject to proteasome degradation. Furthermore, we suggest using a nanoparticle (G4-PAMAM) that can protect peptides and that can be intranasally administered. G4-PAMAM can be useful as both an intranasal peptide transporter and a mucous membrane adjuvant due to its molecular interactions and the consequent formation of water-soluble stable complexes, which confer mucoadhesive properties that enhance bioadhesion to the glycoprotein layer of the mucous membrane.39 The G4-PAMAM dendrimer can improve the polar physicochemical properties of peptides,40 mucous membrane protection against proteases by internalization of peptides into G4-PAMAM dendrimer13 and passive diffusion of complexes mediated by other functional groups on spikes of G4-PAMAM.40 Additionally, it has been reported that the G4-PAMAM can improve antigen cell uptake40,41 in nasal-associated lymphoid tissue by enhancing the pH-dependent stability of the complex42 and it has been suggested that the release of the peptides could be carried out in an acidic endosome microenvironment.40,43 Once the G4-PAMAM releases peptides, these peptides can bind to MHC-I/II, generating a cell-MHC-I/II–peptide complex that elicits cellular/humoral immune responses. Thus, G4-PAMAM dendrimer–peptide complexes administered intranasally are useful for eliciting cellular and humoral responses against Chlamydia trachomatous in genital infection.13,14 This immune response in mammals44,45 is due to i.n. administration, which yields antibody precursor cells that travel to other lymphoid tissues such as the genital tract to elicit immune responses44 that can be favored by the use of nanoparticles as adjuvants.46,47In this work, we identified pPGT122 and pVRC03 from gp120 through the structural dissociation of the gp120 monomeric structure from the VRC03 bNAb and the PGT122 bNAb,48–51 as previously reported for other peptides.14 Quaternary analyses revealed that neither pPGT122 nor pVRC03 was hidden in the gp120–gp120 or gp41–gp120 interface, which allowed them to be recognized by specific antibodies.
According to our results, both peptide epitopes are anionic, could be non-toxic, and can be used to produce water-soluble complexes with G4-PAMAM. Moreover, the amphipathic composition of both peptide epitopes is useful for making hydrophobic contacts with internal G4-PAMAM sites (Fig. 2A and B). We explored the capability to make G4-PAMAM–pPGT122 or G4-PAMAM–pVRC03 complexes by ligand diffusion strategies by using MD simulations (Fig. 2A and B). We explored the conformational space during 250 ns MD simulations of free peptide epitopes diffused into G4-PAMAM until the structural stabilization of the G4-PAMAM–peptide complexes was obtained. The energetic contributions that allow the binding of peptides to G4-PAMAM were then determined by the MMGBSA method. pVRC03 and pPGT122 are capable of forming non-covalent associations with G4-PAMAM by complementary electrostatic interactions; however, determining the total energetic contribution is necessary to explain the formation and stability of G4-PAMAM–peptide complexes by considering all types of non-covalent interactions between G4-PAMAM and peptide epitope molecules and the solvation process.14,26 In this work, the energetic contributions were determined by the MMGBSA method for G4-PAMAM–peptide complexes, thus, the nonpolar energetic contribution mainly explains the capability to form G4-PAMAM–peptide complexes.26 The differences in the affinity of pVRC03 and pPGT122 for G4-PAMAM can be explained by their different binding poses, which are dependent on their peptide sequences. Peptide diffusion on G4-PAMAM during the MD simulations showed that the peptides could be partially encapsulated, allowing interaction with G4-PAMAM methylene groups; specific regions formed intramolecular hydrophobic interactions, whereas proton acceptor groups in amides formed intramolecular hydrogen bonds. van der Waals interactions were formed at close distances with intramolecular groups, and ionic interactions with acidic peptide epitope functional groups were formed between G4-PAMAM protonated amides (internally at branch bifurcations or in outer primary amides). G4-PAMAM can be useful as a reversible transporter because it forms internal cavities that are functional groups capable of undergoing amine protonation at different pH values.14,26 This allows the formation of G4-PAMAM–peptide complexes at an acidic pH, which favors the “open conformation”, whereas a neutral pH favors the closed conformation, allowing the peptide to then be released under acidic conditions by the antigen-presenting cell peptide epitope processing pathway. It is important to note that both peptides have hydrophobic properties that suggest they can be accommodated in MHC-I/II cavities (ESI Fig. 2S†).52 It was useful to select peptide epitopes based on their potential to be transported by G4-PAMAM to confirm the peptide immunogenic properties. The formation of MHC-I/II–peptide complexes and the peptide interactions with MHC-I/II hydrophobic cavities was corroborated by protein molecular docking studies and validated by MD simulations (ESI Fig. 2S†) coupled with MMGBSA (Table 1). Thus, the non-polar energetic contribution mainly explains the capability to form stable MHC-I/II–peptide complexes. However, these in silico predictions must be demonstrated experimentally. Furthermore, both peptides were used to form G4-PAMAM–peptide complexes in aqueous solutions. The G4-PAMAM–peptide complexes were chemically characterized by MALDI-TOF and LC-ESI-QTOF-MS, with MALDI-TOF being the most precise (Fig. 3)53,54 due to the fact that during LC-ESI-QTOF-MS, the peptide was separated during the LC stages. However, LC-ESI-QTOF-MS allowed the characterization of several G4-PAMAM populations according to their protonation patterns, as has been reported for other PAMAM generations.53,54
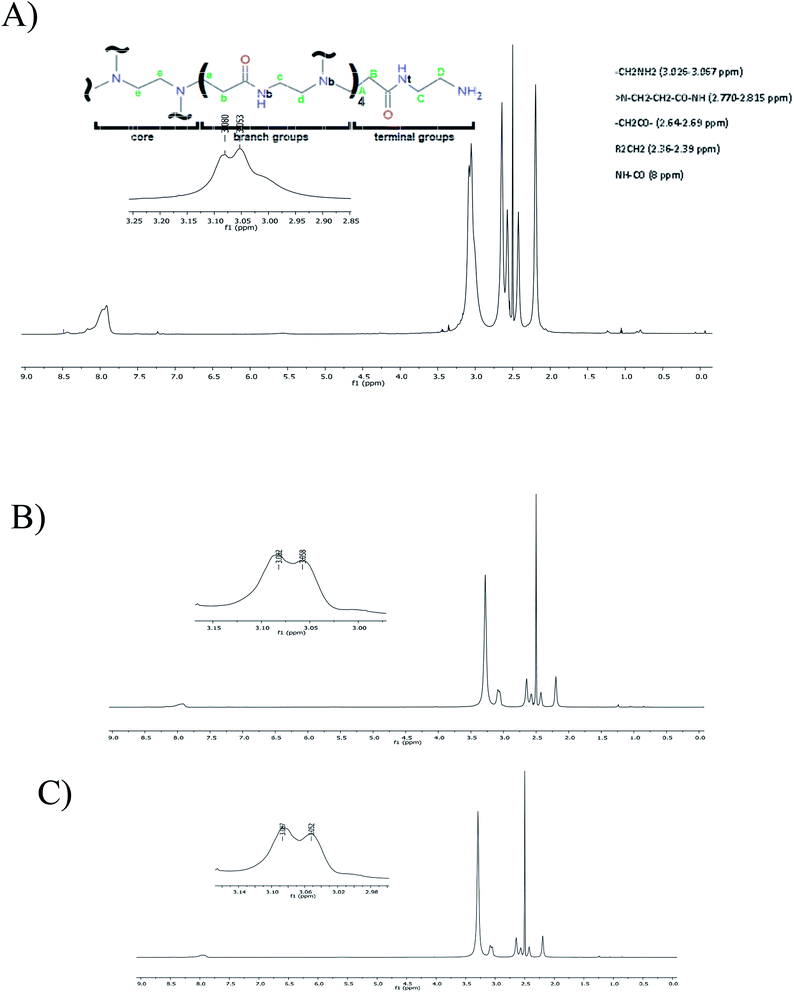
By using 1H NMR, it was possible to depict the non-bonding interactions of the peptide with different protons of G4-PAMAM (Fig. 5). All signals showed a decrease in electron density with a respective change in the chemical shift at high frequency due to the presence of intermolecular interactions around the protons.55 According to the 1H NMR spectra of G4-PAMAM-pVRC03 or G4-PAMAM-pPGT122, there was a decrease in the amide group (≅8 ppm), methylene groups (≅2.36–2.81 ppm) and –NH2 (≅3.026-3.067 ppm). These signal shifts allowed us to deduce the locations of pVRC03 or pPGT122 molecules that may have been entrapped on the outer surfaces of functional groups of G4-PAMAM. As such, some exposed regions of the pVRC03 or pPGT122 molecules, which depend on their conformation, may have interacted with methylene groups and amides in the G4-PAMAM branches near NH2 groups located on most external surfaces of G4-PAMAM, and pVRC03 or pPGT122 molecules may have been entrapped between one or more branches.
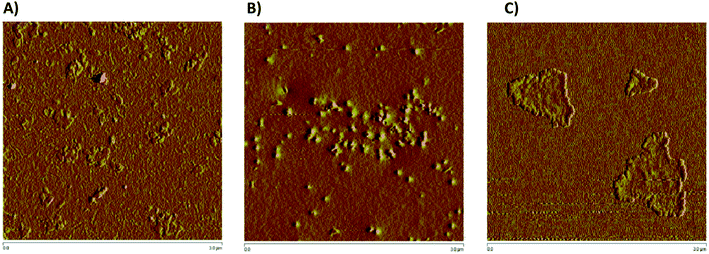
The particle morphological shapes determined by AFM showed different patterns according to the presence or absence of peptides (Fig. 4), as was reported by using a peptide carrier.43,56 The morphological shifts observed by AFM in the G4-PAMAM molecules were dependent on the G4-PAMAM physicochemical properties according to the microenvironment and ligand interactions. All of these interactions can induce conformational rearrangement, which modifies the molecular surface exposure of G4-PAMAM molecules.43,56 Because of this, the interaction of ligands on G4-PAMAM implies that some intermolecular interactions in G4-PAMAM change to favor intermolecular interactions with the ligands, which in turn induces conformational rearrangements in G4-PAMAM molecules and decreases the G4-PAMAM exposed surface that interacts with ligand molecules. Overall, this implies that according to the physicochemical properties of the exposed molecular surface, the complex differs in the supramolecular rearrangement of G4-PAMAM, and the corresponding intermolecular interactions between complexes can explain the supramolecular rearrangements between molecular G4-PAMAM–peptide complexes that result in several complexes.43,56
In this work, G4-PAMAM–peptide complexes were administered via the intranasal route to induce humoral immune responses in the genital tract and systemically, as has been reported previously;13,14 G4-PAMAM has been used as a vaccine adjuvant46,57,58 and as a peptide epitope carrier.47,59 The immune response in the genital mucous membrane has been attributed to Ab-secreting plasma cell precursors elicited by intranasal immunization44 and induction of a proinflammatory cytokine profile.60 However, antibody transudation processes in the mucous membrane,45,61 lymphoid tissue immune responses induced by nasal immunization62 and antigens can elicit variable immune responses in distant mucous membrane such as that in the genital tract.45 G4-PAMAM–peptide complexes induce humoral immune responses systemically and in the genital tract, as previously reported using nasal immunization.13,14 The estral cycle is considered to be involved in immune response variability according to the female sex;63 thus, immune responses can be modulated independently of antigen-elicited immune responses, resulting in variable immune responses.64 The co-administration of G4-PAMAM–peptide complexes and nasal adjuvants could modulate antiviral immune responses along with female sex hormones,63 and the immune innate responses elicited by G4-PAMAM nasal immunization could be tunable. Therefore, common mucous innate62 and adaptive immune system mechanisms could be modulated to improve the production of Ab-secreting plasma cell precursors.65
5. Conclusions
In this work, we identified two peptide epitopes derived from the in silico dissociation of bNAb PGT122 and VRC03. Both epitopes were able to recognize molecules of MHC-I/II and G4-PAMAM according to in silico and experimental studies. Additionally, both G4-PAMAM–peptide complexes produced a humoral response in serum and mucous membranes greater than or equal to that produced by the individual peptide epitope, demonstrating their potential for application in the rational development of future antibodies with recognition profiles similar to bNAb PGT122 and VRC03.Conflicts of interest
The author(s) confirm that this article content has no conflict of interest.Acknowledgements
The work was supported by grants from CONACYT (CB-254600, CB-241339 and PDCPN-782), CYTED: 214RT0842, PIFI-SIP-COFAA and Proyecto insignia-2015 from IPN, and scholarships to Rolando Alberto Rodríguez-Fonseca from CONACYT. We thank the Centro de Nanociencias y Micro y Nanotecnologías del Instituto Politécnico Nacional, México (https://www.nanocentro.ipn.mx by Mass Spectroscopy experiments.References
- UNAIDS, UNAIDS DATA, 2017 http://www.unaids.org/en/resources/documents/2017/<?pdb_no 2017?>2017<?pdb END?>_data_book Search PubMed.
- A. S. Fauci and H. D. Marston, N. Engl. J. Med., 2014, 370, 495–498 CrossRef CAS PubMed.
- N. Romano, L. De Crescenzo, G. Lupo, M. V. Torregrossa, D. Russo Alesi, M. Portera and F. Vitale, Am. J. Epidemiol., 1998, 128, 254–260 CrossRef PubMed.
- J. M. Winchell, H. J. Kruiningen and K. L. Silbart, AIDS Res. Hum. Retroviruses, 1997, 10, 881–889 CrossRef PubMed.
- S. Zolla-Pazner, Nat. Rev. Immunol., 2004, 4, 199–210 CrossRef CAS PubMed.
- M. A. Checkley, B. G. Luttge and E. O. Freed, J. Mol. Biol., 2011, 410, 582–608 CrossRef CAS PubMed.
- J. P. Julien, D. Sok, R. Khayat, J. H Lee, K. J. Doores, L. M. Walker, A. Ramos, D. C. Diwanji, R. Pejchal, A. Cupo, U. Katpally, R. S. Depetris, R. L. Stanfield, R. McBride, A. J. Marozsan, J. C. Paulson, R. W. Sanders, J. P. Moore, D. R. Burton, P. Poignard, A. B. Ward and I. A. Wilson, PLoS Pathog., 2013, 9, e1003342, DOI:10.1371/journal.ppat.1003342.
- A. T. Jones, V. Chamcha, S. Kesavardhana, X. Shen, D. Beaumont, R. Das, L. S. Wyatt, C. C. LaBranche, S. Stanfield-Oakley, G. Ferrari, D. C. Montefiori, B. Moss, G. D. Tomaras, R. Varadarajan and R. R. Amara, J. Virol., 2018, 92(5), e01796-17, DOI:10.1128/JVI.01796-17.
- R. Gottardo, R. T. Bailer, B. T. Korber, S. Gnanakaran, J. Phillips, X. Shen and D. C. Montefiori, PLoS One, 2013, e75665, DOI:10.1371/journal.pone.0075665.
- M. Thali, J. Virol., 1993, 67, 3978–3988 CrossRef CAS PubMed.
- T. W. Chun, D. Murray, J. S. Justement, J. Blazkova, C. W. Hallahan, O. Fankuchen, K. Gittens, E. Benko, C. Kovacs, S. A. Moir and S. Fauci, Proc. Natl. Acad. Sci., 2014, 111, 13151–13156 CrossRef CAS PubMed.
- A. Facciolà, G. Visalli, P. Laganà, V. La Fauci, R. Squeri, G. F. Pellicanò, G. Nunnari, M. Trovato and A. Di Pietro, Eur. Rev. Med. Pharmacol. Sci., 2019, 16, 7163–7182 Search PubMed.
- I. S. Ganda, Q. Zhong, M. Hali, R. L. C. Albuquerque, F. F. Padilha, S. R. P. da Rocha and J. A. Whittum-Hudson, Int. J. Pharm., 2017, 527, 79–91 CrossRef CAS PubMed.
- R. A. Rodríguez-Fonseca, M. Bello, M. A. Muñoz-Fernández, J. L. Jiménez, S. Rojas-Hernández, M. J. Fragoso-Vázquez, M. Gutiérrez-Sánchez, J. Rodrigues, N. Cayetano-Castro, R. Borja-Urby, O. Rodríguez-Cortés, J. García-Machorro and J. Correa-Basurto, Colloids Surf., B, 2019, 177, 77–93 CrossRef PubMed.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
- S. Gupta, P. Kapoor, K. Chaudhary, A. Gautam, R. Kumar and G. P. Raghava, PLoS One, 2013, e73957, DOI:10.1371/journal.pone.0073957.
- Y. Shen, J. Maupetit, P. Derreumaux and P. Tufféry, J. Chem. Theory Comput., 2014, 10, 4745–4758 CrossRef CAS PubMed.
- D. Bhattacharya and J. Cheng, Proteins, 2013, 81, 119–131 CrossRef CAS PubMed.
- G. N. Ramachandran, C. Ramakrishnan and V. Sasisekharan, J. Mol. Biol., 1963, 7, 95–99 CrossRef CAS PubMed.
- D. A. Case, T. E. Cheatham, T. Darden, H. Gohlke, R. Luo, K. M. Merz Jr and R. J. Woods, J. Comput. Chem., 2005, 26, 1668–1688 CrossRef CAS PubMed.
- Y. Duan, C. Wu, S. Chowdhury, M. C. Lee, G. Xiong, W. Zhang and P. Kollman, J. Comput. Chem., 2003, 24, 1999–2012 CrossRef CAS PubMed.
- S. Pronk, S. Páll, R. Schulz, P. Larsson, P. Bjelkmar, R. Apostolov and E. Lindahl, Bioinformatics, 2013, 29, 845–854 CrossRef CAS PubMed.
- B. Hess, H. Bekker, J. C. Herman, J. Berendsen and G. E. M. Fraaije, J. Comput. Chem., 1997, 18, 1463–1472 CrossRef CAS.
- S. Miyamoto and P. A. Kollman, J. Comput. Chem., 1992, 13, 952–962 CrossRef CAS.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089 CrossRef CAS.
- M. Bello and R. A. Rodríguez-Fonseca, J. Mol. Graphics Modell., 2019, 93, 107443 CrossRef CAS PubMed.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 15, 1157–1174 CrossRef PubMed.
- W. L. Jorgensen, J. Chandrasekhar and F. D. Madura, J. Chem. Phys., 1983, 926, 79 Search PubMed.
- W. F. Van Gunsteren and H. J. C. Berendsen, Mol. Phys., 1977, 34, 1311–1327 CrossRef CAS.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- H. J. C. Berendsen, J. P. M. Postma, W. F. van Gunsteren, A. DiNola and J. R. Haak, J. Chem. Phys., 1984, 81, 3684–3690 CrossRef CAS.
- H. Gohlke, C. Kiel and D. A. J. Case, J. Mol. Biol., 2003, 330, 891–913 CrossRef CAS PubMed.
- P. A. Kollman, I. Massova, C. Reyes, B. Kuhn, S. Huo and L. Chong, Acc. Chem. Res., 2000, 33, 889–897 CrossRef CAS PubMed.
- A. Onufriev, V. Bashford and D. A. Case, Proteins, 2004, 55, 383–394 CrossRef CAS PubMed.
- A. Martínez-Muñoz, M. Bello, A. Romero-Castro, R. A. Rodríguez-Fonseca, J. Rodrigues, S. Espinosa and J. Correa-Basurto, J. Mol. Graph. Model., 2017, 76, 330–341 CrossRef PubMed.
- D. Kozakov, D. R. Hall, B. Xia, K. A. Porter, D. Padhorny, C. Yueh, D. Beglov and S. Vajda, Nat. Protoc., 2017, 12, 255–278 CrossRef CAS PubMed.
- D. R. Bolin, A. L. Swain, R. Sarabu, S. J. Berthel, P. Gillespie, N. J. Huby, R. Makofske, L. Orzechowski, A. Perrotta, K. Toth, J. P. Cooper, N. Jiang, F. Falcioni, R. Campbell, D. Cox, D. Gaizband, C. J. Belunis, D. Vidovic, K. Ito, R. Crowther, U. Kammlott, X. Zhang, R. Palermo, D. Weber, J. Guenot, Z. Nagy and G. L. Olson, J. Med. Chem., 2000, 43, 2135–2148 CrossRef CAS PubMed.
- R. Danner, S. N. Chaudhari, J. Rosenberger, J. Surls, T. L. Richie, T. D. Brumeanu and S. Casares, PLoS One, 2011, e19826, DOI:10.1371/journal.pone.0019826.
- N. Raval, R. Maheshwari, K. Kalia and R. K. Tekade, Methods Mol. Biol., 2019, 2000, 93–109 CrossRef CAS PubMed.
- L. Albertazzi, M. Serresi, A. Albanese and F. Beltram, Mol. Pharm., 2010, 3, 680–688 CrossRef PubMed.
- A. E. Gregory, R. Titball and D. Williamson, Front. Cell. Infect. Microbiol., 2013, 25, 3–13 Search PubMed.
- Y. Liu, V. S. Bryantsev, M. S. Diallo and W. A. Goddard, JACS, 2009, 131, 2798–2799 CrossRef CAS PubMed.
- P. Maturavongsadit, X. Bi, A. Togor, G. Yu Zhe and N. Q. Wang, Chin. Chem. Lett., 2016, 27, 1473–1478 CrossRef CAS.
- E. J. Kunkel and E. C. Butcher, Plasma-cell homming, Nat. Rev. Immunol., 2003, 3, 822–829 CrossRef CAS PubMed.
- J. Bouvet, L. Belec, R. Pires and J. Pillot, Infect. Immun., 1994, 62, 3957–3961 CrossRef CAS PubMed.
- M. Zhu, R. Wang and G. Nie, Hum. Vaccines Immunother., 2014, 9, 2761–2774 CrossRef PubMed.
- A. Abdoli, N. Radmehr, A. Bolhassani, A. Eidi, P. Mehrbod, F. Motevalli, Z. Kianmehr, M. Chiani, M. Mahdavi, S. Yazdani, M. S. Ardestani, M. R. Kandi and M. R. Aghasadeghi, Artif. Cells, Nanomed., Biotechnol., 2017, 8, 1762–1768 CrossRef PubMed.
- X. Wu, Z. Y. Yang, Y. Li, C. M. Hogerkorp, W. R. Schief, M. S. Seaman and J. R. Mascola, Science, 2010, 329, 856–861 CrossRef CAS PubMed.
- R. W. Sanders, A. Cupo, J.-P. Julien, A. Yasmeen, N. de Val, H. J. Kim, C. Blattner, A. Torrents de la Peña, J. Korzun, M. Golabek, K. de los Reyes, T. J. Ketas and M. J. van Gils, PLoS Pathog., 2013, e1003618, DOI:10.1371/journal.ppat.1003618.
- D. H. Barouch, J. B. Whitney, B. Moldt, F. Klein, T. Y. Oliveira, J. Liu, K. E. Stephenson, H. W. Chang, K. Shekhar and S. Gupta, Nature, 2013, 503, 224–228 CrossRef CAS PubMed.
- D. R. Burton and L. Hangartner, Annu. Rev. Immunol., 2016, 34, 635–659 CrossRef CAS PubMed.
- V. E. Reyes, L. T. Chin and R. E. Humphreys, Mol. Immunol., 1988, 25, 867–871 CrossRef CAS PubMed.
- C. A. Boswell, P. K. Eck, C. A. Regino, M. Bernardo, K. J. Wong, D. E. Milenic, P. L. Choyke and M. W. Brechbiel, Mol. Pharm., 2008, 4, 527–539 CrossRef PubMed.
- J. R. Lloyd, P. S. Jayasekara and K. A. Jacobson, Anal. Methods, 2016, 2, 263–269 RSC.
- A. Narsireddy, K. Vijayashree, M. G. Adimoolam, S. V. Manorama and N. M. Rao, Int. J. Nanomed., 2015, 10, 6865–6878 CAS.
- M. L. Patil, M. Zhang, O. Taratula, O. B. Garbuzenko, H. He and T. Minko, Biomacromolecules, 2009, 2, 258–266 CrossRef PubMed.
- M. A. Dobrovolskaia, Curr. Pharm. Des., 2017, 21, 3134–3141 Search PubMed.
- F. Lu, A. Mencia, L. Bi, A. Taylor, Y. Yao and H. HogenEsch, J. Controlled Release, 2015, 28, 9–51 Search PubMed.
- E. Vacas-Córdoba, N. Climent, F. J. De La Mata, M. Plana M, R. Gómez, M. Pion, F. García and M. . Á. Muñoz-Fernández, Nanomedicine, 2014, 17, 2683–2702 CrossRef PubMed.
- P. C. Naha, M. Davoren, F. M. Lyng and H. J. Byrne, Toxicol. Appl. Pharmacol., 2010, 246, 91–99 CrossRef CAS PubMed.
- A. Rudin, G. C. Riise and J. Holmgren, Infect. Immun., 1999, 67, 2884–2890 CrossRef CAS PubMed.
- Y. Kurono, et al., J. Infect. Dis., 1999, 180, 122–132 CrossRef CAS PubMed.
- A. Ruggieri, M. C. Gagliardi and S. Anticoli, Front. Immunol., 2018, 9, 2302 CrossRef PubMed.
- J. M. Wessels, A. M. Felker, H. A. Dupont and C. Kaushic, Dis. Models Mech., 2018, 9, 11 Search PubMed.
- C. Czerkinsky, A. M. Svennerholm, M. Quiding, R. Jonsson and J. Holmgren, Infect. Immun., 1991, 59, 996–1001 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra00840k |
| This journal is © The Royal Society of Chemistry 2020 |