Open Access Article
Open Access ArticleDesign, synthesis and biological evaluation of novel amide-linked 18β-glycyrrhetinic acid derivatives as novel ALK inhibitors†
Dong Cai a,
Zhi hua Zhangb,
Yu Chenc,
Chao Ruand,
Sheng qiang Lid,
Shi qin Chend and
Lian shan Chen*d
a,
Zhi hua Zhangb,
Yu Chenc,
Chao Ruand,
Sheng qiang Lid,
Shi qin Chend and
Lian shan Chen*d
aCollege of Public Basic Sciences, Jinzhou Medical University, Jinzhou, 121001, China
bSchool of Chemical and Environmental Engineering, Liaoning University of Technology, Jinzhou, 121001, China
cSchool of Life Science and Biopharmaceutics, Shenyang Pharmaceutical University, Shenyang, 110016, China
dCollege of Pharmacy, Jinzhou Medical University, Jinzhou, 121001, China. E-mail: 509205162@QQ.com
First published on 23rd March 2020
Abstract
A series of novel amide-linked 18β-glycyrrhetinic acid derivatives were developed by incorporating substituted piperazine amide fragments into the C30–COOH of 18β-glycyrrhetinic acid scaffold. The synthesized compounds were evaluated for their anticancer activity against Karpas299, A549, HepG2, MCF-7, and PC-3 cell lines by MTT assay. Besides, some compounds with electron-withdrawing groups on phenyl moieties exhibited noticeable antiproliferative activity. The most potent compound 4a was also found to be non-toxic to normal human hepatocytes LO2 cells. The compound 4a exhibited moderate inhibitory activity against wild-type ALK with an IC50 value of 203.56 nM and relatively weak potent activity to c-Met (IC50 > 1000 nM). Molecular docking studies were performed to explore the diversification in bonding patterns between the compound 4a and Crizotinib.
1. Introduction
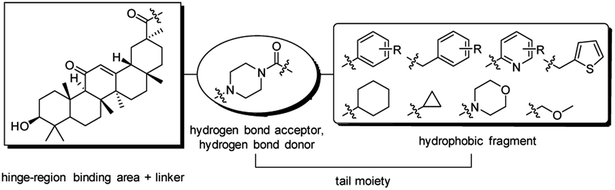
Nowadays, most tyrosine kinase inhibitors bind in a nearly identical position to that of the ATP in kinases by forming hydrogen bonds to hinge residues and by hydrophobic interactions in and around the region taken up by the adenine ring of ATP. Several tyrosine kinase inhibitors can form additional hydrogen bond interactions and hydrophobic interactions with the DFG residues of the activation-loop adjacent to the region taken up by ATP. Those tyrosine kinase inhibitors are termed as typical type I and II inhibitors, respectively.1,2A general pharmacophore model of type II inhibitors covers three regions, namely, hinge-region binding, linker and tail moieties. In particular, a hydrogen bond donor–acceptor pair and a hydrophobic fragment in the tail moiety are capable of selectively taking up allosteric pocket, exhibiting an advantage over type I inhibitors.2,3
18β-Glycyrrhetinic acid (18β-GA, Fig. 1) is an inexpensive and available triterpene extracted from the roots of licorice plants (Glycyrrhiza glabra); its derivatives exhibit remarkable cytotoxic and pharmacological activities, in particular antitumor activity.4–6 The 18β-GA nucleus, a feasible structure for in-depth pharmaceutical exploration and for development of new potential antiproliferative drug candidates, has aroused extensive attention from medicinal chemists over the last decade.7–11 The presence of polar functional groups (C3–OH and C30–COOH) in its structure could impact the biological activities of the mentioned analogues.12
Inspired by the mentioned developments and facts, our previous work was continued in structural modification of 18β-GA.7 In the present study, substituted piperazine amide fragments were introduced into the C30–COOH of 18β-GA, and a novel series of amide-linked derivatives was designed as potential antitumor inhibitors. To assess the effect of different substituents on the piperazine amide fragment, the substitution of phenyl or benzyl group was modified, and the aromatic ring was altered (Fig. 2).
Piperidine carboxamide A (Fig. 3a), an anaplastic lymphoma kinase (ALK) inhibitor, exhibits excellent cytotoxic activity against ALK-positive Karpas-299 cells (IC50 = 0.384 μM). Moreover, such bioactive compound displayed significant activity in the ALK enzyme assay (IC50 = 0.174 μM), probably causing a reduction in the phosphorylation levels of ALK downstream effectors.13
 | ||
| Fig. 3 (a) Piperidine carboxamide A and compound 4a, (b) overlay of piperidine carboxamide A (stick; from cocrystal structure, PDB code: 4DCE) and the energy minimized structure for compound 4a (ball and stick). | ||
Chemical compounds with nearly identical three-dimensional (3D) structures are likely to exhibit similar activities. The similarity can be computed based upon steric, electronic, and/or other physical properties.14 Molecular overlay provided insights into possible improvements in potency and selectivity of the designed compounds. In the present study, the energy minimization conformation of the target compound 4a was aligned to the crystal conformation of piperidine carboxamide A using the molecular overlay option of Discovery Studio 3.5 suite. Fig. 3b suggests that the distance of methoxy groups on the benzyl fragments of two molecules reached 3.0 and 3.5 Å, respectively. Given the instability of linear molecular conformations, it is a relatively acceptable superposition between the mentioned two the lipophilic methoxyphenyl fragments. Moreover, the two nitrogen atom on piperazine ring are very close to the corresponding nitrogen atom on the 2-aminopyrimidine ring. Another overlapping feature was the presence of the alicyclic fragment of A and B rings of the target compound 4a, as well as the p-methyl benzyl group from piperidine carboxamide A. The 3D molecular similarity score was 0.3049. The maximum shape similarity of molecules is expressed by score 1 and the minimum similarity is denoted by score 0.15–17
According to the mentioned results, the novel amide-linked 18β-GA derivatives might exhibit potential antiproliferative activity.
2. Results and discussion
2.1 Chemistry
The synthesis of amide-linked 18β-GA derivatives (4a–4v) was depicted in Scheme 1. To produce compound (2), 18β-GA was reacted with 1-Boc-piperazine in the presence of ethyl-dimethylaminopropyl-carbodiimide hydrochloride (EDCl), 1-hydroxybenzotriazole (HOBt) and triethylamine. Subsequently, the compound (2) was stirred in the solution of trifluoroacetate (TFA) and dichloromethane (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to remove Boc group. Without being further purified, monoamide (3) was acylated with acid chlorides in the presence of triethylamine to produce the target compounds (4a–4v).
1) to remove Boc group. Without being further purified, monoamide (3) was acylated with acid chlorides in the presence of triethylamine to produce the target compounds (4a–4v).
 | ||
| Scheme 1 Synthesis of compound 4. Reagents and conditions: (a) 1-Boc-piperazine, CH3CN, NEt3, EDCl, HOBt, reflux, 24 h; (b) TFA, CH2Cl2, 0/25 °C; (c) substituted acyl chloride, CH2Cl2, Et3N, r.t. | ||
A simple and efficient approach for amide bond formation is based on the reaction of substituted carboxylic acid and amine in the presence of coupling reagents.18–20 Monoamide (3) can also be generated by treating of compound (2) with piperazine in the presence of EDCl, HOBt, and triethylamine. However, the corresponding monoamide (3) could be obtained in relatively low isolated yields, the reaction was complicated by the competing diamidation to form symmetric bisamide (5). As shown in Scheme 2, the amount of the symmetric bisamide (5) generated was determined by the reaction conditions. For instance, no bisamide (5) was identified by treating the compound (2) with piperazine (1.0 equiv.) in the presence of EDCl, HOBt, and triethylamine in CH3CN at room temperature for 24 h. However, an intermediate (6) was largely isolated from the reaction mixture.7 The intermediate (6) did not react with piperazine at low reaction temperatures even after the extended reaction time (up to 48 h). Nevertheless, the above reaction was performed in refluxing CH3CN, and intermediate (6) could be fully converted into bisamide (5) and trace amount of the desired monoamide (3).
The steric hindrance around the C-30 ester group of intermediate (6) significantly impacted the nucleophilic substitution. The lowest-energy conformer of intermediate (6) and piperazine was achieved by MM2 calculations in ChemBio 3D Ultra 12.0. As shown in Fig. 4, the piperazine was difficult to get close to C-30 because of two bulky groups (1H-benzo[d][1,2,3]triazol-1-yl group and scaffold of 18β-GA).
Next, the effect of feeding sequence of above reaction was investigated. To a solution of piperazine (4.0 equiv.) in refluxing CH3CN, intermediate (6) solution (1.0 equiv.) was added dropwise and then stirred for 12 h. As shown in Scheme 2, under the reaction, bisamide (5) and the desired monoamide (3) were formed, and the isolated yields reached 53.7% and 40.8%, respectively.
Condensation of nearly equimolecular amounts of monoamide (3) with substituted acyl chloride in CH2Cl2 with triethylamine at room temperature affords target compounds (4a–4v) with 84.8–94.6% yields. The C3–OH of 18β-GA did not interfere with these amidation reactions. However, the amidation of monoamide (3) with 2-(4-chlorophenyl)acetyl chloride was carried out in the solution of triethylamine and CH2Cl2 at 40 °C, and competitive esterification to form the corresponding compound (7) in 38.4% isolated yield contributed to a lower yield of the target compound (4w). Moreover, if the acid chloride is excessively large and the reaction time is too long at room temperature, it will also lead to competitive esterification (Scheme 3).
 | ||
| Scheme 3 Synthesis of compound 4w, and 7. Reagents and conditions: (a) p-chlorophenylacetyl chloride, CH2Cl2, Et3N, r.t.; (b) p-chlorophenylacetyl chloride, CH2Cl2, Et3N, 40 °C. | ||
2.2 In vitro cell growth inhibitory activity
To test the anticancer activity of synthesized compounds, antiproliferative activity of target compounds (4a–4v, 7) against HepG2 and Karpas299 was assessed by MTT assay. 18β-GA and Crizotinib were used as a positive control. The results are expressed as the growth inhibition, as listed in Table 1. Most compounds exhibited prominent antiproliferative activities at a dose (20 μg mL−1), this was primarily attributed to the fact that the target compounds have a large molecular weight. All the investigated compounds were less activity than Crizotinib.| Compound | HepG2 | Karpas299 | ||
|---|---|---|---|---|
| 2 μg mL−1 | 20 μg mL−1 | 2 μg mL−1 | 20 μg mL−1 | |
| 4a | 0.00 | 93.11 | 0.00 | 96.62 |
| 4b | 0.87 | 50.81 | 0.49 | 58.13 |
| 4c | 0.00 | 63.23 | 0.00 | 84.32 |
| 4d | 0.00 | 68.04 | 0.00 | 91.46 |
| 4e | 0.00 | 11.96 | 0.00 | 18.28 |
| 4f | 0.06 | 29.40 | 0.00 | 12.24 |
| 4g | 0.00 | 14.20 | 0.00 | 8.00 |
| 4h | 0.00 | 24.00 | 0.00 | 17.60 |
| 4i | 0.00 | 19.10 | 0.00 | 12.76 |
| 4j | 0.10 | 34.51 | 0.00 | 6.62 |
| 4k | 0.00 | 25.53 | 0.00 | 4.00 |
| 4l | 4.26 | 51.12 | 0.00 | 69.32 |
| 4m | 0.00 | 40.24 | 0.0 | 53.31 |
| 4n | 0.00 | 32.73 | 0.00 | 26.27 |
| 4o | 0.00 | 30.39 | 0.00 | 20.69 |
| 4p | 0.00 | 12.37 | 0.00 | 0.00 |
| 4q | 0.00 | 27.75 | 0.00 | 45.02 |
| 4r | 0.00 | 63.33 | 6.66 | 82.66 |
| 4s | 0.00 | 47.97 | 0.00 | 42.41 |
| 4t | 1.84 | 31.68 | 0.00 | 23.78 |
| 4u | 3.03 | 78.52 | 0.00 | 91.71 |
| 4v | 2.75 | 45.53 | 0.00 | 27.99 |
| 4w | 1.35 | 19.32 | 0.00 | 11.46 |
| 4x | 0.17 | 15.48 | 0.00 | 17.25 |
| 4y | 1.48 | 6.36 | 0.00 | 0.00 |
| 7 | 0.00 | 0.00 | 0.00 | 0.00 |
| 18β-GA | 0.00 | 10.74 | 0.00 | 15.28 |
| Crizotinib | 27.15 | 97.76 | 75.71 | 96.89 |
Preliminary SAR analyses suggested that the substituent properties and positions of the phenyl ring fragment were critical to modulate their antiproliferative activity. Table 1 presents that electron-withdrawing substituents on the phenyl ring displayed relatively poor antiproliferative activities against HepG2 and Karpas299 cells. Only the 3-fluorophenyl substituted derivative (4l) showed over 50% growth inhibitory activity at 20 μg mL−1 against two test cells as compared with the target compounds (4e, 4f, 4g, 4h, 4i, 4j, 4k, 4m, 4n, 4o, 4p, 4q). The introduction of 6-chloronicotinoyl group at the identical position (4t) also significantly reduced activities against all of the tested cancer cell lines.
In contrast to the compounds with electron-withdrawing groups on the aromatic ring fragment, it was more favorable when the electron-donating group was added to the phenyl ring fragment (e.g. CH3O, CH3). Compound (4a) exhibited nearly the identical cell growth inhibitory activity to Crizotinib against HepG2 and Karpas299 cells at the identical concentration of 20 μg mL−1. 3-Methylphenyl substituted derivative (4d) also displayed good activity, especially for Karpas299 cell. Note that 3,5-dimethoxyphenyl substituted derivative (4b) and 3,5-dimethylphenyl substituted derivative (4c) have drastically reduced inhibitory activity against HepG2 and Karpas299 cells, revealing that the substitution of the 3,5 position on the phenyl ring is not recommended.
When R is a substituted benzyl group, the compound (4v) containing 4-fluorobenzyl fragment exhibited better antiproliferative activity than that of compound (4w) containing 4-chlorobenzyl fragment. However, on the whole, the antitumor inhibition rate of these two compounds was below 50% at 20 μg ⋅mL−1. Nevertheless, compound (4u) with thiophen-2-methylene fragment displayed an outstanding potency against HepG2 and Karpas299 cells.
It is noteworthy that the target compounds (4r, 4s) with naphthenic substituted amides as the side chain showed an evident inhibitory effect against HepG2 and Karpas299 cells. In particular, R is cyclohexyl group, compounds (4r) exhibited notable cell growth inhibitory activity against Karpas299 cell (82.66% at a concentration of 20 μg mL−1). Analogous compounds (4x, 4y) containing morpholine or methoxymethylene group, determined a significant decrease of efficacy. It is therefore indicated that the ether fragment could establish an unfavorable interaction with the receptor.
According to the structural features and corresponding antitumor activities of the compounds reported in the literature,10,21 the antitumor activity of the selected compounds 4a, 4c, 4d, 4r, and 4u was evaluated at the cellular level expressed by IC50 values against five cancer cell lines (Karpas299, A549, HepG2, MCF-7 and PC-3). Karpas299 is a typical anaplastic lymphoma kinase mutant-driven cancer cell line. Table 2 reveals that the most effective compound 4a exhibited superior antiproliferative effect only against Karpas299 and HepG2 cells with IC50 values of 6.51 μM, and 6.93 μM, respectively.
| Compound | Karpas299 | A549 | HepG2 | MCF-7 | PC-3 |
|---|---|---|---|---|---|
| 4a | 6.51 | >40 | 6.93 | 18.85 | 18.18 |
| 4c | 15.59 | >40 | 11.95 | >40 | 27.56 |
| 4d | 9.41 | >40 | 12.92 | >40 | 20.37 |
| 4r | 20.00 | 38.54 | 22.67 | 35.43 | 32.81 |
| 4u | 18.30 | >40 | 11.81 | >40 | 37.84 |
| 18β-GA | >40 | >40 | >40 | >40 | >40 |
| Crizotinib | 2.82 | 1.49 | 10.59 | 4.09 | 7.33 |
To determine whether the cell growth inhibitory effect of compound 4a is associated with a time- and concentration-dependent manner, the cells were treated by MTT cytotoxicity assay, and five concentration gradients of compound 4a were selected. After 24 h incubation, the HepG2 cells were treated for 24, 48 and 72 h by compound 4a at the concentrations of 0.064, 0.32, 1.6, 8.0 and 40 μg∙ mL−1. The result was shown in Fig. 5, compound 4a was observed in a significant time and concentration dependent manner to inhibit the proliferation of HepG2 cell.
 | ||
| Fig. 5 The relationship between different concentrations and time of compound 4a and proliferation inhibitory effect. Data are means ± SD of the inhibition (%) from three independent experiments. | ||
According to the results, the compound 4a with the most potent antiproliferative activity was used for further processing. HepG2 cells and normal human hepatocytes LO2 cells were cultivated with compound 4a at increasing concentrations. A 48 h continuous drug exposure protocol was employed by the MTT assay. As shown in Fig. 6, compound 4a significantly inhibited the proliferation activity of HepG2 cells in a dose-dependent manner. In contrast, compound 4a exhibited slight toxicity towards LO2 cells. As revealed from the results, the compound 4a might exhibit selective antiproliferative activity against human tumor cells.
 | ||
| Fig. 6 Cytotoxicity of compound 4a toward HepG2 and LO2 cells. Data are means ± SD of the inhibition (%) from three independent experiments. | ||
2.3 Kinase activities of compounds 4a and 4d
Given the mentioned results, the compounds 4a and 4d displayed potent, selective inhibitory activity against Karpas299 and HepG2 cells. The two compounds were taken for in-depth evaluation of the enzymatic inhibitory activity against wild-type ALK and c-MET/HGFR. Table 3 lists that the compounds 4a and 4d displayed moderate enzyme inhibitory activities against wild type ALK with IC50 values of 203.56 nM and 686.19 nM, respectively, in comparison with that of the positive control Crizotinib (IC50 = 11.21 nM). Compared with the 3-methylphenyl substituted analog analogs 4d, the 3,4-methoxyphenyl substituted analog 4a were slightly more potent against ALK. In contrast, the two compounds exerted relatively weak inhibitory effect on c-MET/HGFR. As revealed from the results, the inhibition of ALK could be a mechanism for the antitumor effect of these novel carbamate derivatives.| Entry | ALK/IC50 (nM) | c-met/IC50 (nM) |
|---|---|---|
| 4a | 203.56 | >1000 |
| 4d | 686.19 | >1000 |
| Crizotinib | 11.21 | 7.68 |
2.4 Molecular docking study
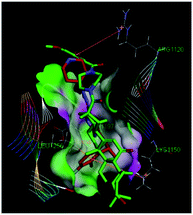
As in a previously published co-crystal structure of Crizotinib with anaplastic lymphoma kinase (ALK),22 the 2-aminopyrimidine ring of Crizotinib formed two hydrogen bonds to the GLU1197 and MET1199 of the kinase hinge region. The lengths of the two hydrogen bonds were 1.87228 and 3.02638 Å, respectively. According to this model, the benzene ring of Crizotinib formed a CH–π interaction with the LEU1256, as well as a cation–π bond with the Lys1150.Like Crizotinib (Fig. 7), the compound 4a possessed a piperazine amide tail fragment can be easily docked into the ATP site of the DFG-out ALK co-crystal structure (PDB code: 2XP2). The docking conformation revealed that the 23,24-dimethylcyclohexan-3-ol fragment of compound 4a was fully buried into the ATP binding site via hydrophobic interactions, compared with the pyridin-2-ylamine fragment of Crizotinib. Nevertheless, for its large size of the linker and the tail fragment, the compound 4a failed to form the expected hydrogen bonding interactions with the kinase hinge region. Moreover, the 3,4-dimethoxybenzoyl fragment formed a CH–π interaction with the ARG1120. The C3–OH of compound 4a pointed to the activation loop (DFG-out conformation). The compound 4a with moderate enzymatic activity was relatively weakly bound to the potential binding sites, probably due to the large steric hindrance tail groups.
2.5 Apoptosis detection by flow cytometry
The effect of compound 4a on the apoptosis was investigated by means of a the 7-amino-actinomycin D (7-AAD) and annexin V-FITC biparametric cytofluorimetric analysis. After treatment with different concentrations of compound 4a (0 μM, 5 μM, 10 μM) in serum-free medium for 48 h, HepG2 cells were stained with 7-AAD and FITC, and then analyzed by the flow cytometry. As illustrated in Fig. 8, the percentage of total apoptotic cells from 10.18 to 24.49% was markedly elevated in a concentration-dependent manner, as compared to 2.30% for the control group.3. Conclusions
In brief, a series of novel amide-linked 18β-GA derivatives was synthesized and evaluated for their anticancer activity against Karpas299, A549, HepG2, MCF-7 and PC-3 cells. Of the compounds screened, some compounds with electron-donating groups on phenyl moiety exhibited evident antiproliferative activity. The most active compound 4a exhibited promising cytotoxicity against Karpas299 (IC50 of 6.51 μM) and HepG2 (IC50 of 6.93 μM) cells. Moreover, the compound 4a exhibited moderate inhibitory activity against wild-type ALK with IC50 value of 203.56 nM and relatively weak potent to c-Met (IC50 > 1000 nM). Furthermore, compound 4a induced apoptosis of HepG2 cells in a concentration-dependent manner, in particular at the late stage of the apoptotic process. Molecular docking was performed to delve into the differences in bonding modes between the compound 4a and Crizotinib. Lastly, the compound 4a would inspire further derivatization and optimization of such scaffold to explore more potent ALK inhibitors.4. Materials and general methods
4.1 Chemistry
Unless otherwise required, all reagents used in the experiment were purchased as commercial analytical grade and used without further purification. Melting points were obtained in open capillary tubes with a WRS-1B melting point apparatus (Shanghai Shenguang Instrument Co., Ltd., Shanghai, CHN) and were uncorrected. The structure of the synthetic compound was confirmed by 1H-NMR and 13C-NMR spectra on 400/54 Premium Shielded NMR Magnet System (Agilent, Santa Clara, USA) with tetramethylsilane (TMS) as an internal standard. HRMS spectra data were collected from an Agilent 6200 Series TOF and 6500 Series Q-TOF LC/MS System B.05.01. (B5125) in positive ion modes (Agilent, Santa Clara, USA).A white solid; yield, 94.3%; mp 224.3–225.7 °C; 1H NMR (400 MHz, chloroform-d) δ 5.66 (s, 1H, CH-12), 3.63–3.52 (m, 4H, piperazinyl CH2 × 2), 3.39 (t, J = 5.2 Hz, 4H, piperazinyl CH2 × 2), 3.22–3.18 (m, 1H, OH-3), 2.79–2.74 (m, 1H, CH-1), 2.31 (s, 1H, CH-9), 2.30–2.23 (m, 1H, CH-16), 1.45 (s, 9H, tert-butyl CH3 × 3), 1.34 (s, 3H, CH3-27), 1.20 (s, 3H, CH3-25), 1.11 (s, 3H, CH3-26), 1.10 (s, 3H, CH3-29), 0.98 (s, 3H, CH3-23), 0.79 (s, 3H, CH3-24), 0.78 (s, 3H, CH3-28), 0.68 (d, J = 11.6 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.10 (C11), 174.13 (C30), 169.40 (C13), 154.53 (Boc C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 128.56 (C12), 80.25 (C3), 78.75 (tert-butyl C), 61.77 (C9), 54.92 (C5), 48.08 (C18), 45.26 (C14), 43.88 (C20), 43.82 (piperazinyl C × 2), 43.26 (C8/C19), 39.12 (C1/C4), 39.10 (piperazinyl C × 2), 37.70 (C22), 37.06 (C10), 33.16 (C7), 32.79 (C17), 31.75 (C21), 28.40 (C29), 28.36 (tert-butyl CH3 × 3), 28.07 (C28), 27.28 (C23), 27.05 (C2), 26.69 (C15), 26.39 (C16), 23.14 (C27), 18.66 (C26), 17.46 (C6), 16.36 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C39H63N2O5: 639.47370, found: 639.47360.
O), 128.56 (C12), 80.25 (C3), 78.75 (tert-butyl C), 61.77 (C9), 54.92 (C5), 48.08 (C18), 45.26 (C14), 43.88 (C20), 43.82 (piperazinyl C × 2), 43.26 (C8/C19), 39.12 (C1/C4), 39.10 (piperazinyl C × 2), 37.70 (C22), 37.06 (C10), 33.16 (C7), 32.79 (C17), 31.75 (C21), 28.40 (C29), 28.36 (tert-butyl CH3 × 3), 28.07 (C28), 27.28 (C23), 27.05 (C2), 26.69 (C15), 26.39 (C16), 23.14 (C27), 18.66 (C26), 17.46 (C6), 16.36 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C39H63N2O5: 639.47370, found: 639.47360.
A white solid; yield, 97.0%; mp 258.5–259.7 °C (literature (ref. 23): 160 °C, decomp.); 1H NMR (400 MHz, chloroform-d) δ 5.67 (s, 1H, CH-12), 3.63 (q, J = 3.7 Hz, 4H, piperazinyl CH2 × 2), 3.20 (dd, J = 10.9, 5.4 Hz, 1H, OH-3), 2.88 (t, J = 5.0 Hz, 4H, piperazinyl CH2 × 2), 2.76 (dt, J = 13.6, 3.6 Hz, 1H, CH-1), 2.31 (s, 1H, CH-9), 2.26 (dd, J = 14.0, 3.9 Hz, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.19 (s, 3H, CH3-25), 1.11 (s, 3H, CH3-26), 1.10 (s, 3H, CH3-29), 0.98 (s, 3H, CH3-23), 0.79 (s, 3H, CH3-24), 0.78 (s, 3H, CH3-28), 0.67 (d, J = 11.4 Hz, 1H); 13C NMR (101 MHz, chloroform-d) δ 200.19 (C11), 173.89 (C30), 169.64 (C13), 128.51 (C12), 78.73 (C3), 61.77 (C9), 54.92 (C5), 48.16 (C18), 45.94 (piperazinyl C × 2), 45.26 (C14), 43.79 (C20), 43.27 (C8), 39.14 (piperazinyl C × 2), 39.11 (C19), 37.72 (C1/4), 37.07 (C22), 33.28 (C10), 32.78 (C7), 31.76 (C17), 28.41 (C21), 28.07 (C29), 27.28 (C28), 27.00 (C23), 26.70 (C2), 26.42 (C15), 23.13 (C27), 18.66 (C26), 17.46 (C6), 16.37 (C25), 15.57 (C24); HRMS (m/z): [M + H]+ calcd for C34H55N2O3: 539.42127, found: 539.42120.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CH2Cl2–methanol).
1 CH2Cl2–methanol).
3β-Hydroxy-30-(4-(3,4-dimethoxybenzoyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4a). A white solid; yield, 94.2%; mp 221.9–222.8 °C; 1H NMR (400 MHz, chloroform-d) δ 7.01 (s, 2H, phenyl-H), 6.88 (s, 1H, phenyl-H), 5.68 (s, 1H, CH-12), 3.92 (s, 6H, –OCH3), 3.66 (s, 8H, piperazinyl-H), 3.23 (dd, J = 10.8, 5.2 Hz, 1H, 3-OH), 2.78 (d, J = 13.2 Hz, 1H, CH-1), 2.34 (s, 1H, CH-9), 2.27 (d, J = 12.8 Hz, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.25 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.01 (s, 3H, CH3-23), 0.82 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.4 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.07 (C11), 174.24 (C30), 170.50 (C13), 169.32 (benzoyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 150.52 (phenyl), 149.04 (phenyl), 128.55 (C12), 127.28 (phenyl), 120.19 (phenyl) 110.85 (phenyl), 110.46 (phenyl), 78.72 (C3), 61.78 (C9), 56.01 (–OCH3), 55.99 (–OCH3), 54.91 (C5), 48.19 (C18), 45.27 (C14), 43.90 (C20), 43.72 (C8), 43.27 (piperazinyl C), 39.10 (piperazinyl C), 37.68 (C4), 37.05 (C22), 33.26 (C10), 32.78 (C7), 31.79 (C17), 29.69 (C21), 28.38 (C29), 28.07 (C28), 27.27 (C23), 27.06 (C2), 26.68 (C15), 26.40 (C16), 23.15 (C27), 18.66 (C26), 17.46 (C6), 16.37 (C25), 15.57 (C24); HRMS (m/z): [M + Na]+ calcd for C43H62N2NaO6: 725.45056, found: 725.45778.
O), 150.52 (phenyl), 149.04 (phenyl), 128.55 (C12), 127.28 (phenyl), 120.19 (phenyl) 110.85 (phenyl), 110.46 (phenyl), 78.72 (C3), 61.78 (C9), 56.01 (–OCH3), 55.99 (–OCH3), 54.91 (C5), 48.19 (C18), 45.27 (C14), 43.90 (C20), 43.72 (C8), 43.27 (piperazinyl C), 39.10 (piperazinyl C), 37.68 (C4), 37.05 (C22), 33.26 (C10), 32.78 (C7), 31.79 (C17), 29.69 (C21), 28.38 (C29), 28.07 (C28), 27.27 (C23), 27.06 (C2), 26.68 (C15), 26.40 (C16), 23.15 (C27), 18.66 (C26), 17.46 (C6), 16.37 (C25), 15.57 (C24); HRMS (m/z): [M + Na]+ calcd for C43H62N2NaO6: 725.45056, found: 725.45778.
3β-Hydroxy-30-(4-(3,5-dimethoxybenzoyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4b). A white solid; yield, 92.5%; mp 224.1–226.0 °C; 1H NMR (400 MHz, chloroform-d) δ 6.55–6.47 (m, 3H, phenyl-H), 5.68 (s, 1H, CH-12), 3.81 (s, 6H, phenyl-CH3), 3.74–3.39 (m, 8H, piperazinyl-H), 3.22 (dd, J = 10.8, 5.4 Hz, 1H, 3-OH), 2.78 (dt, J = 13.4, 3.5 Hz, 1H, CH-1), 2.33 (s, 1H, CH-9), 2.28 (d, J = 12.6 Hz, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.23 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.00 (s, 3H, CH3-23), 0.82 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.8 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.05 (C11), 174.22 (C30), 170.16 (C13), 169.27 (benzoyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 160.91 (phenyl), 137.02 (phenyl), 128.56 (C12), 104.77 (phenyl), 101.90 (phenyl), 78.73 (C3), 61.77 (C9), 55.51 (–OCH3), 54.91 (C5), 48.13 (C18), 45.26 (C14), 43.89 (C20), 43.75 (C8), 43.26 (piperazinyl C), 39.10 (piperazinyl C), 37.67 (C22), 37.05 (C10), 33.18 (C7), 32.78 (C17), 31.78 (C21), 28.38 (C29), 28.07 (C28), 27.28 (C23), 27.04 (C2), 26.67 (C15), 26.39 (C16), 23.14 (C27), 18.65 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C43H63N2O6: 703.46861, found: 703.47552.
O), 160.91 (phenyl), 137.02 (phenyl), 128.56 (C12), 104.77 (phenyl), 101.90 (phenyl), 78.73 (C3), 61.77 (C9), 55.51 (–OCH3), 54.91 (C5), 48.13 (C18), 45.26 (C14), 43.89 (C20), 43.75 (C8), 43.26 (piperazinyl C), 39.10 (piperazinyl C), 37.67 (C22), 37.05 (C10), 33.18 (C7), 32.78 (C17), 31.78 (C21), 28.38 (C29), 28.07 (C28), 27.28 (C23), 27.04 (C2), 26.67 (C15), 26.39 (C16), 23.14 (C27), 18.65 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C43H63N2O6: 703.46861, found: 703.47552.
3β-Hydroxy-30-(4-(3,5-dimethylbenzoyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4c). A white solid; yield, 92.7%; mp 232.4–232.7 °C; 1H NMR (400 MHz, chloroform-d) δ 7.06 (s, 1H, phenyl-H), 7.00 (s, 2H, phenyl-H), 5.68 (s, 1H, CH-12), 3.57 (m, 8H, piperazinyl-H), 3.22 (dd, J = 10.8, 5.4 Hz, 1H, 3-OH), 2.78 (dt, J = 13.5, 3.5 Hz, 1H, CH-1), 2.34 (s, 6H, phenyl-CH3), 2.29 (d, J = 13.1 Hz, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.23 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.00 (s, 3H, CH3-23), 0.82 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.5 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.06 (C11), 174.22 (C30), 170.93 (C13), 169.27 (benzoyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 138.34 (phenyl), 135.13 (phenyl), 131.52 (phenyl), 128.59 (C12), 124.59 (phenyl), 78.75 (C3), 61.78 (C9), 54.92 (C5), 48.10 (C18), 45.27 (C14), 43.90 (C20), 43.78 (C8), 43.26 (piperazinyl C), 39.11 (piperazinyl C), 37.69 (C22), 37.06 (C10), 33.15 (C7), 32.79 (C17), 31.77 (C21), 28.39 (C29), 28.07 (C28), 27.29 (C23), 27.06 (C2), 26.69 (C15), 26.39 (C16), 23.14 (C27), 21.26 (phenyl-CH3), 18.66 (C26), 17.47 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + Na]+ calcd for C43H63N2O4: 671.47878, found: 671.48498.
O), 138.34 (phenyl), 135.13 (phenyl), 131.52 (phenyl), 128.59 (C12), 124.59 (phenyl), 78.75 (C3), 61.78 (C9), 54.92 (C5), 48.10 (C18), 45.27 (C14), 43.90 (C20), 43.78 (C8), 43.26 (piperazinyl C), 39.11 (piperazinyl C), 37.69 (C22), 37.06 (C10), 33.15 (C7), 32.79 (C17), 31.77 (C21), 28.39 (C29), 28.07 (C28), 27.29 (C23), 27.06 (C2), 26.69 (C15), 26.39 (C16), 23.14 (C27), 21.26 (phenyl-CH3), 18.66 (C26), 17.47 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + Na]+ calcd for C43H63N2O4: 671.47878, found: 671.48498.
3β-Hydroxy-30-(4-(3-methylbenzoyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4d). A white solid; yield, 91.5%; mp 263.4–265.8 °C; 1H NMR (400 MHz, chloroform-d) δ 7.35–7.14 (m, 4H, phenyl-H), 5.68 (s, 1H, CH-12), 3.88–3.35 (m, 8H, piperazinyl-H), 3.22 (dd, J = 10.8, 5.4 Hz, 1H, 3-OH), 2.78 (dt, J = 13.5, 3.5 Hz, 1H, CH-1), 2.38 (s, 3H, phenyl-CH3), 2.33 (s, 1H, CH-9), 2.29 (d, J = 10.7 Hz, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.23 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.00 (s, 3H, CH3-23), 0.82 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.6 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.05 (C11), 174.22 (C30), 170.76 (C13), 169.26 (benzoyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 138.59 (phenyl), 135.11 (phenyl), 130.72 (phenyl), 128.59 (C12), 128.41 (phenyl), 127.70 (phenyl), 123.95 (phenyl), 78.75 (C3), 61.78 (C9), 54.92 (C5), 48.10 (C18), 45.26 (C14), 43.91 (C20), 43.78 (C8), 43.26 (piperazinyl C), 39.11 (piperazinyl C), 37.69 (C1/4), 37.06 (C22), 33.15 (C10), 32.79 (C7), 31.77 (C17), 29.69 (C21), 28.39 (C29), 28.07 (C28), 27.29 (C23), 27.05 (C2), 26.69 (C15), 26.39 (C16), 23.14 (C27), 21.37 (phenyl CH3), 18.66 (C26), 17.47 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C42H61N2O4: 657.46313, found: 657.46856.
O), 138.59 (phenyl), 135.11 (phenyl), 130.72 (phenyl), 128.59 (C12), 128.41 (phenyl), 127.70 (phenyl), 123.95 (phenyl), 78.75 (C3), 61.78 (C9), 54.92 (C5), 48.10 (C18), 45.26 (C14), 43.91 (C20), 43.78 (C8), 43.26 (piperazinyl C), 39.11 (piperazinyl C), 37.69 (C1/4), 37.06 (C22), 33.15 (C10), 32.79 (C7), 31.77 (C17), 29.69 (C21), 28.39 (C29), 28.07 (C28), 27.29 (C23), 27.05 (C2), 26.69 (C15), 26.39 (C16), 23.14 (C27), 21.37 (phenyl CH3), 18.66 (C26), 17.47 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C42H61N2O4: 657.46313, found: 657.46856.
3β-Hydroxy-30-(4-(3,4-dichlorobenzoyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4e). A white solid; yield, 93.4%; mp 237.9–239.6 °C; 1H NMR (400 MHz, chloroform-d) δ 7.52 (d, J = 8.5 Hz, 2H, phenyl-H), 7.29–7.24 (m, 2H, phenyl-H), 5.67 (s, 1H, CH-12), 3.68–3.47 (m, 8H, piperazinyl-H), 3.22 (dd, J = 11.0, 5.2 Hz, 1H, 3-OH), 2.78 (d, J = 13.2 Hz, 1H, CH-1), 2.33 (s, 1H, CH-9), 2.27 (d, J = 13.0 Hz, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.24 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.00 (s, 3H, CH3-23), 0.82 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.5 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.09 (C11), 174.28 (C30), 169.24 (C13), 168.09 (benzoyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 134.82 (phenyl), 134.52 (phenyl), 133.19 (phenyl), 130.75 (phenyl), 129.36 (phenyl), 128.58 (C12), 126.42 (phenyl), 78.74 (C3), 61.79 (C9), 54.92 (C5), 48.14 (C18), 45.27 (C14), 43.92 (C20), 43.74 (C8), 43.27 (piperazinyl C), 39.11 (piperazinyl C), 37.67 (C4), 37.06 (C22), 33.19 (C10), 32.79 (C7), 31.79 (C17), 29.69 (C21), 28.39 (C29), 28.07 (C28), 27.39 (C23), 27.05 (C2), 26.67 (C15), 26.39 (C16), 23.15 (C27), 18.66 (C26), 17.45 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + Na]+ calcd for C41H56Cl2N2NaO4: 733.35148, found: 733.35752.
O), 134.82 (phenyl), 134.52 (phenyl), 133.19 (phenyl), 130.75 (phenyl), 129.36 (phenyl), 128.58 (C12), 126.42 (phenyl), 78.74 (C3), 61.79 (C9), 54.92 (C5), 48.14 (C18), 45.27 (C14), 43.92 (C20), 43.74 (C8), 43.27 (piperazinyl C), 39.11 (piperazinyl C), 37.67 (C4), 37.06 (C22), 33.19 (C10), 32.79 (C7), 31.79 (C17), 29.69 (C21), 28.39 (C29), 28.07 (C28), 27.39 (C23), 27.05 (C2), 26.67 (C15), 26.39 (C16), 23.15 (C27), 18.66 (C26), 17.45 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + Na]+ calcd for C41H56Cl2N2NaO4: 733.35148, found: 733.35752.
3β-Hydroxy-30-(4-(2,4-dichlorobenzoyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4f). A white solid; yield, 93.4%; mp 231.9–233.3 °C; 1H NMR (400 MHz, chloroform-d) δ 7.45 (s, 1H, phenyl-H), 7.34 (d, J = 8.3 Hz, 1H, phenyl-H), 7.24 (s, 1H, phenyl-H), 5.67 (d, J = 5.3 Hz, 1H, CH-12), 3.99–3.54 (m, 6H, piperazinyl-H), 3.34–3.23 (m, 2H, piperazinyl-H), 3.21 (d, J = 5.3 Hz, 1H, 3-OH), 2.82–2.74 (m, 1H, CH-1), 2.34 (s, 1H, CH-9), 2.27 (d, J = 13.4 Hz, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.23 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.00 (s, 3H, CH3-23), 0.82 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.5 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.09 (C11), 174.23 (C30), 169.29 (C13), 166.11 (benzoyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 135.90 (phenyl), 133.64 (phenyl), 131.20 (phenyl), 129.68 (phenyl), 128.87 (phenyl), 128.80 (phenyl), 128.59 (C12), 127.85 (phenyl), 78.75 (C3), 61.78 (C9), 54.92 (C5), 48.10 (C18), 46.76 (C14), 45.27 (C20), 43.91 (C8), 43.26 (piperazinyl C), 41.88 (C19), 39.10 (piperazinyl C), 37.68 (C4), 37.05 (C22), 33.26 (C10), 32.78 (C7), 31.79 (C17), 29.69 (C21), 28.38 (C29), 28.07 (C28), 27.27 (C23), 27.06 (C2), 26.68 (C15), 26.40, 41.88 (piperazinyl C), 39.11 (C4), 37.67 (C22), 37.06 (C10), 32.79 (C7), 31.77 (C17), 29.69 (C21), 28.38 (C29), 28.07 (C28), 27.28 (C23), 27.06 (C2), 26.67 (C15), 26.38 (C16), 23.15 (C27), 18.65 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C41H57Cl2N2O4: 711.36954, found: 711.37585.
O), 135.90 (phenyl), 133.64 (phenyl), 131.20 (phenyl), 129.68 (phenyl), 128.87 (phenyl), 128.80 (phenyl), 128.59 (C12), 127.85 (phenyl), 78.75 (C3), 61.78 (C9), 54.92 (C5), 48.10 (C18), 46.76 (C14), 45.27 (C20), 43.91 (C8), 43.26 (piperazinyl C), 41.88 (C19), 39.10 (piperazinyl C), 37.68 (C4), 37.05 (C22), 33.26 (C10), 32.78 (C7), 31.79 (C17), 29.69 (C21), 28.38 (C29), 28.07 (C28), 27.27 (C23), 27.06 (C2), 26.68 (C15), 26.40, 41.88 (piperazinyl C), 39.11 (C4), 37.67 (C22), 37.06 (C10), 32.79 (C7), 31.77 (C17), 29.69 (C21), 28.38 (C29), 28.07 (C28), 27.28 (C23), 27.06 (C2), 26.67 (C15), 26.38 (C16), 23.15 (C27), 18.65 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C41H57Cl2N2O4: 711.36954, found: 711.37585.
3β-Hydroxy-30-(4-(3,5-dichlorobenzoyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4g). A white solid; yield, 92.9%; mp 235.5–236.9 °C; 1H NMR (400 MHz, chloroform-d) δ 7.44 (t, J = 1.9 Hz, 1H, phenyl-H), 7.29 (d, J = 1.9 Hz, 2H, phenyl-H), 5.68 (s, 1H, CH-12), 3.74–3.41 (m, 8H, piperazinyl-H), 3.22 (dd, J = 10.7, 5.5 Hz, 1H, 3-OH), 2.83–2.74 (m, 1H, CH-1), 2.33 (s, 1H, CH-9), 2.27 (d, J = 12.7 Hz, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.24 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.00 (s, 3H, CH3-23), 0.82 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.5 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 199.91 (C11), 174.27 (C30), 171.02 (C13), 169.28 (benzoyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 167.78 (C–Cl), 164.23 (C–Cl), 164.11 (C–Cl), 161.73 (C–Cl), 161.61 (C–Cl), 138.13 (phenyl), 128.53 (C12), 110.54 (phenyl), 110.46 (phenyl), 110.35 (phenyl), 110.27 (phenyl), 105.83 (phenyl), 105.58 (phenyl), 105.33 (phenyl), 80.55 (C3), 61.69 (C9), 54.99 (C5), 48.15 (C18), 45.28 (C14), 43.92 (C20), 43.70 (C8), 43.26 (piperazinyl C), 38.77 (piperazinyl C), 38.01 (C4), 37.66 (C22), 36.90 (10), 33.19 (C7), 32.72 (C17), 31.78 (C21), 29.69 (C29), 28.39 (C28), 28.02 (C23), 27.04 (C2), 26.66 (C15), 26.36 (C16), 21.32 (C27), 18.65 (C26), 17.34 (C6), 16.66 (C25), 16.41 (C24); HRMS (m/z): [M + Na]+ calcd for C41H56Cl2N2NaO4: 733.35148, found: 733.35754.
O), 167.78 (C–Cl), 164.23 (C–Cl), 164.11 (C–Cl), 161.73 (C–Cl), 161.61 (C–Cl), 138.13 (phenyl), 128.53 (C12), 110.54 (phenyl), 110.46 (phenyl), 110.35 (phenyl), 110.27 (phenyl), 105.83 (phenyl), 105.58 (phenyl), 105.33 (phenyl), 80.55 (C3), 61.69 (C9), 54.99 (C5), 48.15 (C18), 45.28 (C14), 43.92 (C20), 43.70 (C8), 43.26 (piperazinyl C), 38.77 (piperazinyl C), 38.01 (C4), 37.66 (C22), 36.90 (10), 33.19 (C7), 32.72 (C17), 31.78 (C21), 29.69 (C29), 28.39 (C28), 28.02 (C23), 27.04 (C2), 26.66 (C15), 26.36 (C16), 21.32 (C27), 18.65 (C26), 17.34 (C6), 16.66 (C25), 16.41 (C24); HRMS (m/z): [M + Na]+ calcd for C41H56Cl2N2NaO4: 733.35148, found: 733.35754.
3β-Hydroxy-30-(4-(3-chlorobenzoyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4h). A white solid; yield, 92.1%; mp 222.9–224.0 °C; 1H NMR (400 MHz, chloroform-d) δ 7.46–7.33 (m, 3H, phenyl-H), 7.29 (dd, J = 7.3, 1.7 Hz, 1H, phenyl-H), 5.68 (s, 1H, CH-12), 3.74–3.44 (m, 8H, piperazinyl-H), 3.23 (dd, J = 10.8, 5.5 Hz, 1H, 3-OH), 2.81–2.76 (m, 1H, CH-1), 2.34 (s, 1H, CH-9), 2.28 (d, J = 11.5 Hz, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.24 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.01 (s, 3H, CH3-23), 0.82 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.6 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.07 (C11), 174.27 (C30), 169.26 (C13), 168.97 (benzoyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 136.81 (phenyl), 134.76 (phenyl), 130.19 (phenyl), 130.02 (phenyl), 128.59 (C12), 127.31 (phenyl), 125.15 (phenyl), 78.75 (C3), 61.78 (C9), 54.92 (C5), 48.11 (C18), 45.27 (C14), 43.92 (C20), 43.77 (C8), 43.26 (piperazinyl C), 39.12 (piperazinyl C), 39.10 (C1), 37.68 (C4), 37.06 (C22), 33.15 (C10), 32.79 (C7), 31.78 (C17), 29.69 (C21), 28.39 (C29), 28.07 (C28), 27.28 (C23), 27.04 (C2), 26.68 (C15), 26.38 (C16), 23.14 (C27), 18.66 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C41H58ClN2O4: 677.40851, found: 677.41461.
O), 136.81 (phenyl), 134.76 (phenyl), 130.19 (phenyl), 130.02 (phenyl), 128.59 (C12), 127.31 (phenyl), 125.15 (phenyl), 78.75 (C3), 61.78 (C9), 54.92 (C5), 48.11 (C18), 45.27 (C14), 43.92 (C20), 43.77 (C8), 43.26 (piperazinyl C), 39.12 (piperazinyl C), 39.10 (C1), 37.68 (C4), 37.06 (C22), 33.15 (C10), 32.79 (C7), 31.78 (C17), 29.69 (C21), 28.39 (C29), 28.07 (C28), 27.28 (C23), 27.04 (C2), 26.68 (C15), 26.38 (C16), 23.14 (C27), 18.66 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C41H58ClN2O4: 677.40851, found: 677.41461.
3β-Hydroxy-30-(4-(4-chlorobenzoyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4i). A white solid; yield, 94.0%; mp 233.9–234.6 °C; 1H NMR (400 MHz, chloroform-d) δ 7.39 (d, J = 16.0 Hz, 3H, phenyl-H), 7.29 (d, J = 7.4 Hz, 1H, phenyl-H), 5.68 (s, 1H, CH-12), 3.69 (s, 6H, piperazinyl-H), 3.45 (s, 2H, piperazinyl-H), 3.22 (dd, J = 10.8, 5.3 Hz, 1H, 3-OH), 2.78 (dt, J = 13.6, 3.5 Hz, 1H, CH-1), 2.33 (s, 1H, CH-9), 2.32–2.25 (m, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.23 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.00 (s, 3H, CH3-23), 0.82 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.5 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.05 (C11), 174.26 (C30), 169.24 (C13), 168.95 (benzoyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 136.82 (phenyl), 134.75 (phenyl), 130.18 (phenyl), 130.02 (phenyl), 128.58 (C12), 127.30 (phenyl), 125.14 (phenyl), 78.72 (C3), 61.78 (C9), 54.91 (C5), 48.10 (C18), 45.26 (C14), 43.91 (C20), 43.77 (C8), 43.26 (piperazinyl C), 39.11 (piperazinyl C), 37.67 (C4), 37.06 (C22), 33.14 (C10), 32.78 (C7), 31.77 (C17), 29.69 (C21), 28.39 (C29), 28.07 (C28), 27.28 (C23), 27.04 (C2), 26.67 (C15), 26.38 (C16), 23.14 (C27), 18.66 (C26), 17.46 (C6), 16.36 (C25), 15.57 (C24); HRMS (m/z): [M + H]+ calcd C41H58ClN2O4: 677.40851, found: 677.41284.
O), 136.82 (phenyl), 134.75 (phenyl), 130.18 (phenyl), 130.02 (phenyl), 128.58 (C12), 127.30 (phenyl), 125.14 (phenyl), 78.72 (C3), 61.78 (C9), 54.91 (C5), 48.10 (C18), 45.26 (C14), 43.91 (C20), 43.77 (C8), 43.26 (piperazinyl C), 39.11 (piperazinyl C), 37.67 (C4), 37.06 (C22), 33.14 (C10), 32.78 (C7), 31.77 (C17), 29.69 (C21), 28.39 (C29), 28.07 (C28), 27.28 (C23), 27.04 (C2), 26.67 (C15), 26.38 (C16), 23.14 (C27), 18.66 (C26), 17.46 (C6), 16.36 (C25), 15.57 (C24); HRMS (m/z): [M + H]+ calcd C41H58ClN2O4: 677.40851, found: 677.41284.
3β-Hydroxy-30-(4-(3-bromobenzoyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4j). A white solid; yield, 90.9%; mp 239.2–240.1 °C; 1H NMR (400 MHz, chloroform-d) δ 7.57 (s, 2H, phenyl-H), 7.32 (q, J = 7.4 Hz, 2H, phenyl-H), 5.68 (s, 1H, CH-12), 3.83–3.38 (m, 8H, piperazinyl-H), 3.22 (dd, J = 10.7, 5.5 Hz, 1H, 3-OH), 2.78 (dt, J = 13.6, 3.6 Hz, 1H, CH-1), 2.33 (s, 1H, CH-9), 2.31–2.23 (m, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.24 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.00 (s, 3H, CH3-23), 0.82 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.6 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.06 (C11), 174.26 (C30), 169.24 (C13), 168.82 (benzoyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 137.05 (phenyl), 133.12 (phenyl), 130.25 (phenyl), 130.16 (phenyl), 128.59 (C12), 125.60 (phenyl), 122.79 (phenyl), 78.75 (C3), 61.79 (C9), 54.92 (C5), 48.11 (C18), 45.27 (C14), 43.92 (C20), 43.77 (C8), 43.26 (piperazinyl C), 39.11 (piperazinyl C), 37.68 (C4), 37.06 (C22), 33.15 (C10), 32.79 (C7), 31.78 (C17), 29.69 (C21), 28.39 (C29), 28.07 (C28), 27.28 (C23), 27.06 (C2), 26.68 (C15), 26.39 (C16), 23.15 (C27), 18.66 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + Na]+ calcd for C41H57BrN2NaO4: 743.33994, found: 743.34621, 745.34582.
O), 137.05 (phenyl), 133.12 (phenyl), 130.25 (phenyl), 130.16 (phenyl), 128.59 (C12), 125.60 (phenyl), 122.79 (phenyl), 78.75 (C3), 61.79 (C9), 54.92 (C5), 48.11 (C18), 45.27 (C14), 43.92 (C20), 43.77 (C8), 43.26 (piperazinyl C), 39.11 (piperazinyl C), 37.68 (C4), 37.06 (C22), 33.15 (C10), 32.79 (C7), 31.78 (C17), 29.69 (C21), 28.39 (C29), 28.07 (C28), 27.28 (C23), 27.06 (C2), 26.68 (C15), 26.39 (C16), 23.15 (C27), 18.66 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + Na]+ calcd for C41H57BrN2NaO4: 743.33994, found: 743.34621, 745.34582.
3β-Hydroxy-30-(4-(4-bromobenzoyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4k). A white solid; yield, 91.1%; mp 233.2–235.4 °C; 1H NMR (400 MHz, chloroform-d) δ 7.57 (d, J = 8.1 Hz, 2H, phenyl-H), 7.30 (d, J = 8.1 Hz, 2H, phenyl-H), 5.67 (s, 1H, CH-12), 3.68 (s, 8H, piperazinyl-H), 3.22 (dd, J = 10.7, 5.5 Hz, 1H, 3-OH), 2.81–2.76 (m, 1H, CH-1), 2.33 (s, 1H, CH-9), 2.27 (d, J = 11.7 Hz, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.23 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.00 (s, 3H, CH3-23), 0.82 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.6 Hz, 1H, CH-5);13C NMR (101 MHz, chloroform-d) δ 200.06 (C11), 174.25 (C30), 169.54 (C13), 169.25 (benzoyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 133.89 (phenyl), 131.88 (phenyl), 128.84 (phenyl), 128.58 (C12), 124.45 (phenyl), 78.75 (C3), 61.79 (C9), 54.92 (C5), 48.15 (C18), 45.27 (C14), 43.91 (C20), 43.74 (C8), 43.27 (piperazinyl C), 39.11 (piperazinyl C), 37.67 (C4), 37.06 (C22), 33.21 (C10), 32.79 (C7), 31.78 (C17), 29.70 (C21), 28.38 (C29), 28.07 (C28), 27.28 (C23), 27.05 (C2), 26.68 (C15), 26.39 (C16), 23.15 (C27), 18.66 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C41H58BrN2O4: 721.35800, found: 721.36354, 723.36266.
O), 133.89 (phenyl), 131.88 (phenyl), 128.84 (phenyl), 128.58 (C12), 124.45 (phenyl), 78.75 (C3), 61.79 (C9), 54.92 (C5), 48.15 (C18), 45.27 (C14), 43.91 (C20), 43.74 (C8), 43.27 (piperazinyl C), 39.11 (piperazinyl C), 37.67 (C4), 37.06 (C22), 33.21 (C10), 32.79 (C7), 31.78 (C17), 29.70 (C21), 28.38 (C29), 28.07 (C28), 27.28 (C23), 27.05 (C2), 26.68 (C15), 26.39 (C16), 23.15 (C27), 18.66 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C41H58BrN2O4: 721.35800, found: 721.36354, 723.36266.
3β-Hydroxy-30-(4-(3-fluorobenzoyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4l). A white solid; yield, 92.1%; mp 231.5–232.9 °C; 1H NMR (400 MHz, chloroform-d) δ 7.41 (td, J = 7.8, 5.5 Hz, 1H, phenyl-H), 7.16 (dd, J = 21.9, 8.1 Hz, 3H, phenyl-H), 5.68 (s, 1H, CH-12), 3.71–3.44 (m, 8H, piperazinyl-H), 3.22 (dd, J = 10.8, 5.5 Hz, 1H, 3-OH), 2.79 (dd, J = 13.4, 3.7 Hz, 1H, CH-1), 2.33 (s, 1H, CH-9), 2.28 (d, J = 12.4 Hz, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.23 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.00 (s, 3H, CH3-23), 0.82 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.5 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.06 (C11), 174.26 (C30), 169.25 (C13), 169.04 (benzoyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 163.78 (C–F), 161.30 (C–F), 137.17 (phenyl), 137.10 (phenyl), 130.52 (phenyl), 130.44 (phenyl), 128.59 (C12), 122.74 (phenyl), 122.71 (phenyl), 117.22 (phenyl), 117.01 (phenyl), 114.56 (phenyl), 114.33 (phenyl), 78.74 (C3), 61.78 (C9), 54.92 (C5), 48.11 (C18), 45.27 (C14), 43.92 (C20), 43.77 (C8), 43.26 (piperazinyl C), 39.11 (piperazinyl C), 37.68 (C4), 37.06 (C22), 33.15 (C10), 32.79 (C7), 31.77 (C17), 29.69 (C21), 28.39 (C29), 28.07 (C28), 27.28 (C23), 27.04 (C2), 26.68 (C15), 26.38 (C16), 23.14 (C27), 18.66 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd C41H58FN2O4: 661.43806, found: 661.44479.
O), 163.78 (C–F), 161.30 (C–F), 137.17 (phenyl), 137.10 (phenyl), 130.52 (phenyl), 130.44 (phenyl), 128.59 (C12), 122.74 (phenyl), 122.71 (phenyl), 117.22 (phenyl), 117.01 (phenyl), 114.56 (phenyl), 114.33 (phenyl), 78.74 (C3), 61.78 (C9), 54.92 (C5), 48.11 (C18), 45.27 (C14), 43.92 (C20), 43.77 (C8), 43.26 (piperazinyl C), 39.11 (piperazinyl C), 37.68 (C4), 37.06 (C22), 33.15 (C10), 32.79 (C7), 31.77 (C17), 29.69 (C21), 28.39 (C29), 28.07 (C28), 27.28 (C23), 27.04 (C2), 26.68 (C15), 26.38 (C16), 23.14 (C27), 18.66 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd C41H58FN2O4: 661.43806, found: 661.44479.
3β-Hydroxy-30-(4-(2,4-difluorobenzoyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4m). A white solid; yield, 92.0%; mp 229.7–231.1 °C; 1H NMR (400 MHz, chloroform-d) δ 7.43 (td, J = 8.2, 6.3 Hz, 1H, phenyl-H), 6.98 (td, J = 8.2, 2.4 Hz, 1H, phenyl-H), 6.87 (td, J = 9.2, 2.4 Hz, 1H, phenyl-H), 5.68 (s, 1H, CH-12), 3.89–3.53 (m, 6H, piperazinyl-H), 3.34 (s, 2H, piperazinyl-H), 3.22 (dd, J = 10.7, 5.5 Hz, 1H, 3-OH), 2.78 (dt, J = 13.5, 3.6 Hz, 1H, CH-1), 2.33 (s, 1H, CH-9), 2.32–2.26 (m, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.24 (s, 3H, CH3-25), 1.13 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.00 (s, 3H, CH3-23), 0.82 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.4 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.05 (C11), 174.22 (C30), 169.24 (C13), 165.13 (C–F), 165.01 (C–F), 164.52 (benzoyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 162.62 (C–F), 162.50 (C–F), 159.77 (C–F), 159.65 (C–F), 157.28 (C–F), 157.16 (C–F), 130.89 (phenyl), 130.84 (phenyl), 130.79 (phenyl), 130.74 (phenyl), 128.60 (C12), 119.80 (phenyl), 119.76 (phenyl), 119.62 (phenyl), 119.58 (phenyl), 112.58 (phenyl), 112.54 (phenyl), 112.36 (phenyl), 112.33 (phenyl), 104.51 (phenyl), 104.26 (phenyl), 104.00 (phenyl), 78.73 (C3), 61.78 (C9), 54.92 (C5), 48.06 (C18), 47.12 (C14), 45.26 (C20), 43.92 (C8), 43.82, 43.26 (C8), 42.31 (piperazinyl C), 39.10 (C4), 37.68 (C22), 37.06 (C10), 33.09 (C7), 32.78 (C17), 31.76 (C21), 28.07 (C28), 27.27 (C23), 27.05 (C2), 26.67 (C15), 26.37 (C16), 23.14 (C27), 18.65 (C26), 17.46 (C6), 16.36 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C41H57F2N2O4: 679.42864, found: 679.43500.
O), 162.62 (C–F), 162.50 (C–F), 159.77 (C–F), 159.65 (C–F), 157.28 (C–F), 157.16 (C–F), 130.89 (phenyl), 130.84 (phenyl), 130.79 (phenyl), 130.74 (phenyl), 128.60 (C12), 119.80 (phenyl), 119.76 (phenyl), 119.62 (phenyl), 119.58 (phenyl), 112.58 (phenyl), 112.54 (phenyl), 112.36 (phenyl), 112.33 (phenyl), 104.51 (phenyl), 104.26 (phenyl), 104.00 (phenyl), 78.73 (C3), 61.78 (C9), 54.92 (C5), 48.06 (C18), 47.12 (C14), 45.26 (C20), 43.92 (C8), 43.82, 43.26 (C8), 42.31 (piperazinyl C), 39.10 (C4), 37.68 (C22), 37.06 (C10), 33.09 (C7), 32.78 (C17), 31.76 (C21), 28.07 (C28), 27.27 (C23), 27.05 (C2), 26.67 (C15), 26.37 (C16), 23.14 (C27), 18.65 (C26), 17.46 (C6), 16.36 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C41H57F2N2O4: 679.42864, found: 679.43500.
3β-Hydroxy-30-(4-(3,5-difluorobenzoyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4n). A white solid; yield, 92.1%; mp 236.3–236.8 °C; 1H NMR (400 MHz, chloroform-d) δ 7.05–6.79 (m, 3H, phenyl-H), 5.67 (d, J = 2.1 Hz, 1H, CH-12), 3.84–3.52 (m, 6H, piperazinyl-H), 3.42 (s, 2H, piperazinyl-H), 3.22 (dd, J = 10.7, 5.4 Hz, 1H, 3-OH), 2.78 (dt, J = 13.5, 3.1 Hz, 1H, CH-1), 2.33 (s, 1H, CH-9), 2.27 (d, J = 13.1 Hz, 1H, CH-16), 1.35 (s, 3H, CH3-27), 1.23 (s, 3H, CH3-25), 1.13 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.00 (s, 3H, CH3-23), 0.81 (s, 3H, CH3-24), 0.80 (s, 3H, CH3-28), 0.70 (d, J = 11.5 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.05 (C11), 174.28 (C30), 169.21 (C13), 167.80 (benzoyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 167.77 (C–F), 167.74 (C–F), 164.23 (C–F), 164.11 (C–F), 161.72 (C–F), 161.60 (C–F), 138.21 (phenyl), 138.13 (phenyl), 138.04 (phenyl), 128.58 (C12), 110.53 (phenyl), 110.45 (phenyl), 110.34 (phenyl), 110.26 (phenyl), 105.82 (phenyl), 105.57 (phenyl), 105.32 (phenyl), 78.72 (C3), 61.78 (C9), 54.91 (C5), 48.10 (C18), 45.26 (C14), 45.21 (20), 43.92 (C8), 43.76 (piperazinyl C), 43.26 (C19), 39.10 (piperazinyl C), 37.66 (C22), 37.06 (C10), 33.12 (C7), 32.78 (C17), 31.77 (C21), 28.38 (C29), 28.07 (C28), 27.27 (C23), 27.02 (C2), 26.67 (C15), 26.37 (C16), 23.14 (C27), 18.66 (C26), 17.46 (C6), 16.36 (C25), 15.57 (C24); HRMS (m/z): [M + H]+ calcd for C41H57F2N2O4: 679.42864, found: 679.43500.
O), 167.77 (C–F), 167.74 (C–F), 164.23 (C–F), 164.11 (C–F), 161.72 (C–F), 161.60 (C–F), 138.21 (phenyl), 138.13 (phenyl), 138.04 (phenyl), 128.58 (C12), 110.53 (phenyl), 110.45 (phenyl), 110.34 (phenyl), 110.26 (phenyl), 105.82 (phenyl), 105.57 (phenyl), 105.32 (phenyl), 78.72 (C3), 61.78 (C9), 54.91 (C5), 48.10 (C18), 45.26 (C14), 45.21 (20), 43.92 (C8), 43.76 (piperazinyl C), 43.26 (C19), 39.10 (piperazinyl C), 37.66 (C22), 37.06 (C10), 33.12 (C7), 32.78 (C17), 31.77 (C21), 28.38 (C29), 28.07 (C28), 27.27 (C23), 27.02 (C2), 26.67 (C15), 26.37 (C16), 23.14 (C27), 18.66 (C26), 17.46 (C6), 16.36 (C25), 15.57 (C24); HRMS (m/z): [M + H]+ calcd for C41H57F2N2O4: 679.42864, found: 679.43500.
3β-Hydroxy-30-(4-(3-(trifluoromethyl)benzoyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4o). A white solid; yield, 90.8%; mp 258.5–259.7 °C; 1H NMR (400 MHz, chloroform-d) δ 7.71 (d, J = 10.3 Hz, 2H, phenyl-H), 7.65–7.53 (m, 2H, phenyl-H), 5.68 (s, 1H, CH-12), 3.70 (m, 6H, piperazinyl-H), 3.44 (s, 2H, piperazinyl-H), 3.22 (dd, J = 10.8, 5.5 Hz, 1H, 3-OH), 2.83–2.73 (m, 1H, CH-1), 2.33 (s, 1H, CH-9), 2.28 (d, J = 11.4 Hz, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.24 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 0.98 (s, 3H, CH3-23), 0.82 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.6 Hz, 1H); 13C NMR (101 MHz, chloroform-d) δ 200.06 (C11), 174.27 (C30), 169.23 (C13), 168.94 (benzoyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 138.91 (phenyl), 131.09 (phenyl), 130.38 (phenyl), 129.27 (CF3), 128.59 (C12), 126.84 (phenyl), 124.21 (phenyl), 124.17 (phenyl), 78.74 (C3), 61.79 (C9), 54.92 (C5), 48.12 (C18), 45.27 (C14), 43.93 (C20), 43.76 (C8), 43.26 (piperazinyl C), 39.11 (piperazinyl C), 37.67 (C4), 37.06 (C22), 33.15 (C10), 32.79 (C7), 31.78 (C17), 29.70 (C21), 28.39 (C29), 28.07 (C28), 27.28 (C23), 27.04 (C2), 26.67 (C15), 26.38 (C16), 23.14 (C27), 18.66 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C42H58F3N2O4: 711.43487, found: 711.44181.
O), 138.91 (phenyl), 131.09 (phenyl), 130.38 (phenyl), 129.27 (CF3), 128.59 (C12), 126.84 (phenyl), 124.21 (phenyl), 124.17 (phenyl), 78.74 (C3), 61.79 (C9), 54.92 (C5), 48.12 (C18), 45.27 (C14), 43.93 (C20), 43.76 (C8), 43.26 (piperazinyl C), 39.11 (piperazinyl C), 37.67 (C4), 37.06 (C22), 33.15 (C10), 32.79 (C7), 31.78 (C17), 29.70 (C21), 28.39 (C29), 28.07 (C28), 27.28 (C23), 27.04 (C2), 26.67 (C15), 26.38 (C16), 23.14 (C27), 18.66 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C42H58F3N2O4: 711.43487, found: 711.44181.
3β-Hydroxy-30-(4-(4-(trifluoromethyl)benzoyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4p). A white solid; yield, 90.8%; mp 237.2–239.2 °C; 1H NMR (400 MHz, chloroform-d) δ 7.72 (d, J = 8.0 Hz, 2H, phenyl-H), 7.55 (d, J = 8.0 Hz, 2H, phenyl-H), 5.68 (s, 1H, CH-12), 3.69 (m, 6H, piperazinyl-H), 3.41 (s, 2H, piperazinyl-H), 3.23 (dd, J = 10.7, 5.6 Hz, 1H, 3-OH), 2.84–2.75 (m, 1H, CH-1), 2.34 (s, 1H, CH-9), 2.30–2.21 (m, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.24 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.13 (s, 3H, CH3-29), 1.01 (s, 3H, CH3-23), 0.83 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.6 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.07 (C11), 174.28 (C30), 169.26 (C13), 169.05 (benzoyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 138.65 (phenyl), 132.15 (phenyl), 131.82 (phenyl), 130.39 (phenyl), 128.57 (C12), 127.48 (phenyl), 125.82 (CF3), 125.78 (CF3), 125.74 (CF3), 125.70 (CF3), 124.94 (phenyl), 122.23 (phenyl), 78.74 (C3), 61.79 (C9), 54.92 (C5), 48.18 (C18), 45.28 (C14), 43.92 (C20), 43.70 (C8), 43.27 (piperazinyl C), 39.10 (piperazinyl C), 37.66 (C4), 37.06 (C22), 33.24 (C10), 32.78 (C7), 31.79 (C17), 29.69 (C21), 28.38 (C29), 28.07 (C28), 27.27 (C23), 27.04 (C2), 26.67 (C15), 26.39 (C16), 23.14 (C27), 18.66 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + Na]+ calcd for C42H57F3N2NaO4: 733.41681, found: 733.42400.
O), 138.65 (phenyl), 132.15 (phenyl), 131.82 (phenyl), 130.39 (phenyl), 128.57 (C12), 127.48 (phenyl), 125.82 (CF3), 125.78 (CF3), 125.74 (CF3), 125.70 (CF3), 124.94 (phenyl), 122.23 (phenyl), 78.74 (C3), 61.79 (C9), 54.92 (C5), 48.18 (C18), 45.28 (C14), 43.92 (C20), 43.70 (C8), 43.27 (piperazinyl C), 39.10 (piperazinyl C), 37.66 (C4), 37.06 (C22), 33.24 (C10), 32.78 (C7), 31.79 (C17), 29.69 (C21), 28.38 (C29), 28.07 (C28), 27.27 (C23), 27.04 (C2), 26.67 (C15), 26.39 (C16), 23.14 (C27), 18.66 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + Na]+ calcd for C42H57F3N2NaO4: 733.41681, found: 733.42400.
3β-Hydroxy-30-(4-(4-cyanobenzoyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4q). A white solid; yield, 86.4%; mp 235.5–237.7 °C; 1H NMR (400 MHz, chloroform-d) δ 7.75 (d, J = 8.2 Hz, 2H, phenyl-H), 7.57–7.50 (m, 2H, phenyl-H), 5.66 (s, 1H, CH-12), 3.84–3.35 (m, 8H, piperazinyl-H), 3.22 (dd, J = 10.7, 5.5 Hz, 1H, 3-OH), 2.78 (dt, J = 13.5, 3.5 Hz, 1H, CH-1), 2.33 (s, 1H, CH-9), 2.28–2.20 (m, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.23 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.00 (s, 3H, CH3-23), 0.82 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.6 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.09 (C11), 174.30 (C30), 169.28 (C13), 168.45 (benzoyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 139.42 (phenyl), 132.56 (phenyl), 128.55 (C12), 127.82 (phenyl), 117.92 (phenyl), 113.89 (CN), 78.72 (C3), 61.80 (C9), 54.92 (C5), 48.21 (C18), 45.28 (C14), 43.93 (C20), 43.66 (C8), 43.27 (piperazinyl C), 39.11 (piperazinyl C), 37.64 (C4), 37.07 (C22), 33.29 (C10), 32.78 (C7), 31.79 (C17), 29.69 (C21), 28.38 (C29), 28.07 (C28), 27.27 (C23), 27.03 (C2), 26.67 (C15), 26.39 (C16), 23.14 (C27), 18.66 (C26), 17.45 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C42H58N3O4: 668.44273, found: 668.44912.
O), 139.42 (phenyl), 132.56 (phenyl), 128.55 (C12), 127.82 (phenyl), 117.92 (phenyl), 113.89 (CN), 78.72 (C3), 61.80 (C9), 54.92 (C5), 48.21 (C18), 45.28 (C14), 43.93 (C20), 43.66 (C8), 43.27 (piperazinyl C), 39.11 (piperazinyl C), 37.64 (C4), 37.07 (C22), 33.29 (C10), 32.78 (C7), 31.79 (C17), 29.69 (C21), 28.38 (C29), 28.07 (C28), 27.27 (C23), 27.03 (C2), 26.67 (C15), 26.39 (C16), 23.14 (C27), 18.66 (C26), 17.45 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C42H58N3O4: 668.44273, found: 668.44912.
3β-Hydroxy-30-(4-cyclohexanecarbonyl-1-piperazinyl)-olean-12-ene-11,30-dione (4r). A white solid; yield, 86.4%; mp 213.6–215.4 °C; 1H NMR (400 MHz, chloroform-d) δ 5.68 (s, 1H, CH-12), 3.69–3.46 (m, 8H, piperazinyl-H), 3.22 (dd, J = 10.7, 5.5 Hz, 1H, 3-OH), 2.78 (dt, J = 13.4, 3.5 Hz, 1H, CH-1), 2.46 (tt, J = 11.6, 3.4 Hz, 1H, cyclohexanecarbonyl-CH), 2.34 (s, 1H, CH-9), 2.29 (dd, J = 12.7, 3.5 Hz, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.30–1.25 (m, 10H, cyclohexane), 1.23 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.01 (s, 3H, CH3-23), 0.82 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.5 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.05 (C11), 174.82 (cyclohexanecarbonyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 174.20 (C30), 169.29 (C13), 128.58 (C12), 78.74 (C3), 61.77 (C9), 54.92 (C5), 48.07 (C18), 45.33 (C14), 45.26 (C20), 43.91 (C8), 43.82 (piperazinyl C), 43.26 (cyclohexane), 41.60 (C19), 40.39 (piperazinyl C), 39.11 (C4), 37.70 (C22), 37.06 (C10), 33.08 (C7), 32.79 (C17), 31.76 (C21), 29.69 (cyclohexane), 29.36 (cyclohexane), 29.29 (C29), 28.39 (C28), 28.07 (C23), 27.28 (C2), 27.03 (C15), 26.69 (C16), 26.38 (cyclohexane), 25.79 (cyclohexane), 25.76 (cyclohexane), 23.15 (C27), 18.66 (C26), 17.46 (C6), 16.36 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C41H65N2O4: 649.49443, found: 649.50050.
O), 174.20 (C30), 169.29 (C13), 128.58 (C12), 78.74 (C3), 61.77 (C9), 54.92 (C5), 48.07 (C18), 45.33 (C14), 45.26 (C20), 43.91 (C8), 43.82 (piperazinyl C), 43.26 (cyclohexane), 41.60 (C19), 40.39 (piperazinyl C), 39.11 (C4), 37.70 (C22), 37.06 (C10), 33.08 (C7), 32.79 (C17), 31.76 (C21), 29.69 (cyclohexane), 29.36 (cyclohexane), 29.29 (C29), 28.39 (C28), 28.07 (C23), 27.28 (C2), 27.03 (C15), 26.69 (C16), 26.38 (cyclohexane), 25.79 (cyclohexane), 25.76 (cyclohexane), 23.15 (C27), 18.66 (C26), 17.46 (C6), 16.36 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C41H65N2O4: 649.49443, found: 649.50050.
3β-Hydroxy-30-(4-cyclopropanecarbonyl-1-piperazinyl)-olean-12-ene-11,30-dione (4s). A white solid; yield, 84.8%; mp 210.7–212.2 °C; 1H NMR (400 MHz, chloroform-d) δ 5.70 (d, J = 5.2 Hz, 1H, CH-12), 3.67 (m, 8H, piperazinyl-H), 3.23 (dd, J = 11.0, 5.4 Hz, 1H, 3-OH), 2.79 (d, J = 13.3 Hz, 1H, CH-1), 2.32 (m, 2H, CH-9/16), 1.37 (s, 3H, CH3-27), 1.24 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.01 (s, 3H, CH3-23), 0.82 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.6 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.06 (C11), 174.24 (C30), 172.34 (cyclopropanecarbonyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 169.31 (C13), 128.59 (C12), 78.73 (C3), 61.78 (C9), 54.92 (C5), 48.07 (C18), 45.26 (C14), 43.93 (C20), 43.84 (C8), 43.26 (piperazinyl C), 42.09 (C19), 39.10 (piperazinyl C), 37.71 (C22), 37.06 (C10), 33.07 (C7), 32.79 (C17), 31.77 (C21), 29.69 (C29), 28.40 (C28), 28.07 (C23), 27.28 (C2), 27.04 (C15), 26.69 (C16), 23.15 (C27), 18.66 (C26), 17.46 (C6), 16.37 (C25), 15.57 (C24), 10.97 (cyclopropane), 7.72 (cyclopropane), 7.70 (cyclopropane); HRMS (m/z): [M + Na]+ calcd for C38H58N2NaO4: 629.42943, found: 629.43642.
O), 169.31 (C13), 128.59 (C12), 78.73 (C3), 61.78 (C9), 54.92 (C5), 48.07 (C18), 45.26 (C14), 43.93 (C20), 43.84 (C8), 43.26 (piperazinyl C), 42.09 (C19), 39.10 (piperazinyl C), 37.71 (C22), 37.06 (C10), 33.07 (C7), 32.79 (C17), 31.77 (C21), 29.69 (C29), 28.40 (C28), 28.07 (C23), 27.28 (C2), 27.04 (C15), 26.69 (C16), 23.15 (C27), 18.66 (C26), 17.46 (C6), 16.37 (C25), 15.57 (C24), 10.97 (cyclopropane), 7.72 (cyclopropane), 7.70 (cyclopropane); HRMS (m/z): [M + Na]+ calcd for C38H58N2NaO4: 629.42943, found: 629.43642.
3β-Hydroxy-30-(4-(6-chloronicotinoyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4t). A white solid; yield, 94.6%; mp 244.5–246.3 °C; 1H NMR (400 MHz, chloroform-d) δ 8.47 (d, J = 2.3 Hz, 1H, pyridyl-H), 7.76 (dd, J = 8.2, 2.4 Hz, 1H, pyridyl-H), 7.43 (d, J = 8.1 Hz, 1H, pyridyl-H), 5.67 (s, 1H, CH-12), 3.71–3.47 (m, 8H, piperazinyl-H), 3.22 (dd, J = 10.7, 5.5 Hz, 1H, 3-OH), 2.78 (dt, J = 13.3, 3.5 Hz, 1H, CH-1), 2.33 (s, 1H, CH-9), 2.30–2.21 (m, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.24 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.00 (s, 3H, CH3-23), 0.82 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.6 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.05 (C11), 174.30 (C30), 169.22 (C13), 166.88 (chloronicotinoyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 153.02 (pyridyl), 148.12 (pyridyl), 138.01 (pyridyl), 129.72 (pyridyl), 128.57 (C12), 124.51 (pyridyl), 78.74 (C3), 61.79 (C9), 54.92 (C5), 48.16 (C18), 45.27 (C14), 43.93 (C20), 43.72 (C8), 43.27 (piperazinyl C), 39.13 (piperazinyl C), 39.11 (C4), 37.65 (C22), 37.06 (C10), 33.20 (C7), 32.78 (C17), 31.79 (C21), 28.38 (C29), 28.07 (C28), 27.28 (C23), 27.04 (C2), 26.67 (C15), 26.39 (C16), 23.15 (C27), 18.66 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C40H57ClN3O4: 678.40376, found: 678.41033.
O), 153.02 (pyridyl), 148.12 (pyridyl), 138.01 (pyridyl), 129.72 (pyridyl), 128.57 (C12), 124.51 (pyridyl), 78.74 (C3), 61.79 (C9), 54.92 (C5), 48.16 (C18), 45.27 (C14), 43.93 (C20), 43.72 (C8), 43.27 (piperazinyl C), 39.13 (piperazinyl C), 39.11 (C4), 37.65 (C22), 37.06 (C10), 33.20 (C7), 32.78 (C17), 31.79 (C21), 28.38 (C29), 28.07 (C28), 27.28 (C23), 27.04 (C2), 26.67 (C15), 26.39 (C16), 23.15 (C27), 18.66 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C40H57ClN3O4: 678.40376, found: 678.41033.
3β-Hydroxy-30-(4-(2-(thiophen-2-yl)acetyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4u). A white solid; yield, 87.5%; mp 218.5–220.2 °C; 1H NMR (400 MHz, chloroform-d) δ 7.22 (dd, J = 5.1, 1.2 Hz, 1H, thiophen-2-yl-H), 6.97 (dd, J = 5.2, 3.5 Hz, 1H, thiophen-2-yl-H), 6.91 (d, J = 3.4 Hz, 1H, thiophen-2-yl-H), 5.67 (s, 1H, CH-12), 3.94 [s, 2H, (thiophen-2-yl)acetyl-CH2], 3.56 (m, 8H, piperazinyl-H), 3.23 (dd, J = 10.7, 5.6 Hz, 1H, 3-OH), 2.79 (dt, J = 13.5, 3.5 Hz, 1H, CH-1), 2.33 (s, 1H, CH-9), 2.31–2.23 (m, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.21 (s, 3H, CH3-25), 1.13 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.00 (s, 3H, CH3-23), 0.82 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.8 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.05 (C11), 174.23 (C30), 169.25 (C13), 168.62 [(thiophen-2-yl)acetyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O], 135.98 (thiophen-2-yl), 128.58 (C12), 126.98 (thiophen-2-yl), 126.13 (thiophen-2-yl), 124.94 (thiophen-2-yl), 78.74 (C3), 61.77 (C9), 54.91 (C5), 48.02 (C18), 46.16 (C14), 45.26 (C20), 43.90 (C8), 43.82 (piperazinyl C), 43.25 (C19), 41.98 (piperazinyl C), 39.11 (C4), 37.68 (C22), 37.05 (C10), 35.17 [(thiophen-2-yl)acetyl-CH2], 33.00 (C7), 32.79 (C17), 31.75 (C21), 29.69 (C29), 28.38 (C28), 28.07 (C23), 27.28 (C2), 27.03 (C15), 26.67 (C16), 23.14 (C27), 18.65 (C26), 17.46 (C6), 16.37 (C25), 15.57 (C24); HRMS (m/z): [M + Na]+ calcd for C40H58N2NaO4S: 685.40150, found: 685.40759.
O], 135.98 (thiophen-2-yl), 128.58 (C12), 126.98 (thiophen-2-yl), 126.13 (thiophen-2-yl), 124.94 (thiophen-2-yl), 78.74 (C3), 61.77 (C9), 54.91 (C5), 48.02 (C18), 46.16 (C14), 45.26 (C20), 43.90 (C8), 43.82 (piperazinyl C), 43.25 (C19), 41.98 (piperazinyl C), 39.11 (C4), 37.68 (C22), 37.05 (C10), 35.17 [(thiophen-2-yl)acetyl-CH2], 33.00 (C7), 32.79 (C17), 31.75 (C21), 29.69 (C29), 28.38 (C28), 28.07 (C23), 27.28 (C2), 27.03 (C15), 26.67 (C16), 23.14 (C27), 18.65 (C26), 17.46 (C6), 16.37 (C25), 15.57 (C24); HRMS (m/z): [M + Na]+ calcd for C40H58N2NaO4S: 685.40150, found: 685.40759.
3β-Hydroxy-30-(4-(2-(4-fluorophenyl)acetyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4v). A white solid; yield, 91.3%; mp 212.7–213.3 °C; 1H NMR (400 MHz, chloroform-d) δ 7.21 (dd, J = 8.3, 5.2 Hz, 2H, phenyl), 7.03 (t, J = 8.3 Hz, 2H, phenyl), 5.66 (s, 1H, CH-12), 3.72 [s, 2H, (4-fluorophenyl)acetyl-CH2], 3.53 (m, 8H, piperazinyl-H), 3.22 (dd, J = 10.7, 5.4 Hz, 1H, 3-OH), 2.79 (d, J = 13.3 Hz, 1H, CH-1), 2.33 (s, 1H, CH-9), 2.26 (d, J = 12.4 Hz, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.20 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.00 (s, 3H, CH3-23), 0.81 (s, 6H, CH3-24/28), 0.70 (d, J = 11.5 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.06 (C11), 174.24 (C30), 169.53 (C13), 169.27 [(4-fluorophenyl)acetyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O], 163.04 (C–F), 160.60 (C–F), 130.26 (phenyl), 130.22 (phenyl), 130.14 (phenyl), 128.56 (C12), 115.81 (phenyl), 115.60 (phenyl), 78.73 (C3), 61.77 (C9), 54.91 (C5), 48.05 (C18), 45.94 (C14), 45.26 [(4-fluorophenyl)acetyl, CH2], 43.89 (C20), 43.79 (C8), 43.25 (piperazinyl C), 41.86 (C19), 39.90 (piperazinyl C), 39.10 (C1), 37.67 (C4), 37.05 (C22), 33.03 (C10), 32.78 (C7), 31.75 (C17), 29.69 (C21), 28.37 (C29), 28.07 (C28), 27.27 (C23), 27.02 (C2), 26.67 (C15), 26.37 (C16), 23.14 (C27), 18.65 (C26), 17.46 (C6), 16.37 (C25), 15.57 (C24); HRMS (m/z): [M + H]+ calcd for C42H60FN2O4: 675.45371, found: 675.46080.
O], 163.04 (C–F), 160.60 (C–F), 130.26 (phenyl), 130.22 (phenyl), 130.14 (phenyl), 128.56 (C12), 115.81 (phenyl), 115.60 (phenyl), 78.73 (C3), 61.77 (C9), 54.91 (C5), 48.05 (C18), 45.94 (C14), 45.26 [(4-fluorophenyl)acetyl, CH2], 43.89 (C20), 43.79 (C8), 43.25 (piperazinyl C), 41.86 (C19), 39.90 (piperazinyl C), 39.10 (C1), 37.67 (C4), 37.05 (C22), 33.03 (C10), 32.78 (C7), 31.75 (C17), 29.69 (C21), 28.37 (C29), 28.07 (C28), 27.27 (C23), 27.02 (C2), 26.67 (C15), 26.37 (C16), 23.14 (C27), 18.65 (C26), 17.46 (C6), 16.37 (C25), 15.57 (C24); HRMS (m/z): [M + H]+ calcd for C42H60FN2O4: 675.45371, found: 675.46080.
3β-Hydroxy-30-(4-(2-(4-chlorophenyl)acetyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4w). A white solid; yield, 90.0%; mp 239.7–240.5 °C; 1H NMR (400 MHz, chloroform-d) δ 7.31 (d, J = 8.3 Hz, 2H, phenyl), 7.18 (d, J = 8.1 Hz, 2H, phenyl), 5.66 (s, 1H, CH-12), 3.71 [s, 2H, (4-chlorophenyl)acetyl-CH2], 3.65–3.39 (m, 8H, piperazinyl-H), 3.22 (dd, J = 10.7, 5.5 Hz, 1H, 3-OH), 2.78 (dt, J = 13.5, 3.6 Hz, 1H, CH-1), 2.33 (s, 1H, CH-9), 2.30–2.22 (m, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.20 (s, 3H, CH3-25), 1.13 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.00 (s, 3H, CH3-23), 0.80 (s, 6H, CH3-24/28), 0.70 (d, J = 11.8 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.05 (C11), 174.24 (C30), 169.26 (C13), 169.22 [(4-chlorophenyl)acetyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O], 133.01 (phenyl), 132.92 (phenyl), 130.01 (phenyl), 128.96 (phenyl), 128.57 (C12), 78.74 (C3), 61.78 (C9), 54.92 (C5), 48.05 (C18), 45.93 (C14), 45.26 [(4-chlorophenyl)acetyl, CH2], 43.89 (C20), 43.79 (C8), 43.25 (piperazinyl C), 41.86 (C19), 40.07 (piperazinyl C), 39.10 (C1), 37.67 (C4), 37.06 (C22), 33.03 (C1), 32.79 (C7), 31.75 (C17), 29.69 (C21), 28.37 (C29), 28.07 (C28), 27.28 (C23), 27.02 (C2), 26.67 (C15), 26.37 (C16), 23.14 (C27), 18.65 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C42H60ClN2O4: 691.42416, found: 691.43121.
O], 133.01 (phenyl), 132.92 (phenyl), 130.01 (phenyl), 128.96 (phenyl), 128.57 (C12), 78.74 (C3), 61.78 (C9), 54.92 (C5), 48.05 (C18), 45.93 (C14), 45.26 [(4-chlorophenyl)acetyl, CH2], 43.89 (C20), 43.79 (C8), 43.25 (piperazinyl C), 41.86 (C19), 40.07 (piperazinyl C), 39.10 (C1), 37.67 (C4), 37.06 (C22), 33.03 (C1), 32.79 (C7), 31.75 (C17), 29.69 (C21), 28.37 (C29), 28.07 (C28), 27.28 (C23), 27.02 (C2), 26.67 (C15), 26.37 (C16), 23.14 (C27), 18.65 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd for C42H60ClN2O4: 691.42416, found: 691.43121.
3β-Hydroxy-30-(4-(morpholine-4-carbonyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4x). A white solid; yield, 88.2%; mp 211.7–213.7 °C; 1H NMR (400 MHz, chloroform-d) δ 5.68 (s, 1H, CH-12), 3.80–3.53 (m, 8H, piperazinyl-H), 3.26 (m, 9H, morpholinyl-H/3-OH), 2.78 (dt, J = 13.4, 3.4 Hz, 1H, CH-1), 2.34 (s, 1H, CH-9), 2.28 (d, J = 12.9 Hz, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.22 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 1.01 (s, 3H, CH3-23), 0.82 (s, 3H, CH3-24), 0.81 (s, 3H, CH3-28), 0.70 (d, J = 11.5 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.10 (C11), 174.13 (C30), 169.41 (C13), 163.56 (morpholine-4-carbonyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 128.54 (C12), 78.72 (C3), 61.78 (C9), 54.91 (C5), 48.13 (C18), 47.14 (morpholinyl-C), 46.91 (morpholinyl-C), 45.26 (C14), 43.88 (C20), 43.77 (C8), 43.26 (piperazinyl C), 39.11 (piperazinyl C), 37.69 (C4), 37.06 (C22), 33.20 (C10), 32.78 (C7), 31.76 (C17), 29.68 (C21), 28.40 (C29), 28.07 (C28), 27.27 (C23), 27.02 (C2), 26.69 (C15), 26.39 (C16), 23.14 (C27), 18.65 (C26), 17.46 (C6), 16.37 (C25), 15.57 (C24); HRMS (m/z): [M + H]+ calcd C39H62N3O5: 652.46895, found: 652.47377.
O), 128.54 (C12), 78.72 (C3), 61.78 (C9), 54.91 (C5), 48.13 (C18), 47.14 (morpholinyl-C), 46.91 (morpholinyl-C), 45.26 (C14), 43.88 (C20), 43.77 (C8), 43.26 (piperazinyl C), 39.11 (piperazinyl C), 37.69 (C4), 37.06 (C22), 33.20 (C10), 32.78 (C7), 31.76 (C17), 29.68 (C21), 28.40 (C29), 28.07 (C28), 27.27 (C23), 27.02 (C2), 26.69 (C15), 26.39 (C16), 23.14 (C27), 18.65 (C26), 17.46 (C6), 16.37 (C25), 15.57 (C24); HRMS (m/z): [M + H]+ calcd C39H62N3O5: 652.46895, found: 652.47377.
β-Hydroxy-30-(4-(2-methoxyacetyl)-1-piperazinyl)-olean-12-ene-11,30-dione (4y). A white solid; yield, 85.4%; mp 221.7–222.5 °C; 1H NMR (400 MHz, chloroform-d) δ 5.67 (d, J = 3.8 Hz, 1H, CH-12), 4.12 (s, 2H, methoxyacetyl-CH2), 3.57 (d, J = 49.5 Hz, 8H, piperazinyl-H), 3.43 (d, J = 3.6 Hz, 3H, methoxyacetyl-CH3), 3.22 (dt, J = 10.4, 4.6 Hz, 1H, 3-OH), 2.77 (dt, J = 13.4, 3.8 Hz, 1H, CH-1), 2.33 (d, J = 3.7 Hz, 1H, CH-9), 2.28 (d, J = 13.1 Hz, 1H, CH-16), 1.35 (s, 3H, CH3-27), 1.22 (s, 3H, CH3-25), 1.12 (s, 3H, CH3-26), 1.11 (s, 3H, CH3-29), 0.99 (s, 3H, CH3-23), 0.81 (s, 3H, CH3-24), 0.80 (s, 3H, CH3-28), 0.69 (d, J = 12.0 Hz, 1H, CH-5); 13C NMR (101 MHz, chloroform-d) δ 200.05 (C11), 174.24 (C30), 169.27 (C13), 167.80 (methoxyacetyl, C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 128.58 (C12), 78.71 (C3), 71.96 (methoxyacetyl, CH2), 61.77 (C9), 59.12 (methoxyacetyl, CH3), 54.91 (C5), 48.06 (C18), 45.26 (C14), 43.90 (C20), 43.82 (C8), 43.26 (piperazinyl C), 39.10 (piperazinyl C), 37.69 (C4), 37.05 (C22), 33.07 (C10), 32.78 (C7), 31.76 (C17), 29.68 (C21), 28.39 (C29), 28.07 (C28), 27.27 (C23), 27.05 (C2), 26.68 (C15), 26.37 (C16), 23.15 (C27), 18.65 (C26), 17.46 (C6), 16.36 (C25), 15.57 (C24); HRMS (m/z): [M + H]+ calcd C37H59N2O5: 611.44240, found: 611.44608.
O), 128.58 (C12), 78.71 (C3), 71.96 (methoxyacetyl, CH2), 61.77 (C9), 59.12 (methoxyacetyl, CH3), 54.91 (C5), 48.06 (C18), 45.26 (C14), 43.90 (C20), 43.82 (C8), 43.26 (piperazinyl C), 39.10 (piperazinyl C), 37.69 (C4), 37.05 (C22), 33.07 (C10), 32.78 (C7), 31.76 (C17), 29.68 (C21), 28.39 (C29), 28.07 (C28), 27.27 (C23), 27.05 (C2), 26.68 (C15), 26.37 (C16), 23.15 (C27), 18.65 (C26), 17.46 (C6), 16.36 (C25), 15.57 (C24); HRMS (m/z): [M + H]+ calcd C37H59N2O5: 611.44240, found: 611.44608.
Bisamide (5) a white solid; yield, 53.7%; mp 211.4–212.0 °C. HRMS (m/z): [M + Na]+ calcd for C64H98N2NaO6: 1013.73226, found: 1013.73499.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CH2Cl2–methanol).
1 CH2Cl2–methanol).A white solid; yield, 38.4%; mp 219.6–220.4 °C. HRMS (m/z): [M + Na]+ calcd for C50H64Cl2N2NaO5: 865.40900, found: 865.41315.
4.2 Primary anticancer assay
All the cell lines used in this study were either purchased from Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (A549, HepG2, MCF-7, PC-3 and LO2) or from Nanjing Cobioer Biosciences Co., Ltd. (Karpas299). Cells were incubated with 5% CO2 at 37 °C for 24 h. Subsequently the compounds and the reference were dissolved into the culture medium. Then, the cells were treated with different concentrations of test compounds and further incubated. After drug treatment, 20 μL of MTT solution at 5 mg mL−1 was added and incubated for 4 h. 100 μL of DMSO was added into each well to dissolve the purple formazan formed. The absorbance was determined at 630 nm using a plate reader. The IC50 was calculated using GraphPad Prism version 6.0 software (San Diego, USA) from the non-linear curve.4.3 Kinase activity determination
The effects of the compounds on the activities of the tyrosine kinase was determined using enzyme-linked immunosorbent assay (ELISA). Briefly, various concentrations of test compounds diluted in DMSO were then added to each well. DMSO was used as the negative control. The kinase reaction initiated at 37 °C for 60 min after the addition of biotinylated detection Ab working solution. After being washed, HRP affinipure goat anti-mouse IgG (H + L) was added. The plate was then incubated at 37 °C for 30 min. After being washed, the substrate reagent was added to each well at 37 °C. The enzyme–substrate reaction is terminated by the addition of stop solution as the color changed, and the plate was analyzed using a multi-well spectrophotometer at 450 nm. The IC50 values were calculated from the inhibition curves in two separate experiments.4.4 Molecular modeling
The prepared compounds were drawn using the ChemBioDraw Ultra 14.0 software, and then were saved in the sdf format, these molecules were imported into the Discovery Studio 3.5 software for the docking study (the Discovery Studio 3.5 software package, Accelrys, Co. Ltd., San Diego, USA). And those better docking conformations obtained from the full minimization protocol. Molecular docking into the 3D X-ray structure of ALK kinase (PDB code: 2XP2) was performed by using CDOCKER protocol. All the water and ligand were removed from the protein and the polar hydrogen was added. Molecular docking was validated by the docking of the co-crystallized inhibitor for enzyme, and root-mean-square deviation (RMSD) value for the backbone atoms between docked pose and crystallographic pose was below 1.5 Å.The molecular overlay program of the Discovery Studio 3.5 software was used to align moleculars and calculate molecular similarity according to the default parameter (50% steric field and 50% electrostatic field).
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Natural Science Foundation of Liaoning Province (No. 2019-ZD-0693 and No. 20170540396), General Research Projects of Liaoning Provincial Department of Education (No. JQL201715410) and Undergraduate Innovation and Entrepreneurship Training Program (No. 2019052).References
- Y. Liu and N. S. Gray, Nat. Chem. Biol., 2006, 2, 358–364 CrossRef CAS PubMed.
- A. Backes, B. Zech, B. Felber, B. Klebl and G. Müller, Expert Opin. Drug Discovery, 2008, 3, 1427–1449 CrossRef CAS PubMed.
- Y. Niu, X. Yao and H. Ji, RSC Adv., 2019, 9, 12441–12454 RSC.
- A. Roohbakhsh, M. Iranshahy and M. Iranshahi, Curr. Med. Chem., 2016, 23, 498–517 CrossRef CAS PubMed.
- H. Hussain, I. R. Green, U. Shamraiz, M. Saleem, A. Badshah, G. Abbas, N. U. Rehman and M. Irshad, Expert Opin. Ther. Pat., 2018, 28, 383–398 CrossRef CAS PubMed.
- X. Li, Y. Liu, N. Wang, Y. Liu, S. Wang, H. Wang, A. Li and S. Ren, RSC Adv., 2019, 9, 27294–27304 RSC.
- D. Cai, Z. Zhang, Y. Chen, Y. Zhang, Y. Sun and Y. Gong, Molecules, 2019, 24, 3631 CrossRef PubMed.
- D. P. Alho, J. A. Salvador, M. Cascante and S. Marin, Molecules, 2019, 24, 766 CrossRef PubMed.
- B. Xu, G.-R. Wu, X.-Y. Zhang, M.-M. Yan, R. Zhao, N.-N. Xue, K. Fang, H. Wang, M. Chen and W.-B. Guo, Molecules, 2017, 22, 924 CrossRef PubMed.
- T. L. Yan, L. F. Bai, H. L. Zhu, W. M. Zhang and P. C. Lv, ChemMedChem, 2017, 12, 1087–1096 CrossRef CAS PubMed.
- R. Sharma, S. K. Guru, S. K. Jain, A. S. Pathania, R. A. Vishwakarma, S. Bhushan and S. B. Bharate, MedChemComm, 2015, 6, 564–575 RSC.
- R. Csuk, S. Schwarz, R. Kluge and D. Ströhl, Eur. J. Med. Chem., 2010, 45, 5718–5723 CrossRef CAS PubMed.
- J. Liu and S. Ma, Curr. Med. Chem., 2017, 24, 590–613 CrossRef CAS PubMed.
- X. Yan, C. Liao, Z. Liu, A. T. Hagler, Q. Gu and J. Xu, Curr. Drug Targets, 2016, 17, 1580–1585 CrossRef CAS PubMed.
- H. Eckert and J. Bajorath, Drug Discovery Today, 2007, 12, 225–233 CrossRef CAS PubMed.
- F. Barbosa and D. Horvath, Curr. Top. Med. Chem., 2004, 4, 589–600 CrossRef CAS PubMed.
- P. J. Ballester and W. G. Richards, Proc. R. Soc. A, 2007, 463, 1307–1321 CAS.
- C. Ferroud, M. Godart, S. Ung, H. Borderies and A. Guy, Tetrahedron Lett., 2008, 49, 3004–3008 CrossRef CAS.
- W.-C. Chou, M.-C. Chou, Y.-Y. Lu and S.-F. Chen, Tetrahedron Lett., 1999, 40, 3419–3422 CrossRef CAS.
- M. H. Abdelrahman, B. G. Youssif, A. H. Abdelazeem, H. M. Ibrahim, A. M. Abd El Ghany, L. Treamblu and S. N. A. Bukhari, Eur. J. Med. Chem., 2017, 127, 972–985 CrossRef CAS PubMed.
- B. Lallemand, F. Chaix, M. Bury, C. Bruyère, J. Ghostin, J.-P. Becker, C. Delporte, M. Gelbcke, V. Mathieu, J. Dubois, M. Prévost, I. Jabin and R. Kiss, J. Med. Chem., 2011, 54, 6501–6513 CrossRef CAS PubMed.
- J. J. Cui, M. Tran-Dubé, H. Shen, M. Nambu, P.-P. Kung, M. Pairish, L. Jia, J. Meng, L. Funk and I. Botrous, J. Med. Chem., 2011, 54, 6342–6363 CrossRef CAS PubMed.
- S. Sommerwerk, L. Heller, C. Kerzig, A. E. Kramell and R. Csuk, Eur. J. Med. Chem., 2017, 127, 1–9 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra00681e |
| This journal is © The Royal Society of Chemistry 2020 |