Open Access Article
Open Access ArticleIron and copper complexes with antioxidant activity as inhibitors of the metastatic potential of glioma cells†
Joana F. Guerreiro‡
 ab,
Marco Antônio G. B. Gomes‡
ab,
Marco Antônio G. B. Gomes‡ a,
Francesca Pagliari
a,
Francesca Pagliari *a,
Jeannette Jansen
*a,
Jeannette Jansen ac,
Maria G. Marafiotia,
Clelia Nistico
ac,
Maria G. Marafiotia,
Clelia Nistico a,
Rachel Hanley
a,
Rachel Hanley ac,
Rafael O. Costa
ac,
Rafael O. Costa d,
Sarah S. Ferreirae,
Filipa Mendes
d,
Sarah S. Ferreirae,
Filipa Mendes b,
Christiane Fernandes
b,
Christiane Fernandes f,
Adolfo Hornf,
Luca Tirinato
f,
Adolfo Hornf,
Luca Tirinato ag and
Joao Seco
ag and
Joao Seco *ac
*ac
aBiomedical Physics in Radiation Oncology, German Cancer Research Center (DKFZ), Im Neuenheimer Feld 223, 69120 Heidelberg, Germany. E-mail: f.pagliari@dkfz-heidelberg.de/j.seco@dkfz-heidelberg.de; Tel: +49 6221 42 2554
bDepartamento de Engenharia e Ciências Nucleares e Centro de Ciências e Tecnologias Nucleares, Instituto Superior Técnico, Universidade de Lisboa, Estrada Nacional 10 (km 139.7), 2695-066 Bobadela LRS, Portugal
cDepartment of Physics and Astronomy, Heidelberg University, Im Neuenheimer Feld 227, 69120 Heidelberg, Germany
dLaboratório de Ciências Química, Universidade Estadual do Norte Fluminense (UENF), Av. Alberto Lamego, 2000, Campos dos Goytacazes, RJ, 28013602, Brazil
eInstituto Federal Fluminense (IFF), R. Dr Siqueira, 273, Campos dos Goytacazes, RJ CEP 28030-130, Brazil
fDepartamento de Química, Universidade Federal de Santa Catarina (UFSC), Campus Universitário Trindade, 88040900 Florianópolis, SC, Brazil
gBioNEM Laboratory, Department of Experimental and Clinical Medicine, Magna Graecia University of Catanzaro, 88100 Catanzaro, Italy
First published on 30th March 2020
Abstract
Gliomas are the most common type of primary brain tumors, presenting high mortality and recurrence rates that highlight the need for the development of more efficient therapies. In that context, we investigated iron(III) (FeL) and copper(II) (CuL) complexes containing the tetradentate ligand 2-{[(3-chloro-2-hydroxy-propyl)-pyridin-2-ylmethyl-amino]-methyl}-phenol (L) as potential antimetastatic compounds in glioma cells. These complexes were designed to act as mimetics of antioxidant metalloenzymes (catalases and superoxide dismutase) and thus interfere with the production of reactive oxygen species (ROS), important signaling molecules that have been linked to the induction of Epithelial–Mesenchymal Transition (EMT) in cancer cells, a process associated with cancer invasion and aggressiveness. The results obtained have revealed that, in vitro, both compounds act as superoxide dismutase or catalase mimetics, and this translated in glioma cells into a decrease in ROS levels in FeL-treated cells. In addition, both complexes were found to inhibit the migration of monolayer-grown H4 cells and lead to decreased expression of EMT markers. More importantly, this behavior was recapitulated in 3D spheroids models, where CuL in particular was found to completely inhibit the invasion ability of glioma cells, with or without cellular irradiation with X-rays, which is suggestive of these compounds' potential to be used in combination with radiotherapy. Overall, the results herein obtained describe the novel use of these complexes as agents that are able to interfere with regulation of EMT and the invasive behavior of glioma cells, an application that deserves to be further explored.
Introduction
The most common primary malignant brain tumors in adults are gliomas, which correspond to about 80% of all the malignant brain tumors diagnosed.1 The treatment of gliomas varies according to the degree of the disease and the patient's condition, but the current standard of treatment includes surgery for maximum resection of the tumor, followed by radiotherapy and chemotherapy.1 However, achieving complete resection of the tumor is often impossible due to its highly infiltrating nature and inaccessible location, leading to recurrence of the disease in the great majority of cases.1 In addition, while metastases outside of the central nervous system are uncommon, when present, they often exhibit increased resistance to treatment, similarly to what is observed for relapsed tumors, leading to a very poor prognosis for these patients.1–3 As such, it is necessary to develop more efficient therapeutic tools that can improve the patients' outcome.Research exploring metal based compounds as chemotherapeutic drugs for the treatment of cancer has increased since the discovery of cisplatin-based chemotherapy.4,5 Metal complexes present many versatile characteristics, such as their redox activity, diverse reactivity with organic substrates, and different coordination modes that make them attractive tools to be explored in the design of new chemotherapeutic drugs.4,5 In addition to the development of cytotoxic chemotherapeutic drugs, the interest on metal complexes that can be used as metastasis inhibitors has also increased in recent years.5–7 For this purpose, most of the studies done so far have largely been focused on promising ruthenium-based compounds,6,8–10 even though complexes containing other metals have also been described.11,12 Currently, however, only two ruthenium compounds have advanced into clinical trials, although they've failed to show the desired therapeutic efficacy that would make them viable alternative to the therapies currently in use.13
Antimetastatic complexes can target different cellular pathways or processes, but have mainly been designed to modulate or interfere with key features necessary for cancer migration or invasion. One such feature is the modulation of the Epithelial–Mesenchymal Transition (EMT) phenomenon,9 a physiological process involved in the cellular developmental program and tissue repair, but which has been also strongly linked to the metastatic process in cancer.14 Namely, during EMT, cancer cells undergo a series of changes (biochemical, morphologic and genetic) that allow them to have a more mesenchymal-like phenotype that is thought to be necessary to promote cancer cell migration and invasion, and their escape from the primary tumor.14 Despite the fact that the search for compounds able to interfere with the EMT process has been increasing in the past years, it is still mainly based in the use of natural compounds isolated from plants,15 while the use of metal-based compounds in this branch of medicinal chemistry remains poorly researched. In addition to targeting EMT, several of these metal-based compounds also aim to affect Reactive Oxygen Species (ROS) equilibrium8,11,12,16 since ROS can act as signaling molecules in many cellular pathways, including those involved in tumor progression.17 Interestingly, the EMT process seems to be connected to cellular ROS levels and different metals have been shown to induce EMT in different cancers through a ROS-dependent mechanism.18–21 As such, modulation of ROS levels in cancer cells has been put forth as another promising strategy to tackle the problem of local invasiveness and metastization of cancer.22
One possible strategy to modulate the levels of cellular ROS, and, consequently, the cancer cells metastatic ability, is the use of metal-based compounds that mimic the superoxide dismutase (SOD) and catalase (CAT) enzymes, important cellular antioxidant proteins that are responsible for maintaining the cellular redox balance.22 For that purpose, in this work, we used two coordination compounds harboring the ligand 2-{[(3-chloro-2-hydroxy-propyl)-pyridin-2-ylmethyl-amino]-methyl}-phenol (L) complexed with iron (FeL)23,24 and copper (CuL).25 We thus describe for the first time the application of these compounds in the frontier of chemistry and human oncology, by assessing their antioxidant and antimetastatic potential in glioma (H4) cells. The results obtained have revealed an impressive ability of the compounds under study to inhibit the migration of H4 glioma cells in both 2D and 3D cellular models. In addition, this effect was maintained after irradiation with X-rays, suggesting that these compounds might be suitable to be used as co-adjuvants for radiotherapeutic treatments.
Results and discussion
Cytotoxicity of FeL and CuL compounds in H4 glioma cells
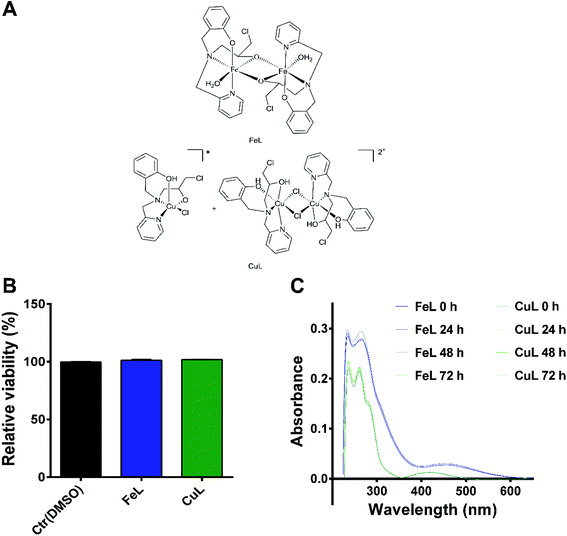
The synthesis of the ligand 2-{[(3-chloro-2-hydroxy-propyl)-pyridin-2-ylmethyl-amino]-methyl}-phenol (L) and of the iron and copper complexes studied here were described previously by us.23–26 The ligand contains four coordinating groups (N2O2) and its coordination behavior depends on the metal center. For example, it forms dinuclear phenoxo bridge complexes with Ni(II),27 while with Fe(III), mononuclear and dinuclear (alkoxo bridge) were already described.23,24 The iron compound described here shows a dinuclear structure (Fig. 1), in which the iron(III) ions are connected by two alkoxo bridges from two ligand molecules. The coordination environment is completed by two nitrogen atoms (the tertiary N atom and one from the pyridyl group), one oxygen from the phenolate unit and a water molecule. It has been shown that this compound is able to promote DNA cleavage.23 Concerning the copper complex, its molecular structure solved by monocrystal X-ray analysis showed the presence of two distinct species in the crystal, a mononuclear and a dinuclear one,25 shown in Fig. 1. The dinuclear species may be considered the dimer of the mononuclear one and studies showed that the dinuclear species is transformed in the mononuclear one in solution, and, therefore, only the mononuclear species remains. It has been previously demonstrated that the copper complex shows cytotoxicity on pathogenic bacteria.25In order to determine if the FeL and CuL complexes (Fig. 1A) exhibited significant antitumoral properties, their cytotoxic activity after 24 hours (h) of treatment was determined in H4 glioma cells using the AlamarBlue assay. Both compounds were found to have IC50 values in the high micromolar range (85 ± 1 and 82 ± 1 μM for FeL and CuL, respectively; ESI Fig. 1†), indicating that they display only moderate cytotoxicity in glioma cells. In fact, these compounds exhibited about 40% less cytotoxicity than the one previously reported for the reference chemotherapeutic drug cisplatin (50 μM) in this same cancer cell line after 24 h of incubation.28 Since we were not interested in evaluating the intrinsic cytotoxic activity of the compounds, but how their antioxidant activity may influence other properties of cancer cell development, the lack of cytotoxic effect is of relevance for the present study. As such, we selected a concentration of the compounds that did not induce significant loss of viability (25 μM; Fig. 1B) to further proceed with the evaluation of these compounds as antimetastatic agents, while minimizing potential interference from cytotoxic effects exerted by the drugs. Due to the lack of a proper non-malignant control brain cell line, this study did not consider the effects of the tested compounds on healthy brain cells. Although we acknowledge that this aspect can be of particular interest, it is currently beyond the scope of this study and, therefore, it will be further investigated in the future.
Stability studies by UV-Vis spectrometry indicated that both compounds were stable at the selected concentration in a PBS solution at physiological pH for up to 72 h of incubation (Fig. 1C), the latest time point used for our assays. For the iron compound, it was also possible, using a higher concentration of FeL (50 μM), to visualize through confocal fluorescence microscopy the presence of a fluorescent species in the lysosomes, suggesting that it was indeed entering the cells under these conditions (ESI Fig. 2†). On the other hand, no signal was observed for the copper compound.
In vitro and cellular antioxidant properties of FeL and CuL
Based on the important role that transition metal ions play in cellular redox mechanisms, several studies having metalloenzymes, such as SOD and CAT, as targets for new mimetic compounds have been performed.29–32 Within this approach, in recent years, our group has developed different ligands and their respective coordination compounds with different transition metals, that exhibit antioxidant properties.29,33–35 For example, we have shown that copper, iron and manganese complexes with the ligand 1-[bis(pyridin-2-ylmethyl)amino]-3-chloropropan-2-ol (L1), similar to the one reported here, but with two pyridine groups instead of a pyridine and a phenol group, present protective antioxidant effects on Saccharomyces cerevisiae cells subjected to oxidative stress.33 These results thus prompted us to investigate the antioxidant activity of this set of compounds (FeL and CuL) and evaluate if their antioxidant activity could exert any influence on biological processes, particularly on the migratory ability of cancer cells.To address if the compounds FeL and CuL displayed antioxidant activities, we first assessed their ability to mimic the SOD enzyme in vitro. Both compounds reacted with the superoxide anion (ESI Fig. 3, 4 and Table 1†) as evidenced by the IC50 values obtained, which represent the concentration of the compound required to inhibit half of the reduction of nitrobluetetrazolium (NBT) by the superoxide anion generated in situ at a constant rate by the enzymatic system xanthine/xanthine oxidase, in comparison to control conditions.
In order to show SOD-like activity, the compounds have to be able to promote the oxidation 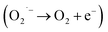 and the reduction
and the reduction 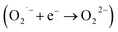 of the superoxide anion. This behavior is shown by systems that catalytically induce the superoxide decomposition. If the system promotes only the reduction or only the oxidation of the superoxide anion, they will work as superoxide reductase or superoxide oxidase, respectively. In light of this, the compounds described here would react only stoichiometrically with the superoxide anion. Thus, considering the difference (71 nmol) between the number of moles of formazan formed in the presence and in the absence of CuL and the number of moles of the copper complex (2.0 nmol) employed in the assay that showed the lower formation of formazan (Fig. 1 ESI†), each molecule of the copper complex was able to react with 35 molecules of superoxide anion after 40 min, clearly suggesting catalytic activity. On the other hand, since the iron complex was less active, the reaction ratio superoxide
of the superoxide anion. This behavior is shown by systems that catalytically induce the superoxide decomposition. If the system promotes only the reduction or only the oxidation of the superoxide anion, they will work as superoxide reductase or superoxide oxidase, respectively. In light of this, the compounds described here would react only stoichiometrically with the superoxide anion. Thus, considering the difference (71 nmol) between the number of moles of formazan formed in the presence and in the absence of CuL and the number of moles of the copper complex (2.0 nmol) employed in the assay that showed the lower formation of formazan (Fig. 1 ESI†), each molecule of the copper complex was able to react with 35 molecules of superoxide anion after 40 min, clearly suggesting catalytic activity. On the other hand, since the iron complex was less active, the reaction ratio superoxide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FeL was only 1.5 after 40 min. Since this ratio is only a little bit higher than the stoichiometric reaction, at the moment it is not possible to conclude if FeL showed SOD or SOO activity. Therefore, CuL was found to possess a higher reactivity on the superoxide anion (almost 50 times higher) than the FeL compound and due to its catalytic activity it might be considered as presenting SOD-like activity. However, the Kcat obtained for CuL is ca. 1.8 × 102 lower than the one observed for the natural SOD. Comparing the data with the complexes synthesized with the ligand L1 (Table 1), the activities obtained here were of the same order of magnitude.
FeL was only 1.5 after 40 min. Since this ratio is only a little bit higher than the stoichiometric reaction, at the moment it is not possible to conclude if FeL showed SOD or SOO activity. Therefore, CuL was found to possess a higher reactivity on the superoxide anion (almost 50 times higher) than the FeL compound and due to its catalytic activity it might be considered as presenting SOD-like activity. However, the Kcat obtained for CuL is ca. 1.8 × 102 lower than the one observed for the natural SOD. Comparing the data with the complexes synthesized with the ligand L1 (Table 1), the activities obtained here were of the same order of magnitude.
| Compound | SOD activity | CAT activity | Ref. | |||
|---|---|---|---|---|---|---|
| IC50 (μM) | Kcat (M−1 s−1) | Kcat (s−1) | KM (mM) | Kcat/KM (M−1 s−1) | ||
| a The kinetic data do not allow to confirm if the compound shows superoxide dismutase or superoxide oxidase activity.b L = N-(2-hydroxybenzyl)-N-(2-pyridylmethyl)[(3-chloro)(2-hydroxy)]propylamine; L1 = 1-[bis(pyridin-2-ylmethyl) amino]-3-chloropropan-2-ol; ND = not determined; NA = not active. | ||||||
| FeLa | 8.946 ± 0.345 | 1.43 × 105 | 0.080 ± 0.003 | 23.2 ± 1.2 | 3.45 ± 0.04 | This work |
| CuL | 0.181 ± 0.016 | 7.07 × 106 | 0.360 ± 0.125 | 41.9 ± 15.7 | 8.25 ± 0.06 | This work |
| FeL1b | 26.8 ± 2.5 | 1.2 × 105 | ND | ND | ND | Ribeiro et al.33 |
| CuL1b | 0.43 ± 0.2 | 7.7 × 106 | NA | NA | NA | Riberio et al.33 |
| Cu, Zn-SOD | 0.03 | 1.3 × 109 | — | — | — | Weser et al.36 |
| CAT (human erythrocytes) | — | — | 5.87 × 105 | 80 | 7.34 × 106 | Switala et al.37 |
Next, the ability of the complexes to mimic the enzyme CAT was evaluated through a direct reaction with hydrogen peroxide (H2O2), which was monitored by measuring H2O2 absorption using electronic spectroscopy at 240 nm. The FeL complex showed CAT mimetic activity in phosphate buffer solution (pH 7.8), while the CuL complex exhibited CAT-like activity only when one co-catalyst (piperazine) was added to the reaction (Table 1). Once again, the kinetic parameters calculated for both compounds, in particular the Kcat, revealed that CuL possesses higher CAT-like activity than FeL, albeit limited by the need of the addition of the mentioned co-catalyst.
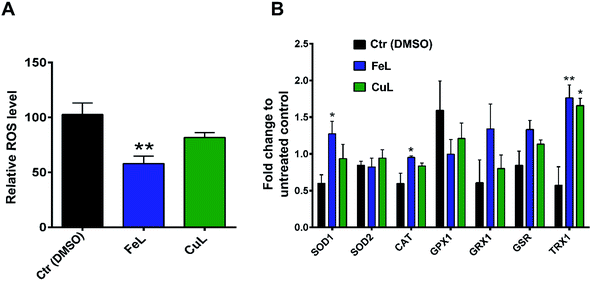
Following the results obtained with the in vitro enzymatic assays, we then proceeded to determine the ROS levels in H4 glioma cells incubated with the 2 complexes. For that, we used CM-H2DCFDA-based flow cytometry which is useful to detect several ROS species, but mainly H2O2, the hydroxyl radical (OH˙) or peroxynitrite.38 Cellular treatment with FeL for 24 h induced a statistically significant reduction in ROS levels (Fig. 2A). In contrast, CuL led to an evident, but not statistically significant, decrease in ROS levels (Fig. 2A). These results indicate that the in vitro antioxidant activity is not translated in the cellular environment. This behavior has been described previously when the in vitro SOD/CAT activity of Fe, Cu and Mn of similar mimetic complexes was not replicated in live cells.33
To assess if the complexes antioxidant effects might also be due to indirect instead of direct effects, we assessed the level of expression of several ROS-related genes by qPCR. The vehicle control sample, treated with DMSO, exhibited a clear effect on the expression of some of the genes analysed (Fig. 2B), which is in agreement with the fact that DMSO has been previously described to be a ROS scavenger, able to interfere with several related cellular processes,39 even if under our experimental conditions we saw no significant changes in ROS levels in DMSO-control cells in the cytometric study (Fig. 2A). From the results obtained, however, it became evident that both treatment with FeL and CuL led to a considerable upregulation of the expression of thiorredoxin (Fig. 2B), Trx1, an important cytosolic detoxifying protein,40 which suggests that these compounds might have an impact on the homeostasis of cytosolic redox status. Additionally, the FeL compound also led to significant changes in SOD1 and CAT expression levels when compared with DMSO-treated cells (Fig. 2B), which is in accordance to the fact that it induced a significant decrease in ROS levels (Fig. 2A) and might contribute to its apparently higher antioxidant effect when compared with CuL. In addition to having an effect on intracellular ROS levels, we cannot rule out the possibility that the complexes used are also altering extracellular ROS levels. These species have been hypothesized to be extremely relevant players in the tumor microenvironment and different aspects of cancer progression, including the development of metastasis,41 and, as such, this possibility is worthy of further investigation in the future.
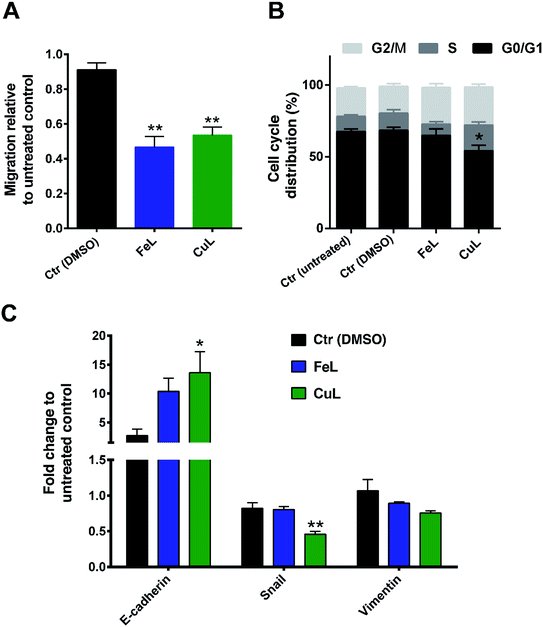
FeL and CuL complexes reduce migration through inhibition of epithelial–mesenchymal transition (EMT) in glioma cells
Since an increase in ROS had been previously implicated in EMT induction in different cells,18–21 we hypothesized that the reduction in ROS levels induced by the compounds could be leading to changes in the metastatic ability of H4 cells. The effect of FeL and CuL on the migration of H4 cells was thus investigated by the transwell migration assay. The number of cells migrated to the bottom of the membrane revealed that both complexes can clearly inhibit the migratory ability of H4 cells (Fig. 3A).To investigate to what extent this observation was related to cell proliferation or cell cycle arrest induction, since ROS has also been shown to be related with regulation of cellular proliferation/cell cycle,42 the effects of the compounds on the cell cycle of H4 cells were investigated by flow cytometry. While FeL showed no effect on the cell cycle of H4 cells, CuL induced a significant decrease in the G0/G1 phase of the cycle (*p ≤ 0.05), with a concomitant increase in the % of cells in the S and G2/M phases (of about 7.7 and 6.2%, respectively) that was, however, statistically not significant (Fig. 3B). This suggests that CuL-treated cells might experience a shift in the cell cycle from the G0/G1 phase to the S and G2/M phases, which could either reflect a slight increase in proliferation, or that cells are arrested during DNA duplication or prior to cell division.43 However, this difference does not seem likely to justify the significant change observed in the migration of glioma cells upon exposure to the complex.
As such, looking for another possible explanation, we next analyzed the expression of several EMT markers in the FeL/CuL treated cells by qPCR. The results evidenced that treatment with the compounds is accompanied by an obvious and statistically significant increase in expression of E-cadherin mRNA, and a slight, but not significant, reduction of Vimentin in the case of CuL (Fig. 3C). The expression of the EMT-related transcription factor Snail was found to also be statistically significantly decreased upon treatment with CuL (Fig. 3C). This gene expression profile is consistent with the hypothesis that cells treated with FeL and CuL had a more epithelial-like phenotype, possibly experiencing an inhibition of the EMT transition process, which should originate cells with a less motile phenotype,14 and is in accordance with the decreased migratory ability observed in complex-treated cells (Fig. 3A), demonstrating that the compounds do seem to possess anti-metastatic properties.
FeL and CuL complexes inhibit 3D spheroids invasion
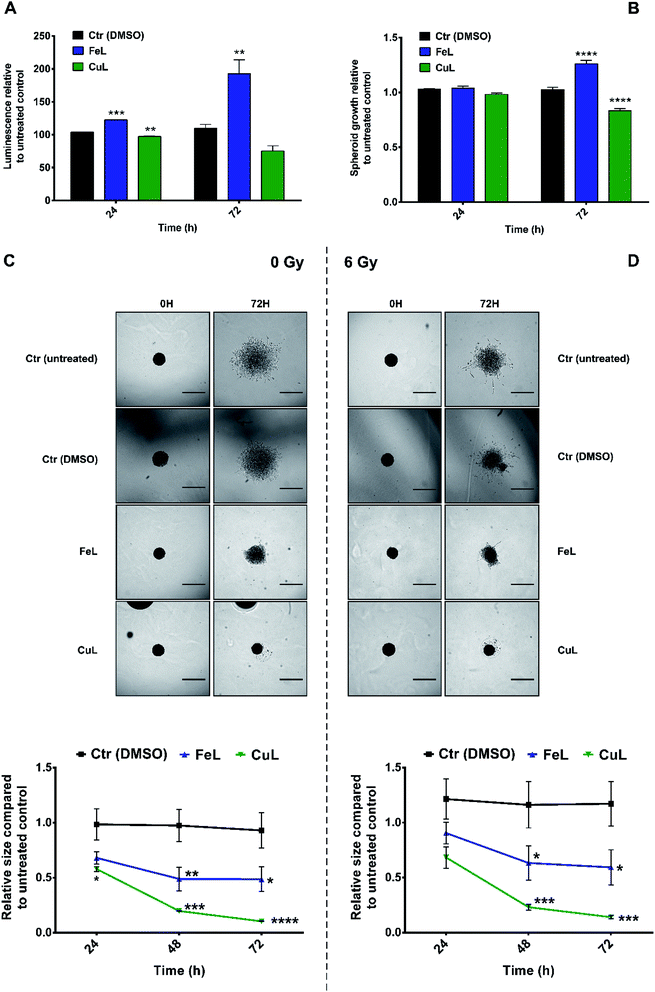
There is mounting evidence that the results obtained in 2D cellular models, where many of the characteristics of the original tumor microenvironment are missing, present several limitations when being transposed into the clinical setting.44 In that context, several 3D cellular models have been developed that present a level of complexity which is much closer and more representative of several aspects of tumor tissues than the ones shown by monolayer cell cultures.44 In particular, matrix-embedded 3D cultures have been increasingly applied to investigate tumor migration and invasion.45As such, and in order to try to better estimate the clinical translational potential of the compounds under evaluation, we extended our studies to H4 multicellular spheroids, which are expected to better recapitulate in vivo tumor properties. For that purpose, spheroids generated in agarose-coated plates were first treated with FeL or CuL for 24 h or 72 h. Then, cell viability was assessed using the CellTiter-Glo® 3D assay, while spheroid size and growth were accompanied using bright field microscopy. Surprisingly, incubation with FeL increased cellular viability (Fig. 4A), both after 24 h and 72 h of incubation. This increase in viability was accompanied by an increase in spheroid size after 72 h of incubation (Fig. 4B). In contrast, CuL induced a decrease in viability as early as 24 h of incubation, along with a concomitant decrease in spheroid size (Fig. 4A and B).
Next, we observed that both complexes were able to interfere with the invasive behavior exhibited by H4 cells embedded in matrigel (Fig. 4C). CuL, in particular, exhibited very encouraging results, completely eliminating H4 cell ability to invade the matrigel matrix, an effect that cannot be attributed solely to the decrease in viability and growth found to occur following incubation with this compound (around 31% and 19% in terms of cellular viability and growth, respectively). FeL also displayed the ability to inhibit the invasive behavior of H4 cells, an effect that was, however, not as striking as the one found for CuL. However, this can be due to the fact that this compound was found to present a stimulatory effect in cellular viability, as described above (Fig. 4A and B), counteracting the desirable effect it seemed to also have as an anti-metastatic compound.
Notably, the effect of the compounds on H4 spheroids' invasive ability was maintained even when cells were irradiated with X-rays (6 Gy). This is highly relevant in the clinical context, since it has been demonstrated that the use of low linear energy transfer (LET) irradiation, which includes X-rays radiation, might, in patient-specific contexts, increase migration and invasion of glioma cells.46 In addition, most glioma relapses occur in an area within 2 cm of the area where the primary tumor initially developed, which impairs tumor removal and local radiotherapy.2 The results obtained in the 3D invasion assays thus clearly demonstrate that both complexes possess an anti-metastatic effect not only in monolayer cells, but also in the more representative spheroids model that has potential to be highly relevant in the clinical context.
FeL and CuL complexes alter glutathione metabolism or oxidative stress in H4 spheroids
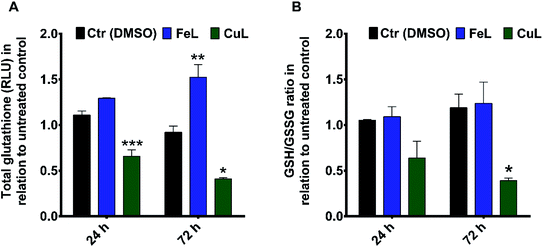
Since we had previously observed an apparent decrease in ROS levels in complex-treated cells that could be related to the decreased migration observed in monolayer-cultured cells, we investigated whether the remarkable effect of the compounds on the inhibition of H4 spheroids' invasion in matrigel could be also related with changes in cellular oxidative stress in this 3D cellular model. For that, the antioxidant ability of the complexes was assessed by determining the relative levels of cellular glutathione and the ratio of reduced glutathione (GSH), an important cellular antioxidant and detoxifying agent, and oxidized glutathione (GSSG) using the GSH/GSSG-Glo™ Assay. The results obtained revealed that treatment with FeL induced an increase in GSH levels, while the GSH/GSSG ratio remained unchanged (Fig. 5A and B, respectively). This suggests that this compound did not change the oxidative stress levels in H4 spheroids, but it seemed to affect cellular glutathione metabolism. Contrastingly, cells treated with the CuL complex showed an evident decrease of the GSH/GSSG ratio, compared with the vehicle control sample, which indicates that CuL was inducing oxidative stress under these conditions (Fig. 5B). In addition, the level of total GSH in these cells was also found to be reduced (Fig. 5A).These observations raise the question of what might be the impact of such metabolic changes on the behavior observed for FeL- and CuL-treated H4 spheroids. One possibility is that the elevated oxidative stress found in CuL-treated cells could underlie the decrease in viability observed under these same conditions (Fig. 4A and B), since several metal-based compounds have been previously described to reduce cancer cell viability through the induction of ROS production.8,11,12 Moreover, increased GSH levels have also been previously correlated with enhanced cancer metastatic ability.47 This could, at least partially, explain the difference in performance observed for the FeL and CuL compounds, since the later significantly decreases GSH levels and is much more efficient at reducing the invasive potential of H4 spheroids, while the former actually increased the GSH levels, exhibiting a less pronounced inhibitory effect. Overall, these results are highly encouraging, since modulation of GSH levels have been proposed as a potential way to sensitize tumor cells to treatment modalities such as chemotherapy,47 and, in particular the CuL complex, seems to be a good candidate to test this goal, while also having been proved herein to have a significant impact on the cell invasive ability.
Conclusion
The highly infiltrative nature of gliomas poses significant therapeutic challenges that result in a high rate of disease recurrence and poor patient prognosis. In this work, we explored the application of two coordination compounds, FeL and CuL in an anticancer therapeutic context. Both complexes showed antioxidant activity (catalase and superoxide dismutase/superoxide oxidase) in vitro and, in the case of FeL, also in H4 glioma cells. Although the complexes did not present significant cytotoxic activity at 25 μM, they exhibited anti-migratory properties in 2D cultures and anti-invasive abilities in 3D multicellular spheroids. While the mechanisms underlying these effects have not been fully elucidated, they seem to be related with cellular oxidative stress and/or glutathione metabolism, particularly in 3D cellular models where the best performing complex, CuL, caused a reduction in GSH levels, which has been previously correlated with increased metastatic properties of cancer cells. Importantly, the concentrations of the compounds tested were not cytotoxic in 2D models or only slightly affected the viability in 3D models, which indicates that the occurrence of extensive cell death is not behind the changes in migratory/invasive ability. Additionally, this suggests that they might also be less toxic to healthy cells, which would result in less treatment side-effects. Considering that recent reports have also proposed that glioma therapy needs to be developed in the context of a potential detrimental enhancement of cancer invasion by radiotherapeutic treatments, our complexes also revealed a decrease in H4 cells invasion when combined with irradiation with x-rays. This is highly relevant, as it indicates that they do have high potential to limit the cancer invasive ability and might be used in combination with other anti-proliferative therapies.Experimental section
Synthesis of complexes, preparation of stock solutions and stability
The ligand 2-{[(3-chloro-2-hydroxy-propyl)-pyridin-2-ylmethyl-amino]-methyl}-phenol (L) and the complexes FeL and CuL used in this work (Fig. 1) were synthesized and characterized as described previously.23–25 Elemental analyses (CHN) and ESI-(+)-MS confirmed the identity and purity of the compounds.A 1.0 mM stock solution of each complex was prepared by dissolving it in a 5% solution of dimethylsulfoxide (DMSO) prepared in ultrapure MiliQ water (H2O). For biological experiments, solutions with the desired concentrations were prepared by diluting the compound's stock in the culture medium used. The stability of the compound's solutions was determined in Dulbecco's Phosphate-Buffered Saline (DPBS) (Gibco™, Thermo Fisher Scientific, Waltham, MA, USA) at physiological pH. For that, a solution of the compounds at a concentration of 25 μM was prepared and the UV-Vis spectrum of the solutions was obtained at different times (0, 24, 48 and 72 h) in a UV-Vis Spectrophotometer (Varian Cary 400).
Cell culture
Human brain neuroglioma (H4) cells (ATCC, Manassas, VA, USA) were grown in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS) and 1% Penicillin/Streptomycin (all from Gibco™, Thermo Fisher Scientific). The cell line was cultured continuously as a monolayer at 37 °C and 5% of CO2.Viability assays
For IC50 determination, H4 cells were seeded at a density of 1.0 × 104 cells in 150 μL of medium in a 96-well black polystyrene microplate (Corning, NY, USA) and allowed to attach for 24 h at 37 °C. Then, the medium was removed and the wells were washed with DPBS before the addition of 150 μL of the 200, 100, 50, 25 and 12.5 μM solutions of the complexes, the respective vehicle controls (DMSO at the same concentration than in the complexes' solutions), or fresh medium (untreated control sample) to the wells. After 24 h of incubation, the medium in each well was removed, the wells washed with DPBS and 150 μL of a 10% solution of AlamarBlue (Thermo Fisher Scientific) in medium were added to each well. The plate was covered with aluminum foil and incubated for 2 h at 37 °C and subsequently read in a CLARIOstar® microplate reader (BMG LABTECH GmbH, Offenburg, Germany) for fluorescence detection.For viability determination by flow cytometry, 7.0 × 105 cells were seeded in 75 cm2 flasks (Greiner Bio-one, Frickenhausen, Germany) and incubated for 24 h at 37 °C. The medium was removed and cells were washed once with DPBS before 10.5 mL of fresh medium, medium with 25 μM of FeL and CuL, or medium with 0.125% of DMSO (as the vehicle control) were added to the flasks. The flasks were incubated for an additional 24 h, after which cells were detached and washed with DPBS. Then, for each sample, 1.0 × 106 cells were resuspended in DPBS and analyzed using a flow cytometer (BD FACS CANTO™ II) (unstained control samples). Then, those same cells were stained with 1 μg mL−1 of propidium iodide (PI) (Sigma Aldrich, St. Louis, MO, USA) and re-analyzed. The percentage of live cells calculated for each sample was normalized to the untreated control sample, and three independent experiments were performed.
Fluorescence study by confocal microscopy
H4 cells were seeded at a density of 5.0 × 104 cells on a 22 mm coverslip placed in a 6-wells plate (CELLSTAR®, Greiner Bio-One), and allowed to attach overnight. Cells were then incubated or not (as a control) with 50 μM of FeL for 24 h at 37 °C. Then, cells were washed once with Hank's Buffered Salt Solution (HBSS; Thermo Fisher Scientific) and stained with 75 nM Lyso-Tracker™ Red DND-99 (Molecular Probes, Thermo fisher Scientific) for 30 min at 37 °C. The staining solution was removed and cells were fixed for 5 min in 4% of paraformaldehyde at room temperature. Samples were washed thrice, coverslips were mounted on HBSS onto a glass slide, and sealed with nail polish. Fluorescence was visualized on a confocal microscope (Zeiss LSM 710) using a standard DAPI filter for visualization of FeL fluorescence, while Lysotracker was visualized using a 561 nm laser for excitation followed by emission detection on the 566–691 nm range.SOD/SOO-like activity
The reactivity on the superoxide anion was evaluated by a methodology described previously, which involves the reduction of nitroblue tetrazolium (NBT) by the superoxide anion.33 Stock solutions of xanthine, nitroblue tetrazolium (NBT) and xanthine oxidase were prepared at the concentrations of 4.5 × 10−4 mol dm−3, 5.6 × 10−5 mol dm−3 and 0.2 U cm−3, respectively, using a 0.05 mol dm−3 phosphate buffer solution at pH 7.8 (all reagents from Sigma-Aldrich).A control solution containing 1000 μL of the xanthine solution, 400 μL of the phosphate buffer solution and 1000 μL of NBT was added to a cuvette followed by the quick addition of 200 μL of the xanthine oxidase solution and then the absorbance was measured over time in a UV-Vis spectrophotometer (Varian Cary 50), thus obtaining the rate of change of the absorption in the absence of the complex.
To evaluate the activity of FeL and CuL, different concentrations of the complexes were employed: for FeL, the concentrations used were 1.92 × 10−6, 3.85 × 10−6, 7.69 × 10−6, 1.15 × 10−5, and 1.54 × 10−5 mol dm−3; for CuL, the concentrations used were 9.62 × 10−8, 1.92 × 10−7, 3.85 × 10−7, 5.77 × 10−7 and 7.67 × 10−7 mol dm−3. The concentration of the compounds which reduced 50% of NBT in relation to the control experiment was calculated, obtaining the IC50, which was then transformed to Kcat using the equation proposed by McCord and Fridovich, Kcat = KNBT × [NBT]/IC50, where KNBT = 5.94 × 104 M−1 s−1.48,49
CAT-like activity
The ability of the compounds in promoting H2O2 degradation was evaluated by the methodology described by Beers and Sizer.50 Initially, the concentration of H2O2 was evaluated by titration with iodide/thiosulfate.51 To determine the CAT-like activity of FeL, solutions of H2O2 at different concentrations (1.64 × 10−2, 1.23 × 10−3, 8.2 × 10−3 and 4.1 × 10−3 mol dm−3) were prepared in a total volume of 2200 μL of a phosphate buffer solution at pH 7.8. Then, each solution was mixed with a FeL solution yielding a final concentration of FeL of 7.69 × 10−5 mol dm−3, and the decrease of the absorbance associated with the reaction with H2O2 was followed by UV-Vis spectroscopy at 240 nm (Varian Cary 50) in a 1 cm path length cell. For CuL, the above protocol was followed but the solutions were prepared on a piperazine solution (0.1 mol dm−3) and the final CuL concentration in the mixture was 9.25 × 10−5 mol dm−3. The experiments were performed in triplicate, and the Michaelis Menten constant (KM) and the turnover number (Kcat) were then calculated for each complex.Intracellular ROS measurements
For determination of intracellular ROS levels, H4 cells were prepared and incubated with the compounds (or respective medium and DMSO controls) as described above for the viability analysis by flow cytometry. Upon detaching and washing, 4.0 × 105 cells were incubated with 5 μM of CM-H2DCFDA (Life Technology, Thermo fisher Scientific) in HBSS for 20 min at 37 °C in the dark. Stained cells were then washed once and resuspended in DPBS. Samples were analyzed in a flow cytometer (BD FACS CANTO™ II) and the average fluorescence intensity of each sample was normalized to the untreated control sample.Cell cycle assay
For the cell cycle assay, cells were grown as described above for the viability determination by flow cytometry. After detaching and washing, 1.0 × 106 cells were fixed through drop by drop addition of 70% cold ethanol (v/v in DPBS) under gently vortexing. Samples were stored at 4 °C for 24 h, centrifuged and the supernatant was removed. Subsequently, 250 μL of RNase A (10 mg mL−1 in PBS; Sigma Aldrich) were added to each sample, which was then incubated at room temperature for 30 min and washed twice with DPBS. In the dark, each sample was stained with 20 μg mL−1 of propidium iodide (PI) (eBioscience, Thermo Fisher Scientific) for 15 min before being analyzed using a flow cytometer (BD FACS CANTO™ II). Three independent experiments were performed.q-PCR
For RNA extraction, cells were prepared and incubated with the complexes (or respective medium and DMSO controls) as described above for the ROS determination and cell cycle analyses. Upon detaching and washing twice with DPBS, 1.0 × 106 cells were centrifuged, the supernatant was removed, and the pellet was stored at −20 °C until further use. Total RNA was extracted using the High Pure RNA isolation kit (Roche, Basel, Switzerland) according to the manufacturer's instructions. All the RNA samples were treated with DNase-1 to remove any contaminating genomic DNA, and the purity of the RNA was checked spectroscopically in a NanoDropND-1000 (NanoDrop Technologies). Then, 1 μg of purified RNA was reverse-transcribed using RT2 First Strand Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Gene expression was assessed by real-time PCR using the cDNA obtained. For that, 25 ng of cDNA was amplified in 15 μL of a reaction mix containing Power SYBR Green PCR Master mix (Thermo Fisher Scientific), 20 pmol of each primer pair (ESI Table 1†) and nuclease-free water. The thermal profile consisted of 1 cycle at 95 °C for 10 min followed by 40 cycles at 95 °C for 15 s, 60 °C for 1 min. The human GAPDH cDNA fragment was amplified as the internal control. Data analysis was performed using the 2−ΔΔCt method.Transwell migration assay
Cells starved overnight were detached and seeded onto cell culture inserts in 24-well plates (Millipore transwell PET filters, 8 μm pore; Merck, Kenilworth, NJ, USA) at a density of 1.0 × 104 cells in 150 μL of FBS-free medium, or FBS-free medium containing 0.125% DMSO, 25 μM of FeL or 25 μM of CuL. The lower transwell chambers were filled with 600 μL of media without FBS (negative control) or with medium containing 10% FBS. After 24 h of incubation at 37 °C, the inserts were washed with DPBS, fixed with 4% paraformaldehyde, washed again, and stained with 1 μg mL−1 of Hoechst 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 342 (Thermo Fisher Scientific) for 20 min at room temperature. Cells were then imaged using a 20× objective on a confocal microscope (Zeiss LSM 710). Seven random fields were photographed per insert, with at least two inserts being analyzed for each condition per experiment. The results shown were calculated based on three independent experiments.
342 (Thermo Fisher Scientific) for 20 min at room temperature. Cells were then imaged using a 20× objective on a confocal microscope (Zeiss LSM 710). Seven random fields were photographed per insert, with at least two inserts being analyzed for each condition per experiment. The results shown were calculated based on three independent experiments.
Spheroid viability assay
For spheroids formation, 2.5 × 103 cells were seeded in 100 μL per well in 96-well plates coated with 1.5% agarose (w/v in PBS). After 1 day of incubation, spheroids were fully formed, and 100 μL of fresh medium or medium with DMSO or the complexes was added to a final concentration of 0.125% and 25 μM, respectively. Cells were incubated for 24 h or 72 h at 37 °C before cell viability was estimated using the CellTiter-Glo® 3D assay (Promega, Madison, WI, USA) according to the manufacturer's instructions. Luminescence was read in a CLARIOstar® microplate reader and the average luminescence of 8 spheroids per condition was normalized to the average luminescence of the untreated control sample, for at least two independent experiments.In addition, spheroid viability was also estimated based on spheroid growth. For that, the total area of each spheroid was determined using the INSIDIA macro in FIJI,52 and then normalized to the area of the spheroid at day 0 (to account for possible differences in the spheroids' initial size) and to the size of the untreated spheroids at each time point (to assess the effect of the DMSO and the compounds on spheroids' growth). Several spheroids (at least 7) were analyzed per condition and time point, for at least two independent experiments.
Spheroid invasion assay
Each one-day old spheroid, formed as described above, was collected into a tube, washed once with FBS-free medium, and resuspended in 40 μL of a 4.5 mg mL−1 Matrigel (Cat. Number 356231; Corning) solution in FBS-free medium. Then, each spheroid-containing suspension was spotted onto the centre of a well of a 24-well plate and incubated as a hanging drop for 1 h until the matrigel had polymerized. Complete medium, complete medium with 0.125% DMSO, or complete medium containing 25 μM of the complexes were added and the spheroids were incubated for 24 h at 37 °C before being irradiated (or not as a control) with 6 Gy X-rays on a Faxitron MultiRad225 and further incubated at 37 °C. Images of spheroids and invading cells were acquired immediately after embedment and every 24 h after that, using an Eclipse Ts2 microscope (Nikon). At each time point (24 h, 48 h, and 72 h) the total area of the spheroid and invading cells was determined as described above.Spheroid GSH/GSSG assay
Spheroids were formed and incubated with the compounds or respective controls as described above for the viability assessment. Then, the spheroids were carefully transferred to a white 96-wells polystyrene plate (Greiner Bio-One) and the media was aspirated. Total glutathione and the ratio of GSH/GSSG were then estimated using the GSH/GSSG-Glo™ Assay (Promega) according to the manufacturer's instructions with one minor change: after addition of the lysis buffer, the plate was shaken for 30 min to allow for proper lysis of the spheroids. Luminescence was then read in a CLARIOstar® microplate reader. Three spheroids were analyzed per day and condition, and the average luminescence of those spheroids was normalized to the average luminescence of the untreated control sample. Three independent experiments were performed.Statistics
All data are shown as mean values ± standard error of the mean (S.E.M.) of the DMSO-treated or complex treated samples relative to the untreated control. Statistical and data analysis was carried out using GraphPad Prism 6 software. Statistical differences between treatment and control samples were assessed by one-way ANOVA or two-way ANOVA followed by Dunnett's test. The threshold for statistical significance was set to P = 0.05.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Coordenação de aperfeiçoamento de pessoal de nível superior (CAPES-Brazil) through Project Probral CAPES-DAAD 88881.143979/2017-01. This work was also supported by the Department of Biomedical Physics in Radiation Oncology at the DKFZ. C2TN/IST authors gratefully acknowledge FCT support through the UID/Multi/04349/2019 and PTDC/BTM-TEC/29256/2017 projects.References
- C. Alifieris and D. T. Trafalis, Pharmacol. Ther., 2015, 152, 63–82 CrossRef CAS PubMed.
- Q. Sun, R. Xu, H. B. Xu, G. M. Wang, X. M. Shen and H. Jiang, World J. Surg. Oncol., 2017, 15, 181 CrossRef PubMed.
- J. Rosen, T. Blau, S. J. Grau, M. T. Barbe, G. R. Fink and N. Galldiks, Case Rep. Oncol., 2018, 11, 591–600 CrossRef PubMed.
- D. L. Ma, C. Wu, S. S. Cheng, F. W. Lee, Q. B. Han and C. H. Leung, Int. J. Mol. Sci., 2019, 20, 341 CrossRef PubMed.
- U. Ndagi, N. Mhlongo and M. E. Soliman, Drug Des., Dev. Ther., 2017, 11, 599–616 CrossRef CAS PubMed.
- N. Muhammad and Z. Guo, Curr. Opin. Chem. Biol., 2014, 19, 144–153 CrossRef CAS PubMed.
- A. Bergamo and G. Sava, Chem. Soc. Rev., 2015, 44, 8818–8835 RSC.
- B. Tang, D. Wan, S. H. Lai, H. H. Yang, C. Zhang, X. Z. Wang, C. C. Zeng and Y. J. Liu, J. Inorg. Biochem., 2017, 173, 93–104 CrossRef CAS PubMed.
- Y. H. He, H. Y. Xue, W. D. Zhang, L. Wang, G. Y. Xiang, L. Li and X. M. Shang, J. Organomet. Chem., 2017, 842, 82–92 CrossRef CAS.
- M. C. Ruiz, J. Kljun, I. Turel, A. L. Di Virgilio and I. E. Leon, Metallomics, 2019, 11, 666–675 RSC.
- S. S. Gu, P. Yu, J. N. Hu, Y. N. Liu, Z. W. Li, Y. Qian, Y. Wang, Y. Gou and F. Yang, Eur. J. Med. Chem., 2019, 164, 654–664 CrossRef CAS PubMed.
- B. Tang, D. Wan, Y. J. Wang, Q. Y. Yi, B. H. Guo and Y. J. Liu, Eur. J. Med. Chem., 2018, 145, 302–314 CrossRef CAS PubMed.
- J. P. C. Coverdale, T. Laroiya-McCarron and I. Romero-Canelon, Inorganics, 2019, 7, 31 CrossRef CAS.
- E. D. Williams, D. Gao, A. Redfern and E. W. Thompson, Nat. Rev. Cancer, 2019, 19, 716–732 CrossRef CAS PubMed.
- L. Avila-Carrasco, P. Majano, J. A. Sanchez-Tomero, R. Selgas, M. Lopez-Cabrera, A. Aguilera and G. G. Mateo, Front. Pharmacol., 2019, 10 Search PubMed.
- A. Sánchez-Mora, H. Valdés, M. T. Ramírez-Apan, A. Nieto-Camacho, S. Hernández-Ortega, D. Canseco-González and D. Morales-Morales, Inorg. Chim. Acta, 2019, 496, 119061 CrossRef.
- W. S. Wu, Cancer Metastasis Rev., 2006, 25, 695–705 CrossRef CAS PubMed.
- C. H. Wu, S. C. Tang, P. H. Wang, H. Lee and J. L. Ko, J. Biol. Chem., 2012, 287, 25292–25302 CrossRef CAS PubMed.
- W. Lv, L. L. Sui, X. N. Yan, H. Y. Xie, L. P. Jiang, C. Y. Geng, Q. J. Li, X. F. Yao, Y. Kong and J. Cao, Chem.-Biol. Interact., 2018, 279, 136–144 CrossRef CAS PubMed.
- C. Ninsontia, P. P. Phiboonchaiyanan and P. Chanvorachote, Cancer Cell Int., 2016, 16, 48 CrossRef PubMed.
- S. H. Jeong, Y. J. Jeon and S. J. Park, Mol. Med. Rep., 2016, 14, 5148–5154 CrossRef CAS PubMed.
- J. W. Jiang, K. Wang, Y. Chen, H. N. Chen, E. C. Nice and C. H. Huang, Signal Transduction Targeted Ther., 2017, 2, 17036 CrossRef PubMed.
- A. Horn, I. Vencato, A. J. Bortoluzzi, R. Horner, R. A. N. Silva, B. Spoganicz, V. Drago, H. Terenzi, M. C. B. de Oliveira, R. Werner, W. Haase and A. Neves, Inorg. Chim. Acta, 2005, 358, 339–351 CrossRef CAS.
- A. Horn, I. Vencato, A. J. Bortoluzzi, V. Drago, M. A. Novak and A. Neves, J. Braz. Chem. Soc., 2006, 17, 1584–1593 CrossRef CAS.
- C. Fernandes, A. Horn, O. Vieira-da-Motta, V. M. de Assis, M. R. Rocha, L. D. Mathias, E. S. Bull, A. J. Bortoluzzi, E. V. Guimaraes, J. C. A. Almeida and D. H. Russell, J. Inorg. Biochem., 2010, 104, 1214–1223 CrossRef CAS PubMed.
- A. Horn, A. Neves, I. Vencato, V. Drago, C. Zucco, R. Werner and W. Haase, J. Braz. Chem. Soc., 2000, 11, 7–10 CrossRef CAS.
- A. Horn, L. Firn, A. J. Bortoluzzi, B. Szpoganicz, M. D. Silva, M. A. Novak, M. B. Neto, L. S. Eberlin, R. R. Catharino, M. N. Eberlin and C. Fernandes, J. Mol. Struct., 2006, 797, 154–164 CrossRef CAS.
- J. Fan, Y. W. Ou, C. Y. Wu, C. J. Yu, Y. M. Song and Q. M. Zhan, Acta Pharmacol. Sin., 2012, 33, 1301–1310 CrossRef CAS PubMed.
- T. D. Ribeiro, F. L. Fonseca, M. D. C. de Carvalho, R. M. D. Godinho, F. P. de Almeida, T. D. Saint'Pierre, N. A. Rey, C. Fernandes, A. Horn and M. D. Pereira, Biochem. J., 2017, 474, 301–315 CrossRef CAS PubMed.
- K. A. Mapuskar, C. M. Anderson, D. R. Spitz, I. Batinic-Haberle, B. G. Allen and R. E. Oberley-Deegan, Semin. Radiat. Oncol., 2019, 29, 72–80 CrossRef PubMed.
- F. De Lazzari, L. Bubacco, A. J. Whitworth and M. Bisaglia, Aging Dis., 2018, 9, 716–728 CrossRef PubMed.
- I. Batinic-Haberle, A. Tovmasyan and I. Spasojevic, Redox Biol., 2015, 6, 656 CrossRef CAS.
- T. P. Ribeiro, C. Fernandes, K. V. Melo, S. S. Ferreira, J. A. Lessa, R. W. A. Franco, G. Schenk, M. D. Pereira and A. Horn, Free Radicals Biol. Med., 2015, 80, 67–76 CrossRef CAS PubMed.
- R. O. Costa, S. S. Ferreira, C. A. Pereira, J. R. Harmer, C. J. Noble, G. Schenk, R. W. A. Franco, J. A. L. C. Resende, P. Comba, A. E. Roberts, C. Fernandes and A. Horn, Front. Chem., 2018, 6, 491 CrossRef CAS PubMed.
- A. Horn, G. L. Parrilha, K. V. Melo, C. Fernandes, M. Horner, L. D. Visentin, J. A. S. Santos, M. S. Santos, E. C. A. Eleutherio and M. D. Pereira, Inorg. Chem., 2010, 49, 1274–1276 CrossRef CAS PubMed.
- U. Weser and L. M. Schubotz, J. Mol. Catal., 1981, 13, 249–261 CrossRef CAS.
- J. Switala and P. C. Loewen, Arch. Biochem. Biophys., 2002, 401, 145–154 CrossRef CAS PubMed.
- B. Kalyanaraman, V. Darley-Usmar, K. J. A. Davies, P. A. Dennery, H. J. Forman, M. B. Grisham, G. E. Mann, K. Moore, L. J. Roberts and H. Ischiropoulos, Free Radicals Biol. Med., 2012, 52, 1–6 CrossRef CAS PubMed.
- S. Tuncer, R. Gurbanov, I. Sheraj, E. Solel, O. Esenturk and S. Banerjee, Sci. Rep., 2018, 8, 14828 CrossRef PubMed.
- J. Lu and A. Holmgren, Free Radicals Biol. Med., 2014, 66, 75–87 CrossRef CAS PubMed.
- Z. Liao, D. Chua and N. S. Tan, Mol. Cancer, 2019, 18, 65 CrossRef PubMed.
- E. H. Verbon, J. A. Post and J. Boonstra, Gene, 2012, 511, 1–6 CrossRef CAS PubMed.
- G. K. Schwartz and M. A. Shah, J. Clin. Oncol., 2005, 23, 9408–9421 CrossRef CAS PubMed.
- T. Ishiguro, H. Ohata, A. Sato, K. Yamawaki, T. Enomoto and K. Okamoto, Cancer Sci., 2017, 108, 283–289 CrossRef CAS PubMed.
- S. Nath and G. R. Devi, Pharmacol. Ther., 2016, 163, 94–108 CrossRef CAS PubMed.
- M. Wank, D. Schilling, T. E. Schmid, B. Meyer, J. Gempt, M. Barz, J. Schlegel, F. Liesche, K. A. Kessel, B. Wiestler, S. Bette, C. Zimmer and S. E. Combs, Cancers, 2018, 10, 456 CrossRef CAS PubMed.
- A. Bansal and M. C. Simon, J. Cell Biol., 2018, 217, 2291–2298 CrossRef CAS PubMed.
- M. Grau, F. Rigodanza, A. J. P. White, A. Soraru, M. Carraro, M. Bonchio and G. J. P. Britovsek, Chem. Commun., 2014, 50, 4607–4609 RSC.
- G. N. Ledesma, H. Eury, E. Anxolabehere-Mallart, C. Hureau and S. R. Signorella, J. Inorg. Biochem., 2015, 146, 69–76 CrossRef CAS PubMed.
- R. F. Beers and I. W. Sizer, J. Biol. Chem., 1952, 195, 133–140 CAS.
- J. P. N. Ribeiro, M. A. Segundo, S. Reis and J. L. F. C. Lima, Talanta, 2009, 79, 1169–1176 CrossRef CAS PubMed.
- C. Moriconi, V. Palmieri, R. Di Santo, G. Tornillo, M. Papi, G. Pilkington, M. De Spirito and M. Gumbleton, Biotechnol. J., 2017, 12 DOI:10.1002/biot.201700140.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra00166j |
| ‡ Equally contributing authors. |
| This journal is © The Royal Society of Chemistry 2020 |