Open Access Article
Open Access ArticleScreening and characterisation of CdTe/CdS quantum dot-binding peptides for material surface functionalisation†
Thanawat Suwatthanarak‡
 a,
Masayoshi Tanaka‡
a,
Masayoshi Tanaka‡ a,
Taisuke Minamidea,
Andrew J. Harvie
a,
Taisuke Minamidea,
Andrew J. Harvie bc,
Abiral Tamangb,
Kevin Critchleyb,
Stephen D. Evansb and
Mina Okochi
bc,
Abiral Tamangb,
Kevin Critchleyb,
Stephen D. Evansb and
Mina Okochi *a
*a
aDepartment of Chemical Science and Engineering, Tokyo Institute of Technology, 2-12-1, O-okayama, Meguro-ku, Tokyo 152-8552, Japan. E-mail: okochi.m.aa@m.titech.ac.jp
bSchool of Physics and Astronomy, University of Leeds, Leeds, LS2 9JT, UK
cDepartment of Chemistry, Norwegian University of Science and Technology (NTNU), Trondheim 7491, Norway
First published on 26th February 2020
Abstract
Quantum dots (QDs) are promising nanomaterials due to their unique photophysical properties. For them to be useful in biological applications, the particle surface generally needs to be conjugated to biological molecules, such as antibodies. In this study, we screened CdTe/CdS QD-binding peptides from a phage display library as linkers for simple and bio-friendly QD modification. Among five QD-binding peptide candidates, a series of truncated peptides designed from two high-affinity peptides were subjected to an array-based binding assay with QDs to assess their functional core sequences and characteristics. Linking these isolated, shortened peptides (PWSLNR and SGVYK) with an antibody-binding peptide (NKFRGKYK) created dual-functional peptides that are capable of QD surface functionalisation by antibodies. Consequently, the dual-functional peptides could mediate anti-CD9 antibody functionalisation onto CdTe/CdS QD surface; CD9 protein imaging of cancer cells was also demonstrated. Our proposed peptides offer an effective vehicle for QD surface functionalisation in biological applications.
Introduction
Quantum dots (QDs) are nanoscale luminescent semiconductor crystals with diameters in the range of 1–20 nm.1 QDs have attracted much attention because of their unique electronic and photophysical properties,2 leading to a wide range of applications, such as in solar cells,3 sensing,4 therapy,5 and imaging.6 In particular, QDs are superior to common organic and biological fluorophores (e.g., fluorescein, green fluorescent protein) in terms of their broad absorption spectrum and excellent photostability.7,8 For the majority of QD-based biological applications, the materials must be coupled to functional biological molecules (e.g., antibodies, proteins).9,10 Conventional QD functionalisation with biological molecules is occurred via covalent and non-covalent interactions, and each method has some drawbacks.11 For example, covalent crosslinks, such as 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) coupling, require additional chemical reagents, as well as several preparation steps, and may decrease photoluminescence yield.12,13 Non-covalent bonds, such as electrostatic force, also can cause non-specific and unstable conjugates.14,15Peptides are increasingly being used as recognition probes for both organic and inorganic molecules owing to several reasons, including their small molecular size and good stability.16,17 Because peptides can be chemically synthesised in large batch scales, many functional peptides have been discovered from rational and systematic studies for a wide range of applications (e.g., targeting, sensing, imaging).18–20 Importantly for the study of nanoparticle functionalisation, solid-binding peptides can bind or recognise material surfaces and offer simple and bio-friendly alternatives to conventional surface modification techniques.21,22 So far, a few peptides have been reported to possess the capacity to bind to QDs based on histidine (His or H)-metal affinity.23,24 These His-rich peptides have the potential to bind to QD surfaces; however, they are limited to ZnS-shelled QDs.23,24 Since CdTe/CdS core/shell QDs with high quantum yield can be aqueously synthesised, this QD type is one of the widely used QDs for biological fluorescence probes.25 Despite its potent material, no peptide binders to CdTe/CdS QDs have been reported up to date.
Herein, we explored CdTe/CdS QD-binding peptides from a phage display peptide library. Subsequently, re-confirmation of the binding between screened peptides and QDs, evaluation of truncated peptides, and determination of the dissociation constant (KD) were investigated using the peptide array technique, a method for customising peptide synthesis on a cellulose membrane. To shed light on a potential application of the QD-binding peptides, they were connected to an antibody-binding peptide to form dual-functional peptides. These dual-functional peptides were functionally evaluated by surface plasmon resonance (SPR) and were applied to direct QD-antibody bioconjugation for immunofluorescence imaging without additional chemicals. These CdTe/CdS QD binding peptides provide a way for simple, effective, and bio-friendly functionalisation of QD surfaces for bio-applications.
Results and discussion
Screening of CdTe/CdS QD-binding peptides
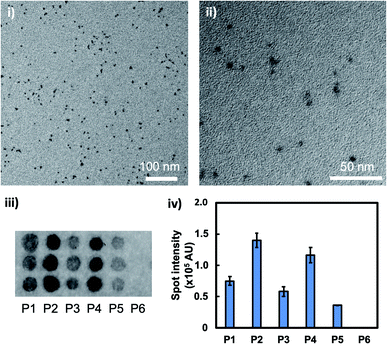
Aqueously synthesised, thioglycolic acid (TGA)-capped CdTe/CdS core/shell QDs had uniform shape, diameters of around 5 nm (mean ± standard deviation = 4.9 ± 2.5 nm, n = 118), and a bright fluorescence emission spectrum peak at 600 nm (Fig. 1i and ii and S1†). In the phage display screening, five peptide sequences (P1–P5) were identified from seven plaques as CdTe/CdS QD-binding peptides (Table 1). P1–P5 exhibited hydrophilic properties (grand average of hydropathy (GRAVY) < 0), correlating with the hydrophilic layer of TGA. Additionally, P1–P5 all carried a net negative charge under neutral conditions (isoelectric point (pI) < 7). In combination with the fact that TGA also carries a net negative charge under neutral conditions, this suggested that the peptide–QD interaction is complex, and the peptide may have an affinity not only with the TGA layer, but also the CdS surface. Interestingly, P1–P5 had no, or a very low, proportion of His residue, implying a different binding mechanism to that of previously reported His-rich QD-binding peptides.23,24 To confirm the peptide binding activity to the QDs, the binding assay was carried out using a peptide array. Fig. 1iii displays the results of the binding assay, which confirmed the binding activity; each peptide spot from P1–P5 displayed strong QD fluorescence, while no QD signal was found for the negative control, peptide P6 (AAAA). Since P2 (TLTSETPWSLNR) and P4 (SGVYKVAYDWQH) gave higher spot intensities than the others (Fig. 1iv), these sequences were used for the following experiments.| Peptide | Sequence | pI | GRAVY | Note |
|---|---|---|---|---|
| a pI and GRAVY were calculated from ProtParam tool (http://web.expasy.org/protparam/).26 | ||||
| P1 | GDGNSVLKPGNW | 5.84 | −0.908 | Screened with 3/7 frequency |
| P2 | TLTSETPWSLNR | 5.66 | −0.842 | Screened with 1/7 frequency |
| P3 | YPSSLNINASSF | 5.52 | −0.017 | Screened with 1/7 frequency |
| P4 | SGVYKVAYDWQH | 6.46 | −0.717 | Screened with 1/7 frequency |
| P5 | SHALQGPEGTAT | 5.22 | −0.617 | Screened with 1/7 frequency |
| P6 | AAAA | 3.69 | 1.800 | Negative control |
| P7 | PWSLNR | 10.18 | −1.250 | Truncated from P2 |
| P8 | SGVYK | 8.31 | −0.440 | Truncated from P4 |
| P9 | NKFRGKYKGGGPWSLNR | 11.17 | −1.606 | Dual-functionalised from P7 |
| P10 | NKFRGKYKGGGSGVYK | 10.30 | −1.375 | Dual-functionalised from P8 |
Characterisation of CdTe/CdS QD-binding peptides
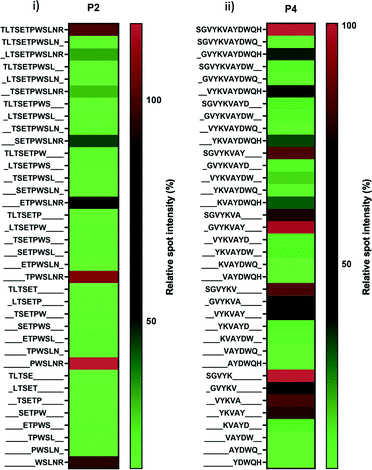
Two peptide candidates (P2 and P4) were subjected to the peptide array-based truncation assay to identify the key motif and amino acid for their binding. As shown in Fig. 2 and Table 1, P7 (PWSLNR) and P8 (SGVYK) were the shortest fragments; they were derived from P2 and P4, respectively, and played a key role in QD binding. Several truncated peptides derived from P2 (TLTSETPWSLNR) with a short series of amino acids lacking from the N-terminus (including TPWSLNR, PWSLNR, and WSLNR) revealed relatively high binding activity, while those lacking amino acids from the C-terminus showed only weak binding activity (Fig. 2i). This result indicates that arginine residue at the C-terminus is crucial to the binding process for the P2-derived peptides.On the other hand, peptides derived from P4 (SGVYKVAYDWQH) exhibited a more complex binding profile (Fig. 2ii). All truncated peptides showed gradual decreases in binding affinity to QDs except P8 (SGVYK). The binding profile appears to indicate that the N-terminus region is more important for binding than the C-terminus. To clarify the key motifs and characterise the binding behaviour of the sequence, we focused on the comparison of eight peptides comprising five amino acids (SGVYK, GVYKV, VYKVA, YKVAY, KVAYD, VAYDW, AYDWQ, and YDWQH). Four of these peptides (SGVYK, GVYKV, VYKVA, and YKVAY), which consist of sequences located towards the N-terminus of P4, exhibited high-binding activity. These data suggest that the combined sequence of these four peptides (i.e. SGVYKVAY) is the main contributor to QD binding, and the partial sequence SGVYK (P8), is the core region. The two peptides P7 and P8 have a weakly positive charge. Despite there being more charged peptides in the evaluated sequences, these two peptides were identified as the core region for QD binding. This observation suggests that the peptides' interaction with the QDs is not simple; electrostatic and other interactions (for instance hydrogen bonding and hydrophobic interactions) are likely to contribute; further research is required to understand this.
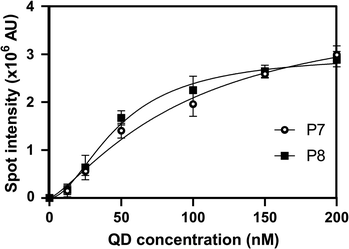
The KD of P7 and P8, as determined by Hill equation fitting, was around 96.0 and 48.5 nM, respectively (Fig. 3). These results indicate that the truncated peptides have high affinity to CdTe/CdS QDs and will be useful for further QD surface functionalisation.
Dual-functional peptides for QD-antibody bioconjugation
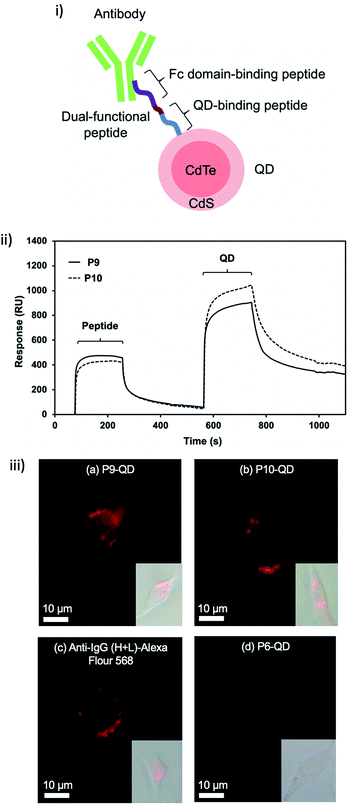
To demonstrate the potential of these peptides for functionalisation of QDs, the conjugation of another functional peptide for antibody binding was evaluated. To create the dual-functional peptides (P9 and P10), P7 and P8 were linked with an antibody fragment crystallisable (Fc) domain-binding peptide (NKFRGKYK, and KD of 8.9 μM)27,28 using GGG as a linker (Fig. 4i and Table 1). By quantifying a bound amount from an unbound excess in an ultra-centrifuged supernatant, the molar binding ratio of QDs, peptides and antibodies synthesised in this study was estimated to be 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 44
44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4. There was therefore a high ratio of short-chain peptides that mediated the linkage between QD and antibody. To further confirm the dual-functionality of P9 and P10, an SPR assay was conducted through real-time monitoring of the SPR response, which indicates the mass of analytes (peptide and QD) bound to a ligand (antibody) on a sensor chip. An injection of peptide (at time t = 70 s) and then QDs (t = 560 s) over the antibody-immobilised sensor chip (Fig. S2†) led to increases in SPR response (Fig. 4ii). Small peptides caused a slight increase in the SPR response, while the relatively large QDs caused a greater increase, corresponding to the difference in their sizes (Fig. 4ii). It illustrates that P9 and P10 have the dual-functionality of binding to QDs as well as antibodies. The QD quenching was also investigated by comparing the fluorescence emission spectrum. As the result of antibody functionalisation, a slight decrease (about 20%) in fluorescence emission peak at 600 nm was observed (Fig. S3†). This result showed that the antibody was immobilised on the QD surface, resulting in the change of surface state and the fluorescence reduction.
4. There was therefore a high ratio of short-chain peptides that mediated the linkage between QD and antibody. To further confirm the dual-functionality of P9 and P10, an SPR assay was conducted through real-time monitoring of the SPR response, which indicates the mass of analytes (peptide and QD) bound to a ligand (antibody) on a sensor chip. An injection of peptide (at time t = 70 s) and then QDs (t = 560 s) over the antibody-immobilised sensor chip (Fig. S2†) led to increases in SPR response (Fig. 4ii). Small peptides caused a slight increase in the SPR response, while the relatively large QDs caused a greater increase, corresponding to the difference in their sizes (Fig. 4ii). It illustrates that P9 and P10 have the dual-functionality of binding to QDs as well as antibodies. The QD quenching was also investigated by comparing the fluorescence emission spectrum. As the result of antibody functionalisation, a slight decrease (about 20%) in fluorescence emission peak at 600 nm was observed (Fig. S3†). This result showed that the antibody was immobilised on the QD surface, resulting in the change of surface state and the fluorescence reduction.
To verify the potential of the dual-functional peptide functionalised QDs, these nanomaterials were applied in the imaging of CD9, a membrane protein linked to the stage and metastasis of cancer cells,29 on fixed MDA-MB-231 cancer cells. P9 and P10-mediated QD-antibody conjugates provided similar CD9 immuno-images to commercial secondary antibodies conjugated with Alexa Fluor 568, whereas poly-alanine P6, the negative control, gave only a weak fluorescent image (Fig. 4iii). Although the quenching was weakly observed by the antibody immobilisation (Fig. S3†), the fluorescence characteristic was still suitable for the imaging application (Fig. 4iii). This finding shows that P9 and P10 can be used for simple conjugation of CdTe/CdS QDs to CD9 antibodies for use in bioimaging. Taken together, the QD-binding peptides can be integrated with other functional molecules to modify or functionalise QDs.
Conclusions
Discovery of high-affinity peptide binders to CdTe/CdS QDs was accomplished using a phage display peptide library and a peptide array, providing a new tool for QD surface engineering. Dual-functional peptides binding to CdTe/CdS QDs and antibodies' Fc-regions were designed and applied to achieve a reagent-free, one-step QD-antibody conjugation. This alternative and simple strategy for construction of antibody-functionalised QDs by using a dual-functionality peptide would be a powerful tool for nanomaterial surface functionalisation for various biotechnological applications such as biosensors, bioimaging and therapy.Experimental
QD preparation
TGA-capped CdTe/CdS core/shell QDs were prepared with a similar method to an earlier work.25 The CdTe core was grown by refluxing a solution of Cd(ClO4)2 and TGA (molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.3), which was introduced with H2Te gas and deoxygenated by N2 gas, at 100 °C for 2 h. To grow the CdS core, thiourea (CH4N2S) was added to the core QD dispersion before the mixture was further refluxed overnight. QDs were cleaned by adding two parts of isopropanol to one part aqueous QD solution and agitating. The dispersion was then centrifuged for 5 min at 3000 g to produce a pellet. The supernatant was decanted and the QDs were then dispersed in ultrapure water (Direct-Q 3 UV-R; Millipore Co., USA) by sonication.
1.3), which was introduced with H2Te gas and deoxygenated by N2 gas, at 100 °C for 2 h. To grow the CdS core, thiourea (CH4N2S) was added to the core QD dispersion before the mixture was further refluxed overnight. QDs were cleaned by adding two parts of isopropanol to one part aqueous QD solution and agitating. The dispersion was then centrifuged for 5 min at 3000 g to produce a pellet. The supernatant was decanted and the QDs were then dispersed in ultrapure water (Direct-Q 3 UV-R; Millipore Co., USA) by sonication.
Phage display screening
The Ph.D.™-12 Phage Display Peptide Library Kit (New England Biolabs, USA) was used according to its user guide. The peptide library solution was diluted with TBST (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.1% v/v Tween-20) and incubated with CdTe/CdS QDs for 1 h. NaCl solution (2 M) was added to the mixture before centrifuging (4 °C, 4000 g, 2 min) to remove the unbound phages. The QD-bound phages were washed twice with TBST, eluted by glycine–HCl (pH 2.2, 0.2 M, 20 min), and neutralised with Tris–HCl (pH 9.1). Then, the eluted phages were amplified and counted for the next round of biopanning. The biopanning was repeated three times, followed by DNA sequence analysis (7 clones).Preparation of peptide array
The peptide array was prepared using Fmoc chemistry and a peptide auto-spotter (MultiPep RSi; Intavis AG, Germany) as described in previous work.30 In each elongation, the Fmoc group at the N-terminus was first deprotected by 20% piperidine in N,N-dimethylformamide (DMF). Next, Fmoc amino acid at 0.5 M (Watanabe chemical, Japan) was activated by 1.1 M hydroxybenzotriazole and 1.1 M N,N′-diisopropylcarbodiimide and spotted on a membrane (grade 542; Whatman, UK). Prior to use, side chain-protecting groups were deprotected using a mixture of ultrapure water, triisopropylsilane and trifluoroacetic acid (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 95), followed by washing with dichloromethane, DMF, ethanol and phosphate-buffered saline (PBS, pH 7.4), respectively.
95), followed by washing with dichloromethane, DMF, ethanol and phosphate-buffered saline (PBS, pH 7.4), respectively.
Binding assay using peptide array
The peptide array was submerged into CdTe/CdS QD solution in ultrapure water for 2 h at room temperature with slow shaking ahead of washing with ultrapure water. The peptide array was scanned by a fluorescence imager (Typhoon FLA 9500; GE healthcare, Sweden) and spot intensity was quantified by Image Quant software (GE healthcare).Evaluation of binding ratio
Peptide powder (Sigma-Aldrich Inc., USA), diluted with TBS to 155 μM, was incubated with CdTe/CdS QDs (0.15 μM, 25 °C, 1 h) and ultra-centrifuged (25 °C, 102![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 5 h) to collect peptide-conjugated QDs. FITC-conjugated anti-IgG (H+L) antibodies (B12113C; Funakoshi, Japan) at 0.5 μM were incubated with the peptide-QD conjugate (25 °C, 1 h) and ultra-centrifuged (25 °C, 435
000g, 5 h) to collect peptide-conjugated QDs. FITC-conjugated anti-IgG (H+L) antibodies (B12113C; Funakoshi, Japan) at 0.5 μM were incubated with the peptide-QD conjugate (25 °C, 1 h) and ultra-centrifuged (25 °C, 435![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 680g, 1 h) to collect antibody-functionalised QDs. To estimate the binding ratio, the excess peptides and antibodies in ultra-centrifuged supernatants were quantified by high performance liquid chromatography (TSKgel ODS-100Z column, 214 nm absorbance detection) and a fluorescence microplate reader, respectively.
680g, 1 h) to collect antibody-functionalised QDs. To estimate the binding ratio, the excess peptides and antibodies in ultra-centrifuged supernatants were quantified by high performance liquid chromatography (TSKgel ODS-100Z column, 214 nm absorbance detection) and a fluorescence microplate reader, respectively.
SPR
The CD9 antibody solution (ab92726; Abcam, UK), diluted to 4 μM with acetate buffer (10 mM sodium acetate, pH 5.5), was immobilised as a ligand onto a CM5 sensor chip using the amine coupling kit in the SPR system (Biacore X100 Plus Package; GE healthcare). Peptide (44 μM) and CdTe/CdS QDs (1 μM) as analytes were subsequently flowed over the immobilised surface. The running buffer was HBS-EP (10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% v/v surfactant P20) and the regenerating solution was 10 mM glycine–HCl at pH 2.0.CD9 imaging
MDA-MB-231 human breast cancer cells (ATCC® HTB-26TM; American type culture collection, USA) were cultured in a 96-well plate using Dulbecco's modified Eagle's medium with 10% fetal bovine serum (Life technologies, USA) at 37 °C under 5% CO2 conditions in a humidified atmosphere. After a 24 h culture, cells were fixed with 4% formaldehyde and blocked with 2% bovine serum albumin. CD9 antibody (50 nM in PBS) was added to the well for 1 h at room temperature. Peptide (550 nM in PBS) was added to the well for 45 min at room temperature. CdTe/CdS QDs (12.5 nM in PBS) was added to the well for 15 min at room temperature. For a positive control result, Alexa Fluor® 568 Goat Anti-Rabbit (IgG) secondary antibody (ab175471; Abcam) at 50 nM was added to the well for 1 h at room temperature instead of peptide and QDs. After each addition, the solutions were removed, and the cells were washed in triplicate with PBS. The cells were observed under fluorescence microscope (DMi8; Leica Camera AG, Germany).Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (18H01795, 18K18970, and 18K04848). This work was partially supported by the Cross-ministerial Strategic Innovation Promotion Program (SIP) from the Cabinet Office, Government of Japan. M. T. thanks for funds from the Iketani Science and Technology Foundation and the Royal Society UK under the Newton International Fellowships scheme. This work was also supported in part by the international collaboration research projects sponsored by the JSPS and the Royal Society (IEC/R3/170038). In addition, the authors thank Suzukakedai Materials Analysis Division, Technical Department, Tokyo Institute of Technology, for mass spectrometry and DNA sequencing analyses. The authors would like to thank Prof. Tsuyoshi Tanaka at Tokyo University of Agriculture and Technology for supporting ultracentrifugation.References
- O. E. Semonin, J. M. Luther and M. C. Beard, Mater. Today, 2012, 15, 508–515 CrossRef CAS.
- L. Tong, F. Qiu, T. Zeng, J. Long, J. Yang, R. Wang, J. Zhang, C. Wang and T. Sun, RSC Adv., 2017, 7, 47999–48018 RSC.
- M. Eck, C. V. Pham, S. Züfle, M. Neukom, M. Sessler, D. Scheunemann, E. Erdem, S. Weber, H. Borchert, B. Ruhstaller and M. Krüger, Phys. Chem. Chem. Phys., 2014, 16, 12251–12260 RSC.
- M. Chern, J. C. Kays, S. Bhuckory and A. M. Dennis, Methods Appl. Fluoresc., 2019, 7, 012005 CrossRef CAS PubMed.
- S. S. Wu, J. Z. Zhang, X. H. Yu, Y. Cao and H. J. Wang, RSC Adv., 2014, 4, 63502–63507 RSC.
- Y. Fu, M. S. Jang, T. Wu, J. H. Lee, Y. Li, D. S. Lee and H. Y. Yang, Carbohydr. Polym., 2019, 224, 115174 CrossRef CAS PubMed.
- M. Masteri-Farahani, K. Khademabbasi and N. Mollatayefeh, Mater. Lett., 2018, 228, 68–71 CrossRef CAS.
- G. Azzellino, F. S. Freyria, M. Nasilowski, M. G. Bawendi and V. Bulović, Adv. Mater. Technol., 2019, 4, 1800727 CrossRef CAS.
- L. Liu, L. Mirandola, M. Chiriva-Internati and J. Chaudhuri, Mater. Lett., 2017, 199, 5–8 CrossRef CAS.
- T. Umakoshi, H. Udaka, T. Uchihashi, T. Ando, M. Suzuki and T. Fukuda, Colloids Surf., B, 2018, 167, 267–274 CrossRef CAS PubMed.
- A. Banerjee, T. Pons, N. Lequeux and B. Dubertret, Interface Focus, 2016, 6, 20160064 CrossRef PubMed.
- K. E. Sapsford, W. R. Algar, L. Berti, K. B. Gemmill, B. J. Casey, E. Oh, M. H. Stewart and I. L. Medintz, Chem. Rev., 2013, 113, 1904–2074 CrossRef CAS PubMed.
- O. Koniev and A. Wagner, Chem. Soc. Rev., 2015, 44, 5495–5551 RSC.
- A. Foubert, N. V. Beloglazova, A. Rajkovic, B. Sas, A. Madder, I. Y. Goryacheva and S. D. Saeger, Trac. Trends Anal. Chem., 2016, 83, 31–48 CrossRef CAS.
- A. S. Karakoti, R. Shukla, R. Shanker and S. Singh, Adv. Colloid Interface Sci., 2015, 215, 28–45 CrossRef CAS PubMed.
- L. Rong, S. Y. Qin, C. Zhang, Y. J. Cheng, J. Feng, S. B. Wang and X. Z. Zhang, Mater. Today Chem., 2018, 9, 91–102 CrossRef CAS.
- A. Lampel, R. V. Ulijn and T. Tuttle, Chem. Soc. Rev., 2018, 47, 3737–3758 RSC.
- M. Tanaka, A. W. L. Alvin and M. Okochi, RSC Adv., 2018, 11, 5953–5959 RSC.
- W. Wang and Z. Hu, Adv. Mater., 2019, 31, 1804827 CrossRef CAS PubMed.
- M. Tanaka, T. Minamide, Y. Takahashi, Y. Hanai, T. Yanagida and M. Okochi, Chem. Lett., 2019, 48, 978–981 CrossRef CAS.
- M. Tanaka, I. H. Harlisa, Y. Takahashi, N. A. Ikhsana and M. Okochi, RSC Adv., 2018, 8, 8795–8799 RSC.
- A. Care, P. L. Bergquist and A. Sunna, Trends Biotechnol., 2015, 33, 259–268 CrossRef CAS PubMed.
- L. Qiu, C. Zhang, T. Gu, Z. Zhu, J. Wang, L. Liu, S. Ding, X. Liu, J. Wang and P. Jiang, Chromatographia, 2018, 81, 41–46 CrossRef CAS.
- J. B. Delehanty, I. L. Medintz, T. Pons, F. M. Brunel, P. E. Dawson and H. Mattoussi, Bioconjugate Chem., 2006, 17, 920–927 CrossRef CAS PubMed.
- K. Virzbickas, L. Rimkute, A. J. Harvie and K. Critchley, J. Exp. Nanosci., 2017, 12, 94–103 CrossRef CAS.
- E. Gasteiger, C. Hoogland, A. Gattiker, S. Duvaud, M. R. Wilkins, R. D. Appel and A. Bairoch, The Proteomics Protocols Handbook ed. J. M. Walker, 2005, pp. 571–607 Search PubMed.
- T. Sugita, M. Katayama, M. Okochi, R. Kato, T. Ichihara and H. Honda, Biochem. Eng. J., 2013, 79, 33–40 CrossRef CAS.
- M. Okochi, T. Kamiya, T. Omasa, M. Tanaka and H. Honda, Anal. Sci., 2016, 32, 93–97 CrossRef CAS PubMed.
- G. Rappa, T. M. Green, J. Karbanová, D. Corbeil and A. Lorico, Oncotarget, 2015, 6, 7970–7991 CrossRef PubMed.
- M. Tanaka, T. Suwatthanarak, A. Arakaki, B. R. G. Johnson, S. D. Evans, M. Okochi, S. S. Staniland and T. Matsunaga, Biotechnol. J., 2018, 13, e1800087 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra00460j |
| ‡ The first two authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |