Open Access Article
Open Access ArticleThree pairs of luminescent coordination polymers based on CoII and CdII clusters for the detection of antibiotics, pesticides and chiral nitro aromatic compounds†
Li-Qing Zhanga,
Xiu-Wen Wanga,
Lei Gua,
Ying-Hui Yu *a and
Jin-Sheng Gao*ab
*a and
Jin-Sheng Gao*ab
aSchool of Chemistry and Materials Science, Heilongjiang University, Harbin 150080, China. E-mail: yuyinghui@hlju.edu.cn; gaojinsheng@hlju.edu.cn; Fax: +86-451-86609151; Tel: +86-451-86609001
bAgricultural College, Heilongjiang University, Harbin 150080, China
First published on 4th March 2020
Abstract
Three couples of coordination polymers (CPs), namely, [Co((R/S)-Hcna)2]n (1-D/L), [Cd6((1R,2R/1S,2S)-cpba)4(phen)6(H2O)3]n (2-D/L) and [Cd2((1R,2R/1S,2S)-Hcpba)2(phen)2]n (3-D/L) {(R/S)-H2cna = (R/S)-6-(1-carboxyethoxy)-2-naphthoic acid, (1R,2R/1S,2S)-H3cpba = (1R,2R/1S,2S)-2,2′-((5-carboxy-1,3-phenylene)bis(oxy))dipropionic acid, phen = 1,10-phenanthroline} are successfully synthesized under hydrothermal conditions. Structural analysis shows that CP 1 has a 3D 3,6-c net structure with a point symbol of (4·62)2(42·610·83). CPs 2 and 3 are obtained under very similar reaction conditions except using different solvent ratios. The presence of the planar chelating ligand phen in CPs 2 and 3 limited the spatial growth of the structure, resulting in the formation of different 1D structures. All CPs crystallized in the chiral space group P21, CPs 1–3 are all SHG active. Their luminescence sensing activities for organics such as antibiotics, pesticides and nitro aromatics are also investigated. The results showed that CP 1 can effectively identify trace amounts of nitrofurans (NFs) and CP 3 has obvious recognition ability toward nitrofurans (NFs) and nitroimidazoles (NMs). Both CPs 1 and 3 could selectively detect 2,6-dichloro-4-nitroaniline (DCN). The luminescence of CPs 1 and 3 can also be quenched by (D/L)-4-nitrophenylalanine ((D/L)-NPA) and (1R,2R/1S,2S)-2-amino-1-(4-nitrophenyl)propane-1,3-diol ((1R,2R/1S,2S)-ANPO).
Introduction
Since the advent of penicillin, there have been thousands of antibiotics, which are widely used in medicine and animal husbandry. The large-scale production and use of antibiotics as well as the discharge of related wastewater have made antibiotic pollution a public concern.1,2 At the same time, pesticides with high toxicity and non-degradability also posed great threat to human health and environment.3 At present, antibiotics and pesticides are detected by liquid chromatography-tandem mass spectrometry,4 chromatography-diode array detection,5 gas chromatography-nitrogen phosphorus detection,6 ion mobility spectrometry7 and so on. However, these methods are time-consuming, expensive, and require complicated equipment and technical personnel. As a basic feature of nature, chirality is closely related to life activities. In the clinic, 62% of the drugs are chiral, with different enantiomeric toxicity and medicinal properties. Chiral recognition has been of great importance, however, a continuing challenge.8–10 Exploring fast, simple and effective ways for the detection of these compounds are fully meaningful.The luminescence sensors have attracted the attention of scientists due to their characteristics of simple, fast and high sensitivity.11,12 Among many luminescence materials, coordination polymers (CPs) represent an attractive way for the efficient detection of such chemicals because of their rationally designed structures and versatile properties. Actually, many luminescent metal coordination polymers have found various applications in molecular recognition,13–15 asymmetric catalysis,16,17 nonlinear optics,18,19 fluorescent sensors,20–22 photoelectric performance,23–26 and so forth. According to Zhang et al., the amino-modified Mg-LMOF exhibits excellent luminescence sensing performance in the specific detection of Fe3+ ions, antibiotics NFs and pesticides DCN.27 In 2019, Yu reported the detection of nitrofurans and quinolone antibiotics with RhB@Tb-dcpcpt, which were achieved through luminescence quenching and luminescent discoloration processes, respectively.28 Zhu research group found that salan-based chiral polymers could identify enantiomers such as chiral phenylglycinol and α-hydroxyl carboxylic acid.29–32 Lin et al. reported that a BINOL-based MOF with chiral tetracarboxylic acid bridging ligand and cadmium ion were highly selective and sensitive to chiral amino alcohols.33
The design and preparation of CPs with desired specific properties have always been an ambitious goal for chemists. As one ongoing research work, the luminescence CPs with sensing activities toward versatile molecules have been continuously studied in our group.34,35 In view of various coordination modes, strong coordination ability of carboxylic groups and the large π-conjugated system of aromatic ligands, aromatic polycarboxylic acid ligands are considered to be favorable candidates for the construction of luminescent complexes.36 Herein, we adopted (R/S)-6-(1-carboxyethoxy)-2-naphthoic acid (R/S-H2cna) and (1R,2R/1S,2S)-2,2′-((5-carboxy-1,3-phenylene)bis(oxy))dipropionic acid (1R,2R/1S,2S-H3cpba) as building blocks (Scheme 1), obtaining three pairs of CPs, namely, [Co((R/S)-Hcna)2]n (1-D/L), [Cd6((1R,2R/1S,2S)-cpba)4(phen)6(H2O)3]n (2-D/L) and [Cd2((1R,2R/1S,2S)-Hcpba)2(phen)2]n (3-D/L). Where CPs 2 and 3 are obtained under different solvent ratio. Their structures are characterized by single-crystal X-ray diffraction analysis, elemental analysis, IR analysis and thermal gravimetric analysis. Their luminescence recognition properties toward antibiotics, pesticides and chiral nitro aromatic compounds were studied.
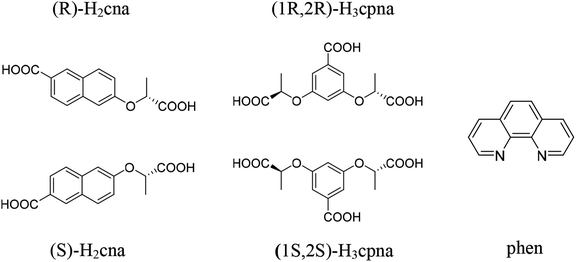 | ||
| Scheme 1 Schematic representation of the ligands (R/S)-H2cna, (1R,2R/1S,2S)-H3cpna and auxiliary ligand phen. | ||
Experimental section
Materials and physical measurements
(R/S)-H2cna and (1R,2R/1S,2S)-H3cpba ligands were prepared based upon previously reported works.37,38 All the chemical reagents and solvents during the synthesis processes were commercially available without further purification. The IR spectra (KBr pellets) were recorded on a PerkinElmer Spectrum 100 FT-IR spectrometer equipped with a DGTS detector (32 scans) from 4000 to 500 cm−1. Elemental analyses (C, H, N) were performed on a PerkinElmer 2400 elemental analyzer. Thermal gravimetric analysis (TGA) experiments were carried out on a PerkinElmer STA 6000 under air atmosphere at a heating rate of 20 °C min−1 scanning from 50 to 800 °C. Powder X-ray diffraction (PXRD) patterns were obtained on a Bruker D8 Advance X-ray powder diffractometer equipped with Cu Kα radiation (λ = 1.54178 Å). The solid CD spectra of CPs were measured at room temperature with a Olis DM245 spectrofluorimeter. The UV-vis spectra for powder samples were recorded on a PerkinElmer Lambda 950 spectrometer in the range of 200–800 nm and the solution UV-vis absorption spectra were collected using a Shimadzu UV-2700 spectrophotometer. The second-order nonlinear optical intensities were approximated by measuring the crystallite sample relative to urea at a wavelength of 1064 nm and a pulse width of 10 ns using a Nd: YAG pulsed laser of the American Spectral Physics Corporation. These luminescence spectra were taken the pass width is 10 nm by using the PerkinElmer LS 55 luminescence spectrophotometer. Solid-state luminescent spectra and luminescence lifetimes were measured with an Edinburgh FLS 1000 luminescence spectrophotometer. Electrochemical testing was performed on an electrochemical analyzer potentiostat model 600E from CH instruments.Synthesis
X-ray crystallography
Picking the right crystal of CPs 1–3 installed onto thin glass fibers. The X-ray diffraction data of CPs 1–3 were collected on a Bruker APEX CCD diffractometer with graphite-monochromated Mo Kα radiation (λ = 0.71073 Å). Absorption corrections were applied by using the multi-scan program SADABS. These structures were solved by direct methods and refined by a full-matrix least-squares method against F2. Refinement makes all non-hydrogen atoms on organic matter anisotropic and adds hydrogen atoms at their geometric positions. CPs 1–3 of crystallographic data and structure refinement parameters are summarized in Table S1† (ESI†). Selected bond lengths and angles of CPs 1–3 are listed in Table S2.† Topology analysis was run by TOPOS software. CCDC numbers 1867800 (1-D) and 1867801 (1-L), 1965710 (2-D) and 1965708 (2-L), 1965712 (3-D) and 1965711 (3-L).Results and discussion
Structural description
The crystallographic analysis indicated that 1-D and 1-L were a couple of enantiomers and crystallized in the same chiral space group P21 with the Flack parameters of 0.003(10) and 0.019(10), so the structure of 1-L is taken as an example for detailed description. The asymmetric unit contains one crystallographic independent Co2+ ion and two deprotonated (S)-Hcna− ions. As shown in Fig. 1(a), the Co2+ center in 1-L is coordinated by six carboxylic oxygen atoms (O1, O5II, O4I, O6, O10III, O9IV) from six different (S)-Hcna− ligands, resulting the formation of a six-coordinate metal center with a distorted octahedron geometry. In 1-L, each (S)-Hcna− ligand and Co2+ ion bridge to form a 1D chain as secondary building unit (SBU). Six SBUs are intertwined with each other by sharing Co2+ ions to generate a 3D framework eventually (Fig. 1(b)). Topological analysis was carried out to better comprehend the framework of 1-L. If the (S)-Hcna− ligands are regarded as a 3-connected node and the Co2+ are viewed as a 6-connected node, the 3D structure of 1-L pertains to a 3,6-c net with a point symbol of (4·62)2(42·610·83) (Fig. 1(c)). The Co–O bond lengths are between 2.0703(10) Å and 2.1309(11) Å, and the O–Co–O bond angles are from 86.22(4)° to 177.09(4)°, which are all within the reasonable limits.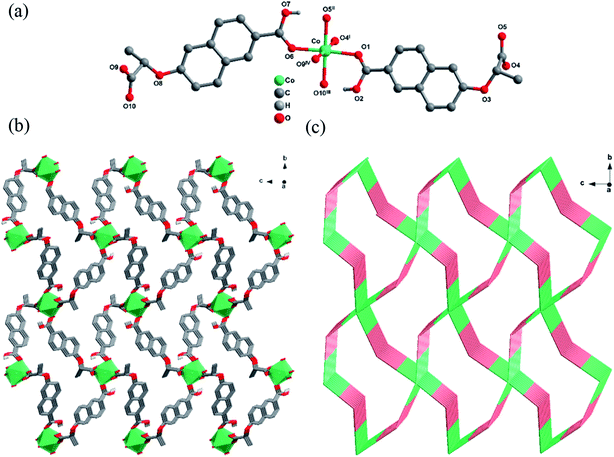 | ||
| Fig. 1 (a) Stick-ball representation of the asymmetric unit of 1-L. (b) 3D framework of 1-L. (c) Schematic view of 3,6-c net with a point symbol of (4·62)2(42·610·83). | ||
Both 2-D and 2-L are crystallized in the monoclinic with chiral space group P21, and the Flack parameters are 0.10(3) and 0.050(17), respectively. Single crystal X-ray diffraction results reveal that they are isomorphic, so only the structure of 2-L is described in detail. As shown in Fig. 2(a), each asymmetric unit contains six crystallographic independent Cd2+ ions, four deprotonated (1S,2S)-cpba3− ligands, six phen and three coordinated water molecules, in which Cd1, Cd2, Cd4 are six-coordinated, Cd3, Cd5, Cd6 are seven-coordinated. Interestingly, none of Cd atoms share same coordination environment in an asymmetric unit. Cd1 ion is the center of a six-coordinated distorted octahedron, with two N atoms from phen and four O atoms from two different (1S,2S)-cpba3− ligands. Cd2 ion is coordinated with two N atoms from phen and four O atoms from three (1S,2S)-cpba3− molecules, forming an octahedron geometric configuration. Cd3 ion is surrounded by two N atoms from phen and five O atoms from three (1S,2S)-cpba3− ligands, showing a twisted decahedron geometry as a whole. In a twisted octahedral environment, Cd4 ion is defined by two N atoms from phen and four O atoms from one coordinated water and two (1S,2S)-cpba3− ligands. Both Cd5 ion and Cd6 ion are coordinated by two N atoms from phen and five O atoms from four carboxylate anions from two (1S,2S)-cpba3− ligands and one coordination water molecule, however, the slight difference of coordination environment between Cd5 ion and Cd6 ion lies in the different carboxylate anions involved. One (1S,2S)-cpba3− ligand in 2-L presents three coordination modes as a result (Fig. S1†). The Cd–O and Cd–N bond lengths are in the range of 2.187(5)–2.551(5) Å, 2.302(6)–2.394(5) Å, respectively. The O–Cd–O, N–Cd–N, and N–Cd–O bond angles range from 53.8(2) to 145.68(16)°, 69.00(17)–73.2(2)°, 77.07(17)–174.53(17)°. Which are within a reasonable range. It is worth noting that the existence of terminal ligands such as phen and H2O limits extension of the crystal structure, and 2-L finally forms a 1D-band structure that extends infinitely along the a axis (Fig. 2(b)).
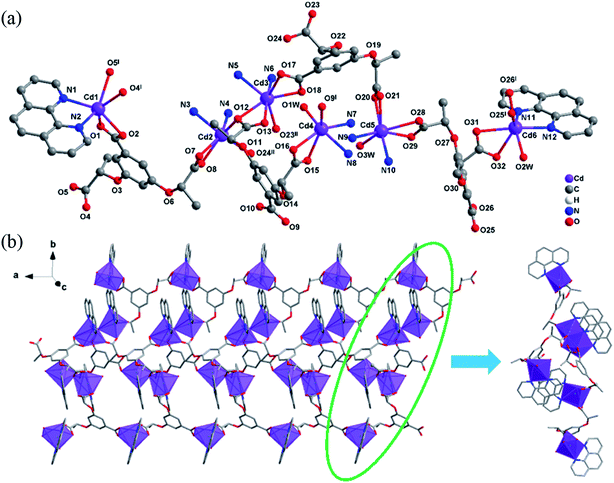 | ||
| Fig. 2 (a) One asymmetric unit of 2-L, the C atoms on some phen are omitted for clarity. (b) 1D-band structure of 2-L extends along the a axis. | ||
3-D and 3-L own the same crystal system and space group as that of 2-D and 2-L, with the Flack parameters of −0.001(19), −0.03(3). Single crystal X-ray diffraction shows that they are isomorphic, so only the structure of 3-L is described. As shown in Fig. 3(a), the asymmetric unit contains two Cd2+ ions, two (1S,2S)-Hcpba2−, two phen. Compared with the 2-L, the structure of the 3-L is relatively simple. Cd1 and Cd2 share the same coordination mode and environment. The Cd2+ ion is located in the center of a twisted octahedron defined by four O atoms from three different (1S,2S)-Hcpba2− and two N atoms from phen molecule. The Cd–O bond lengths range from 2.208(5) to 2.521(7) Å, and the bond lengths of Cd–N lie in the range 2.324(7)–2.354(8) Å. The bond angle for O–Cd–O, N–Cd–O ranges are 53.2(2)–150.6(2)° and 83.7(2)–169.4(3)°, respectively. N–Cd–N bond angles are 70.6(3) and 72.0(2)°, which all are within the normal range. The presence of the planar chelating ligand phen limited the spatial growth of structure, resulting the formation of a 1D-chain structure extending infinitely along c axis (Fig. 3(b)).
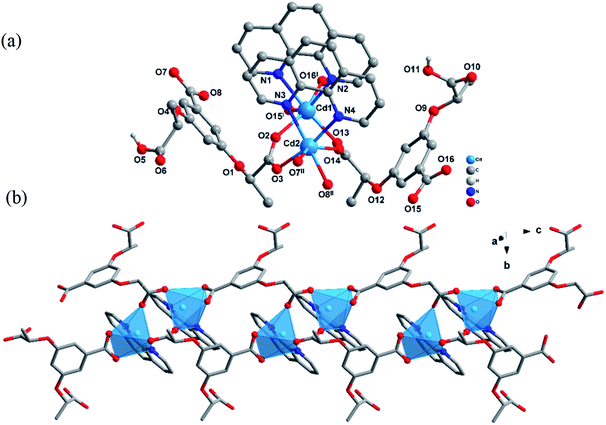 | ||
| Fig. 3 (a) Stick-ball representation of the structure unit of 3-L. (b) 1D-chain structure of 3-L along the c axis. | ||
PXRD and thermal stability analysis
To further demonstrate whether the crystal structure represents the majority of the products, the powder X-ray diffraction of CPs 1–3 was performed. The experimental PXRD patterns are matched to those simulated by single crystal data (Fig. S2†), indicating the purity of the synthesized samples. The preferred orientation of these crystal samples resulted in intensity difference in the experimental and calculated PXRD patterns.The thermal stabilities of CPs were measured in an air atmosphere in a temperature range of 50–800 °C. The thermal gravimetric curve (Fig. S4†) demonstrates that the weight loss process of CPs 1–3 are similar and divided into two stages. For 1-L, the first weight loss was in the range of 218–387 °C attributed to the loss of propionate group of the (S)-Hcna− ligand (obsd 24.72%, calcd 25.05%). The second weight loss was in the range of 388–586 °C due to the loss of 6-hydroxy-2-naphthoic acid group in (S)-Hcna− ligands (obsd 61.05%, calcd 61.00%). The remaining weight was the residue of Co3O4 (calcd 13.95%). In the case of 2-L, the first stage of weight loss was 1.78% (calcd 1.81%) in the range of 50 to 178 °C due to the clearing of three coordinated water molecules; the organic frameworks began to collapse at 382 °C, followed by six phen and four (1S,2S)-cpba3− ligands disintegration (obsd 73.14%, calcd 72.43%). After 560 °C, the remaining weight corresponds to the generation of CdO (obsd 25.08%, calcd 25.76%). The TGA curve of 3-L, gradual weight loss occurs between 300 °C and 400 °C which corresponds to the disappearance of phen (obsd 32.04%, calcd 30.60%). Further structural decomposition of 566 °C is derived from the departure of the two (1S,2S)-Hcpba2− ligands (obsd 47.36%, calcd 47.59%), resulting the formation of CdO (obsd 20.6%, calcd 21.81%).
Circular dichroism (CD) analysis
CPs 1–3 are all crystallized in the chiral space group P21, which confirms that they are chiral compounds. Solid-state circular dichroism (CD) were conducted to further define their homochirality. As shown in Fig. S5,† the CD spectrum of 1-D, 2-D, 3-D exhibits an obvious positive Cotton effect at 304 nm, 273 nm, 272 nm, separately, while 1-L, 2-L, 3-L illustrates an obvious negative Cotton effect at 305 nm, 272 nm, 275 nm, respectively. The results of the CD spectrum further demonstrate that 1, 2 and 3-D/L are three couples of enantiomers.Second-harmonic generation efficiency
With the development and promotion of laser technology, nonlinear optical (NLO) crystal materials have been extensively studied. Second harmonic generation (SHG), as the most ordinary NLO behavior, has drawn lots of attention,17,18,39 Studies have shown that molecular aggregation and chirality can enhance nonlinear optical activities. A structurally variable chiral coordination polymer is believed to be promising to produce higher SHG activity. Therefore, CPs 1–3 crystallized in a centrally symmetric chiral space group were studied for SHG activity. Their SHG activities were tested using the powder by the Kurtz–Perry method. The results show that 1-D, 1-L, 2-D, 2-L, 3-D and 3-L are both SHG active with efficiencies approximately 0.6, 0.5, 0.2, 0.3, 0.2 and 0.3 times of the urea value, respectively. Among the three pairs of CPs investigated, 1-D and 1-L demonstrates better SHG activity than others, the reason of such difference may be due to the tighter accumulation of CP 1.Luminescence properties
It is well known that the CPs based on electron-rich conjugated organic ligands and electron-deficient metal ions with 3d and 4f outer layers might act as potential functional materials with optical activity.40 The CPs 1–3 containing (R/S)-H2cna, (1R,2R/1S,2S)-H3cpba rigid conjugated backbone and Co2+, Cd2+ with d6, d10 are also expected to demonstrate certain optical activities. Therefore, we studied the solid luminescence properties of (S)-H2cna, (1S,2S)-H3cpba, and 1-L, 2-L, 3-L as the representative at room temperature (Fig. S6†). Compared with the free ligands, the luminescence intensity of 1-L, 2-L, 3-L exhibit an obvious increase, which presumably owing to the bending of ligand bonds caused by the coordination. Compared with 2-L and 3-L, the skeleton of 1-L has a larger conjugated π bond structure, which makes the delocalized π bond easier to be excited, and the rigid planar structure reduces the molecular vibration, resulting in stronger luminescence of 1-L. The luminescence recognition ability of CPs 1 and 3 are explored, considering the different luminescence activities of CPs 1–3.Luminescence sensing of antibiotics
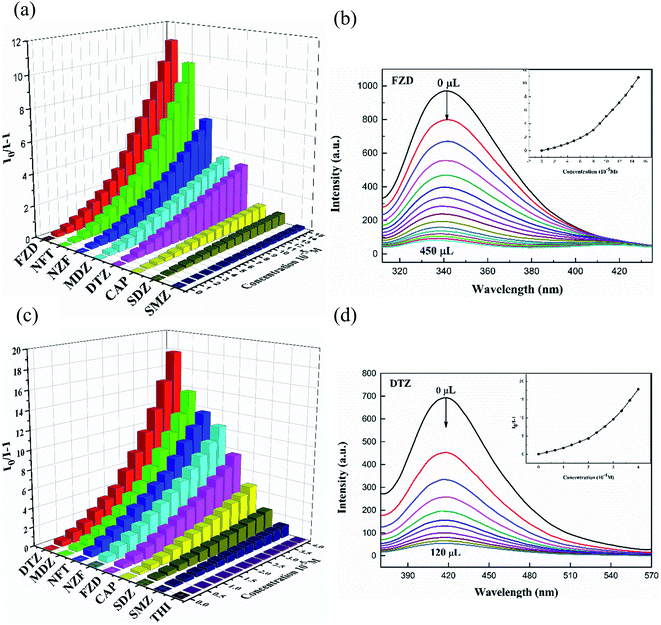
Abuse of antibiotics has been threatening global health and environmental pollution, simple and convenient way to effectively detect antibiotics is of great importance. Considering the good photoluminescence properties of CPs 1 and 3, their sensing activities toward trace antibiotics were studied. In this paper, luminescence recognition studies were performed on nine common antibiotics belonged to four types (Fig. S7†), namely, nitrofurans (NFs) (nitrofurazone, NZF; nitrofurantoin, NFT; furazolidone, FZD), nitroimidazoles (NMs) (metronidazole, MDZ; dinitrazole, DTZ), sulfonamides (sulfadiazine, SDZ; sulfamethazine, SMZ), chloramphenicols (chloramphenicol, CAP; thiamphenicol, THI). According to the experimental results, 1-D, 3-D and 1-L, 3-L all exhibited luminescence sensing ability to antibiotics, only 1-L, 3-L were described in detail. In the sensing experiments, different antibiotics with certain concentration (0.01 mol L−1, 0.10 mol L−1) were added gradually to the 1-L (0.07 mg) or 3-L (0.40 mg) solution in DMF (3 mL). The results showed that the luminescence of 1-L, 3-L were quenched differentially after adding antibiotics (Fig. 4). The order of quenching effect was FDZ > NZF > NFT > DTZ > MDZ > CAP > SMZ > SDZ for 1-L, DTZ > MDZ > NFT > NZF > FZD > CAP > SDZ > SMZ for 3-L. Obviously, 1-L displays good sensing activities toward NFs antibiotics and 3-L could recognize two types of antibiotics, namely NMs and NFs. The luminescence quenching efficiency of NZF, NFT and FZD toward 1-L are 89.7%, 91.2%, 92.1%, and those of MDZ, DTZ, NZF, NFT and FZD toward 3-L are 95.0%, 95.6%, 91.8%, 92.7%, 85.6%, respectively.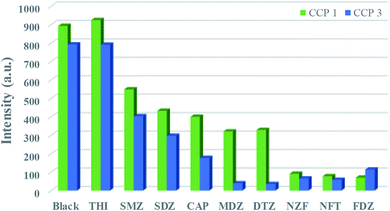 | ||
| Fig. 4 Luminescence intensities of 1-L, 3-L with the presence of 0.10 mM, 0.33 mM different antibiotics. | ||
More interestingly, the antibiotics not only affect the luminescence intensity but also the position of the emission band of the CPs (Fig. S8†). A significant red shift was observed in the luminescence spectra of the 3-L after the addition of NFs. The peak position of the emission spectrum is expected to serve as an important hint for the detection of NFs and NMs by 3-L. In order to estimate the detection sensitivity of 1-L and 3-L, further luminescence titration experiments were performed (Fig. 5(a) and (c)). The Stern–Volmer (SV) equation (I0/I = 1 + Ksv [M]) is applied to quantitatively explain the luminescence quenching efficiency, where I0 and I represent the luminescence intensity of blanks and additives, and Ksv is the quenching constant (M−1), [C] is the concentration of the additive. As shown in Fig. 5(b), (d), S9, and S10,† the luminescence intensity is linearly correlated with the concentration at low concentrations and deviates from linearity at high concentrations, which may be due to energy transfer or self-absorption effects. The KSV value is obtained by linearly fitting the SV chart, and the corresponding detection limit is calculated (LOD = 3σ/KSV, where σ is the standard deviation of repeated measurements of the three blank solutions) (Table S3†).
In addition, anti-interference is an important part of measuring the performance of the detector. As shown in Fig S11(a),† other antibiotics hardly interfere with the specific recognition of FZD by 1-L. In the presence of other antibiotics, DTZ and NFT show excellent quenching effect toward the luminescence of 3-L, and the presence of NFT causes red shift of the spectral line of 3-L. When the competing antibiotics DTZ and NFT coexist, the luminescence will be quenched more thoroughly (Fig S11(b)†). In summary, 1-L and 3-L have good anti-interference ability as the antibiotics detector. Finally, the recyclability of the CPs were also conducted to further confirm their application as antibiotics detector. After the luminescence sensing experiments, the CPs were collected and washed with fresh DMF, then centrifuged and reused again. It is found that the samples could be reused for five times and still maintains its original luminescence sensing activities, which further proved its potential application in antibiotics detection (Fig. S12†).
Identify pesticides behavior
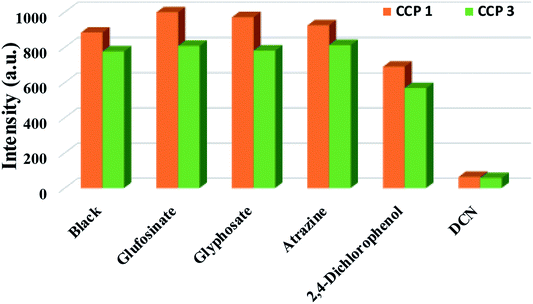
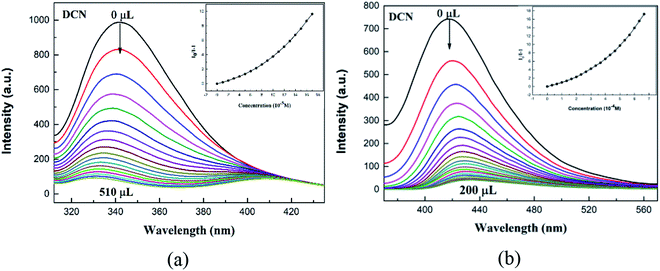
Pesticides play an important role in agricultural production, but residual pesticides threaten the balance of human health and ecosystems. In view of the application of luminescent CPs in the rapid detection of trace substances,41 we investigated the sensing activities of CPs 1, 3 in pesticides detection. In this work, five common pesticides, glufosinate, glyphosate, 2,4-dichlorophenol, atrazine, 2,6-dichloro-4-nitroaniline (DCN), were examinated (Fig. S13†). The selected pesticide (0.10 mol L−1, 10 μL) was added to a DMF (3 mL) solution containing finely ground crystals 1-L (0.07 mg) or 3-L (0.40 mg), and the luminescence signal was recorded the sample after being thoroughly mixed (Fig. 6). The data indicates that the order of the luminescent quenching efficiency of different pesticides toward 1-L and 3-L are as the following: DCN > 2,4-dichlorophenol > atrazine > glyphosate > glufosinate for 1-L, DCN > 2,4-dichlorophenol > glyphosate > glufosinate > atrazine represents 3-L. Evidently, both 1-L and 3-L demonstrate best sensing abilities toward DCN. The high luminescent quenching efficiency produced by DCN is 92.8%, 92.6% for 1-L, 3-L, calculated by (1 − I/I0) × 100%. Afterwards, we performed an antibiotic-like titration experiment for DCN (Fig. 7). At low concentrations, the Stern–Volmer equation is followed between luminescence intensity and concentration, and the curve is bent upward at a high concentration. The calculated KSV values of DCN were 2.20 × 104 M−1 for 1-L and 2.17 × 104 M−1 for 3-L. The detection limits of DCN are 212 ppb and 233 ppb, respectively (Table S3†). In view of the importance of anti-interference ability to sensors, the ability of 1-L, 3-L to identify DCN in the presence of other pesticides were further explored (Fig. S14†). The results show that the interference of other pesticides are negligible. In addition, recovery tests have shown that 1-L, 3-L still maintain the original sensing activities toward DCN after five cycles (Fig. S15†).Possible sensing mechanisms
Previous studies have shown that structural collapse, weak interactions between host and guest, electron and energy transfer are important causes of luminescence quenching.42,43 Based on this, we conducted several experiments to explore the possible luminescence quenching mechanisms of CPs 1 and 3. First, the PXRD of 1-L, 3-L after sensing experiments were fitted well to the initial patterns, demonstrating that the quenching effect was not caused by structural collapse (Fig. S16†). Given that electron-rich complex backbones can provide electrons to electron-deficient quenchers, photo-induced electron transfer (PET) may trigger luminescence quenching, revealing that lower LUMO levels in analytes are more susceptible to electrons.27,28 Therefore, the HOMO and LUMO levels of the analytes were tested electrochemically (Fig. S17†). The results demonstrated that NFs, DCN did have relatively low LUMO energy levels, while the LUMO energy levels did not exactly match their quenching efficiency order, meaning that PET is not the only mechanism for the luminescence quenching.7,44 In addition, the red shift phenomenon during the quenching process and the upward bending of the SV curve suggest the existence of a fluorescence resonance energy transfer (FRET) mechanism. The emission bands of 1-L and 3-L have obvious overlap with the UV-vis bands of NFs and DCN (Fig. S18†). Under a certain wavelength of light excitation, 1-L and 3-L could transfers energy without radiation to nearby NFs and DCN through dipole–dipole interaction, resulting in luminescence quenching. In summary, the combination of PET and FRET mechanisms led to luminescence quenching of 1-L and 3-L toward NFs and DCN.45,46 Furthermore, the excitation spectrum of 3-L effectively overlap the UV-vis absorption spectrum of NMs, the competitive absorption excitation energy between NMs and 3-L might also lead to luminescence quenching.47Luminescence recognition of chiral nitro aromatic compounds
Inspired by previous studies,29,30 we selected two pairs of chiral nitro aromatic compounds, namely, (D/L)-4-nitrophenylalanine ((D/L)-NPA) and (1R,2R/1S,2S)-2-amino-1-(4-nitrophenyl)propane-1,3-diol ((1R,2R/1S,2S)-ANPO), to investigate the discriminated recognition effect of CPs 1, 3 (Fig. S19†). The same procedure as that of pesticides/antibiotics luminescence sensing was performed except that (D/L)-NPA and (1R,2R/1S,2S)-ANPO were substituted for pesticides/antibiotics (10 mM, 10 μL addition each time). Unfortunately, the selected chiral quenchers only exhibit obvious luminescence quenching effect for CPs 1, 3, however, without any enantiomeric discrimination (Fig. S20 and S21†). We will continue this research in future work, expecting to fulfil the enantiomeric recognition for other chiral enantiomers. As 1-D/3-D demonstrate very similar recognization effect to 1-L/3-L on the selected amphoteric nitro aromatics, only the sensing mechanism of 1-L and 3-L toward (D/L)-NPA and (1R,2R/1S,2S)-ANPO are described in detail. In order to explore the above-mentioned luminescence quenching mechanism, we compared the PXRD patterns of the CPs samples before and after sensing experiments, no obvious changes are found, indicating that the crystal structure 1-L and 3-L did not change during the sensing process (Fig. S22(a) and S23(a)†). Considering the presence of hydrogen bond donors and acceptors, it was speculated that there may be weak hydrogen bond interactions between CPs and analytes. To verify this possibility, we examined the luminescence lifetime before and after sensing experiments to find that no change occurs for 1-L, however, a significant change for 3-L (Fig. S22(b)†). We speculate that the 1-L and selected quenchers are combined by hydrogen bonding, resulting in static quenching of the luminescence.33 According to UV-vis spectra of two chiral nitro aromatic compounds (Fig. S23(b)†), it shows that the excitation spectra of 3-L and the UV-vis absorption spectra of (D/L)-NPA or (1R,2R/1S,2S)-ANPO overlap widely, indicating that the luminescence quenching might resulted from the absorption of 3-L excitation energy by the analytes.48,49Conclusion
Three pairs of enantiomeric CPs 1, 2 and 3-D/L were successfully synthesized based on (R/S)-6-(1-carboxyethoxy)-2-naphthoic acid and (1R,2R/1S,2S)-2,2′-((5-carboxy-1,3-phenylene)bis(oxy))dipropionic acid, respectively. Crystallized in the monoclinic system with chiral space group P21, the CPs are all SHG active. CPs 1 and 3 demonstrate good sensing activities toward the antibiotics NFs and pesticide DCN, the mechanism of which might due to the occurrence of PET and FRET phenomena. CP 3 could also effectively recognize NMs as a result of competitive absorption of excitation energy. CPs 1 and 3 also exhibit obvious sensing abilities toward nitro aromatic compounds and the possible mechanism is discussed. The sensing abilities of CPs 1, 3 toward different organics indicate that they could act as potential functional materials for the detecting of certain compounds.Conflicts of interest
There are no conflicts to declare.References
- Q.-Q. Zhang, G.-G. Ying, C.-G. Pan, Y.-S. Liu and J.-L. Zhao, Environ. Sci. Technol., 2015, 49, 6772–6782 CrossRef CAS PubMed.
- H. Wang, N. Wang, B. Wang, Q. Zhao, H. Fang, C. Fu, C. Tang, F. Jiang, Y. Zhou, Y. Chen and Q. Jiang, Environ. Sci. Technol., 2016, 50, 2692–2699 CrossRef CAS PubMed.
- E. Szöcs, M. Brinke, B. Karaoglan and R. B. Schäfer, Environ. Sci. Technol., 2017, 51, 7378–7385 CrossRef PubMed.
- W. Cheng, L. Jiang, N. Lu, L. Ma, X. Sun, Y. Luo, K. Lin and C. Cui, Anal. Methods, 2015, 7, 1777–1787 RSC.
- M. Abbaspour, M. A. Farajzadeh, S. M. Sorouraddin and A. Mohebbi, Anal. Methods, 2019, 11, 4022–4033 RSC.
- J. Fenoll, P. Hellín, C. M. Martínez, M. Miguel and P. Flores, Food Chem., 2007, 105, 711–719 CrossRef CAS.
- X.-Y. Guo, Z.-P. Dong, F. Zhao, Z.-L. Liu and Y.-Q. Wang, New J. Chem., 2019, 43, 2353–2361 RSC.
- V. Schurig, Ann. Pharm. Fr., 2010, 68, 82–98 CrossRef CAS PubMed.
- Y. Wang, S. Zhuo, J. Hou, W. Li and Y. Ji, ACS Appl. Mater. Interfaces, 2019, 11, 48363–48369 CrossRef CAS PubMed.
- M. Liu, L. Chen, T. Tian, Z. Zhang and X. Li, Anal. Chem., 2019, 91, 13803–13809 CrossRef CAS PubMed.
- X.-D. Zhu, K. Zhang, Y. Wang, W.-W. Long, R.-J. Sa, T.-F. Liu and J. Lü, Inorg. Chem., 2018, 57, 1060–1065 CrossRef CAS PubMed.
- J. Li, C. Yu, Y.-n. Wu, Y. Zhu, J. Xu, Y. Wang, H. Wang, M. Guo and F. Li, Environ. Int., 2019, 125, 135–141 CrossRef CAS PubMed.
- Z.-J. Li, J. Yao, Q. Tao, L. Jiang and T.-B. Lu, Inorg. Chem., 2013, 52, 11694–11696 CrossRef CAS PubMed.
- J. Zhang, Z. Li, W. Gong, X. Han, Y. Liu and Y. Cui, Inorg. Chem., 2016, 55, 7229–7232 CrossRef CAS PubMed.
- J. Navarro-Sánchez, A. I. Argente-García, Y. Moliner-Martínez, D. Roca-Sanjuán, D. Antypov, P. Campíns-Falcó, M. J. Rosseinsky and C. Martí-Gastaldo, J. Am. Chem. Soc., 2017, 139, 4294–4297 CrossRef PubMed.
- K. Tanaka, K. Sakuragi, H. Ozaki and Y. Takada, Chem. Commun., 2018, 54, 6328–6331 RSC.
- V. Gupta and S. K. Mandal, Inorg. Chem., 2019, 58, 3219–3226 CrossRef CAS PubMed.
- L. Li, J. Ma, C. Song, T. Chen, Z. Sun, S. Wang, J. Luo and M. Hong, Inorg. Chem., 2012, 51, 2438–2442 CrossRef CAS PubMed.
- H.-T. Ye, C.-Y. Ren, G.-F. Hou, Y.-H. Yu, X. Xu, J.-S. Gao, P.-F. Yan and S.-W. Ng, Cryst. Growth Des., 2014, 14, 3309–3318 CrossRef CAS.
- S. Halder, A. Barma, C. Rizzoli, P. Ghosh and P. Roy, ACS Appl. Nano Mater., 2019, 2, 5469–5474 CrossRef CAS.
- Z.-W. Zhai, S.-H. Yang, M. Cao, L.-K. Li, C.-X. Du and S.-Q. Zang, Cryst. Growth Des., 2018, 18, 7173–7182 CrossRef CAS.
- Y. Zhao, Y.-J. Wang, N. Wang, P. Zheng, H.-R. Fu, M.-L. Han, L.-F. Ma and L.-Y. Wang, Inorg. Chem., 2019, 58, 12700–12706 CrossRef CAS PubMed.
- Y.-P. Wu, J.-W. Tian, S. Liu, B. Li, J. Zhao, L.-F. Ma, D.-S. Li, Y.-Q. Lan and X. Bu, Angew. Chem., Int. Ed., 2019, 58, 12185–12189 CrossRef CAS PubMed.
- Y.-J. Cheng, R. Wang, S. Wang, X.-J. Xi, L.-F. Ma and S.-Q. Zang, Chem. Commun., 2018, 54, 13563–13566 RSC.
- X.-G. Yang, X.-M. Lu, Z.-M. Zhai, Y. Zhao, X.-Y. Liu, L.-F. Ma and S.-Q. Zang, Chem. Commun., 2019, 55, 11099–11102 RSC.
- Y. Zhao, X.-G. Yang, X.-M. Lu, C.-D. Yang, N.-N. Fan, Z.-T. Yang, L.-Y. Wang and L.-F. Ma, Inorg. Chem., 2019, 58, 6215–6221 CrossRef CAS PubMed.
- N. Xu, Q. Zhang, B. Hou, Q. Cheng and G. Zhang, Inorg. Chem., 2018, 57, 13330–13340 CrossRef CAS PubMed.
- M. Yu, Y. Xie, X. Wang, Y. Li and G. Li, ACS Appl. Mater. Interfaces, 2019, 11, 21201–21210 CrossRef CAS PubMed.
- Y. Xu, L. Zheng, X. Huang, Y. Cheng and C. Zhu, Polymer, 2010, 51, 994–997 CrossRef CAS.
- F. Song, G. Wei, L. Wang, J. Jiao, Y. Cheng and C. Zhu, J. Org. Chem., 2012, 77, 4759–4764 CrossRef CAS PubMed.
- W. Zhang, G. Wei, Z. Wang, J. Ma, C. Zhu and Y. Cheng, Polymer, 2016, 101, 93–97 CrossRef CAS.
- L. Wang, F. Song, J. Hou, J. Li, Y. Cheng and C. Zhu, Polymer, 2012, 53, 6033–6038 CrossRef CAS.
- M. M. Wanderley, C. Wang, C.-D. Wu and W. Lin, J. Am. Chem. Soc., 2012, 134, 9050–9053 CrossRef CAS PubMed.
- Q. Huang, J.-H. Huang, L. Gu, J.-x. Ruan, Y.-H. Yu and J.-S. Gao, RSC Adv., 2018, 8, 557–566 RSC.
- P. Hao, Q. Huang, L. Gu, Y.-H. Yu and D.-S. Ma, Polyhedron, 2018, 149, 117–125 CrossRef CAS.
- R. Wang, L. Liu, L. Lv, X. Wang, R. Chen and B. Wu, Cryst. Growth Des., 2017, 17, 3616–3624 CrossRef CAS.
- J.-H. Huang, G.-F. Hou, D.-S. Ma, Y.-H. Yu, W.-H. Jiang, Q. Huang and J.-S. Gao, RSC Adv., 2017, 7, 18650–18657 RSC.
- T. Liu, Y.-H. Yu, H.-Z. Zhang, W.-H. Jiang, J.-S. Gao and G.-F. Hou, Cryst. Growth Des., 2017, 17, 1788–1795 CrossRef CAS.
- L. Gu, H.-Z. Zhang, W.-H. Jiang, G.-F. Hou, Y.-H. Yu and D.-S. Ma, RSC Adv., 2017, 7, 45862–45868 RSC.
- N. Xu, Q. Zhang and G. Zhang, Dalton Trans., 2019, 48, 2683–2691 RSC.
- C.-L. Tao, B. Chen, X.-G. Liu, L.-J. Zhou, X.-L. Zhu, J. Cao, Z.-G. Gu, Z. Zhao, L. Shen and B. Z. Tang, Chem. Commun., 2017, 53, 9975–9978 RSC.
- Q.-Q. Zhu, H. He, Y. Yan, J. Yuan, D.-Q. Lu, D.-Y. Zhang, F. Sun and G. Zhu, Inorg. Chem., 2019, 58, 7746–7753 CrossRef PubMed.
- J.-M. Li, R. Li and X. Li, CrystEngComm, 2018, 20, 4962–4972 RSC.
- X. S. Li, J. D. An, H. M. Zhang, J. J. Liu, Y. Li, G. X. Du, X. X. Wu, L. Fei, J. D. Lacoste, Z. Cai, Y. Y. Liu, J. Z. Huo and B. Ding, Dyes Pigm., 2019, 170, 107631 CrossRef CAS.
- D. Zhao, X.-H. Liu, Y. Zhao, P. Wang, Y. Liu, M. Azam, S. I. Al-Resayes, Y. Lu and W.-Y. Sun, J. Mater. Chem. A, 2017, 5, 15797–15807 RSC.
- H. He, Q.-Q. Zhu, F. Sun and G. Zhu, Cryst. Growth Des., 2018, 18, 5573–5581 CrossRef CAS.
- S. Pal and P. K. Bharadwaj, Cryst. Growth Des., 2016, 16, 5852–5858 CrossRef CAS.
- J.-K. Wang, X.-W. Wang, Z.-S. Wang, L.-S. Yao, L.-Z. Niu, Y.-H. Yu and J.-S. Gao, Polyhedron, 2019, 167, 85–92 CrossRef CAS.
- X.-W. Wang, J.-H. Huang, J.-K. Wang, Y.-H. Yu and D.-S. Ma, Z. Anorg. Allg. Chem., 2019, 645, 863–869 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 1867800, 1867801, 1965710–1965712 and 1965708. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d0ra00329h |
| This journal is © The Royal Society of Chemistry 2020 |