Open Access Article
Open Access ArticleTwo novel ACE inhibitory peptides isolated from longan seeds: purification, inhibitory kinetics and mechanisms†
Atthasith Nuchpraphaa,
Supawee Paisansaka,
Papassara Sangtanoob,
Piroonporn Srimongkolb,
Tanatorn Saisavoeyb,
Onrapak Reamtongc,
Kiattawee Choowongkomond and
Aphichart Karnchanatat *b
*b
aProgram in Biotechnology, Faculty of Science, Chulalongkorn University, 254 Phayathai Road, Pathumwan, Bangkok 10330, Thailand
bResearch Unit in Bioconversion/Bioseparation for Value-Added Chemical Production, Institute of Biotechnology and Genetic Engineering, Chulalongkorn University, 254 Phayathai Road, Pathumwan, Bangkok 10330, Thailand. E-mail: Aphichart.K@chula.ac.th
cDepartment of Molecular Tropical Medicine and Genetics, Faculty of Tropical Medicine, Mahidol University, 420/6 Ratchawithi Road, Ratchathewi, Bangkok 10400, Thailand
dDepartment of Biochemistry, Faculty of Science, Kasetsart University, Bangkok 10903, Thailand
First published on 30th March 2020
Abstract
Angiotensin converting enzyme (ACE) inhibition offers a useful means of managing hypertension, because ACE inhibitors (ACEIs) are known to serve as agents with antihypertensive properties in addition to generating positive metabolic and cardioprotective outcomes. However, current ACEIs are linked to adverse consequences, and so there is a requirement for effective but safer compounds, which might be achieved through chemical synthesis or the isolation of naturally obtained bioactive molecules. Protein hydrolysates with ACEI activity can be produced by the combined pepsin and pancreatin proteolysis (to mimic gastrointestinal digestion) of longan seed protein. This study examined longan seed protein hydrolysates, obtained from a sequential 3 h digestion with pepsin and then pancreatin. The resulting hydrolysate underwent sequential ultrafiltration membrane fractionation with a 10, 5, and 3 kDa molecular weight cut-off (MWCO). The permeate derived from the <3 kDa MWCO demonstrated the highest ACEI activity. This permeate subsequently underwent separation by reverse-phase high performance liquid chromatography to give the main fractions on the basis of differing elution times. The ACEI IC50 values for these fractions were then identified. Quadrupole time-of-flight tandem mass spectrometry was employed to determine the peptide mass for the major peak (F5), which was shown to be Glu–Thr–Ser–Gly–Met–Lys–Pro–Thr–Glu–Leu (ETSGMKPTEL) and Ile–Ser–Ser–Met–Gly–Ile–Leu–Val–Cys–Leu (ISSMGILVCL). These two peptides were stable over a temperature and pH range of −20 to 90 °C and 2–12, respectively, for 60 min. From the Lineweaver–Burk plot, both peptides inhibited ACE non-competitively. Molecular docking simulation of the peptides with ACE supported the formation of hydrogen bonds by the peptides with the ACE active pockets. This research indicates that it may be possible to use both of these peptides or longan seed protein hydrolysates in order to create ingredients for functional foods, or to produce pharmaceutical products, capable of lowering hypertension.
1. Introduction
Hypertension, or high blood pressure, accounts for more than 20 million visits to doctors annually. The condition afflicts all age groups, although the elderly tend to be more significantly affected, with the incidence rising in those over 50 years old. It is believed to be the most common of all the cardiovascular disorders, with around a billion people globally affected to some degree, and is a significant contributor to both morbidity and mortality. There are no recognizable symptoms, and so it can be difficult to diagnose hypertension, which makes management of the condition difficult.1 Numerous cardiovascular diseases (CVD) have been linked to hypertension, including arteriosclerosis, congestive heart failure, end-stage renal diseases, coronary heart disease, stroke, and myocardial infarction. In addition, it can also be a cause of abnormal renal function and ultimately lead to renal failure.2,3Blood pressure can be regulated by angiotensin I-converting enzyme (ACE; EC 3.4.15.1) in vivo operating via the renin–angiotensin system (RAS) along with the kallikrein–kinin system (KKS). Within the RAS, the release of the vasopressor angiotensin II from the cleavage of the C-terminal dipeptide from angiotensin I is performed by ACE. Meanwhile, in the KKS, ACE serves to inactivate the vasodilator bradykinin by removing the two C-terminal dipeptides.4,5 As a result, ACE-inhibitors (ACEIs) can be employed for the purpose of reducing blood pressure. There are several synthetic ACEIs that have been employed to treat hypertension in human patients, but the drawback is that they can induce various side effects, including coughing, hypotension, heightened serum levels of potassium, and abnormalities of the fetus.6,7 Accordingly, it is essential to develop new ACEIs that will provide safe and effective hypertension treatment and, because of the need for safety, there is growing interest in ACEI peptides that can be derived from food.8–10
Both the food and non-food sectors are taking an increased interest in proteins derived from plant sources. In the food sector, proteins from soy, legumes, and wheat gluten are very appealing, and the agricultural industry is of interest to other sectors who wish to investigate the extraction of proteins from agricultural by-products. Plant processed waste is often rich in proteins, and so there is an increasing interest in developing nutritional supplements or nutraceuticals from this type of waste, since it can offer strong nutritional qualities along with a number of important amino acids.11 Agricultural by-products might be readily converted into products with applications in other industries. One advantage of using such agricultural by-products to useful ends is that the environmental issues associated with the disposal of such by-products can be eliminated.12
Thai fruit is ingrained in the country's cultural identity. Fruit is both plentiful and geographically widespread in Thailand,13 and is highly valued as a healthy food product. However, when fruit is processed by the food industry, such as during the canning or drying process, there are waste by-products, including seeds. Those seeds are usually disposed of if they are without value, or are sometimes sold as fuel at minimal prices. Earlier studies, however, have shown that useful nutrients can be stored in seeds.14 One such example is the longan (Dimocarpus longan Lour.), which is a Chinese evergreen plant from the Sapindaceae family. Many tropical countries grow this tree for the fruit and so studies have been conducted to assess whether it offers potential pharmacological benefits to human health, especially through the antimicrobial and antioxidant properties of its seeds.15–17
Previous works have focused on non-edible seeds from lychee (Litchi chinensis Sonn.), which produce a protein hydrolysate offering anti-inflammatory activity,18 but it is important to determine the active ingredient(s) in such protein hydrolysates, since the findings may be of interest in the both the pharmaceutical sector and food industry. When plant proteins are digested in vitro with pepsin and pancreatin, it may be the case that bioactive peptides are produced that can play effective roles in developing certain food products. These enzymes, which are required in the process of human gastrointestinal digestion, might prove informative when considering the production of bioactive peptides when these proteins are digested. Accordingly, this study sought to examine the generation of ACEI peptides resulting from the digestion of longan seed protein isolates with pepsin and then pancreatin.
The enriched ACEI peptides from the longan seed source was performed via membrane filtration and RP-HPLC followed by mass spectrometry sequencing for identification. It was also considered possible that the purified peptide might undergo binding at the ACE active site, if the outcomes of molecular docking experiments are interpreted correctly. It is demonstrated that longan seeds, which have hitherto found few applications, might prove to be nutritionally valuable by yielding bioactive peptides that could treat hypertension. The results obtained in this research study may augment the current information available in the database detailing medicinal plants.
2. Materials and methods
2.1 Materials
The longan seeds used in this study were provided by the Malee Group Public Company Limited (Nakhon Pathom, Thailand). Having obtained the samples, they were held in cold (4 °C) and dark conditions until used. The ACE (E.C. 3.4.15.1) from the lungs of rabbits was supplied by Sigma Chemical Co. (St. Louis, MO, USA). The same supplier also provided the acetonitrile (ACN), bovine serum albumin (BSA), captopril, hippuric acid, hippuryl-L-histidyl-L-leucine (HHL), pancreatin from porcine pancreas, pepsin from porcine gastric mucosa, and trifluoroacetic acid (TFA). All materials used in the study were of analytical grade.2.2 Preparation of crude protein from longan seeds
The preparation process followed the guidance of Saisavoey et al.18 commencing with the cleaning and blending of 1.5 kg of longan seeds using 5 L of phosphate buffered saline (PBS; 20 mM phosphate buffer with 0.15 M NaCl pH 7.2). At first, a mixer was employed, followed by overnight stirring at 40 °C with a magnetic stirrer. Filtration was performed using a double-layered cheesecloth in order to obtain a suspension that could be isolated from the liquid. The filtrate was clarified by centrifugation (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 × g, 30 min, 4 °C) before harvesting of the supernatant. Ammonium sulfate was then introduced to 80% saturation and left overnight with stirring at 4 °C followed by centrifugation (15
900 × g, 30 min, 4 °C) before harvesting of the supernatant. Ammonium sulfate was then introduced to 80% saturation and left overnight with stirring at 4 °C followed by centrifugation (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 × g, 30 min, 4 °C) to harvest the insoluble pellet. The pellet was solubilized in PBS, dialyzed against excessive quantities of double deionized water, and then freeze dried in order to generate the required crude protein.
900 × g, 30 min, 4 °C) to harvest the insoluble pellet. The pellet was solubilized in PBS, dialyzed against excessive quantities of double deionized water, and then freeze dried in order to generate the required crude protein.
2.3 Amino acid composition in crude protein from longan seeds
The AOAC 994.12 (ref. 19) and 988.15 (ref. 20) techniques were employed to determine the crude protein amino acid content, whereby 25 mg (dry weight) of longan seed crude protein was dissolved in 5 mL of 6 M hydrochloric acid (HCl) and heated for 24 h at 110 °C. Then 10 μL of 2.5 mM L-amino-n-butyric acid in 0.1 M HCl was added as an internal standard followed by deionized water to a volume of 250 μL. This was heated at 55 °C for 10 min prior to analysis using reversed-phase high-performance liquid chromatography (RP-HPLC) with a Hypersil GOLD column C18 (4.6 mm × 150 mm, 3 μM). Elution was performed with a sodium acetate buffer at pH 4.9 and 60% (v/v) ACN at 0.3 mL min−1. The amino acid content was then established by ALS Laboratory Group (Thailand) Co., Ltd., Suan Luang, Bangkok, Thailand.2.4 Hydrolysis of crude protein by pepsin and pancreatin
Following the technique of Inthuwanarud et al.21 the crude proteins obtained from the longan seeds served as the substrate for forming the protein hydrolysate. The crude protein preparation (1 mg mL−1) was added to pepsin to a final substrate: enzyme concentration ratio of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, while the pH was set at 2.5 using 1 M HCl. Hydrolysis was performed for 3 h at 37 °C with shaking at 180 rpm. The activity was then halted by raising the pH to 7.5 through the introduction of 1 M (NaOH). This procedure was then repeated except using pancreatin instead of pepsin, and the reaction was halted by heating at 80 °C for 20 min. The hydrolysates were clarified by centrifugation (15
1, while the pH was set at 2.5 using 1 M HCl. Hydrolysis was performed for 3 h at 37 °C with shaking at 180 rpm. The activity was then halted by raising the pH to 7.5 through the introduction of 1 M (NaOH). This procedure was then repeated except using pancreatin instead of pepsin, and the reaction was halted by heating at 80 °C for 20 min. The hydrolysates were clarified by centrifugation (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 × g, 30 min, 4 °C) and the supernatant was evaluated for ACEI activity. These enzymic processes are broadly similar to those which would occur naturally inside the human gastro-intestinal tract.
900 × g, 30 min, 4 °C) and the supernatant was evaluated for ACEI activity. These enzymic processes are broadly similar to those which would occur naturally inside the human gastro-intestinal tract.
2.5 Fractionation of the hydrolysate via ultrafiltration
Suttisuwan et al.22 noted that ultrafiltration can be employed to fractionate pepsin and pancreatin hydrolysates using a high-performance ultrafiltration cell (Pellicon XL Filter; Merck Millipore, Billerica, MA). In this study, three molecular weight cut-off (MWCO) membranes were sequentially employed (10, 5, and 3 kDa MWCO) to create four fractions (>5–10, 3–5, and <3 kDa). Having clarified the fractions by centrifugation (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 × g, 30 min, 4 °C), they were then passed through the 10 kDa MWCO membrane first, recycling the retentate into a second 10 kDa MWCO membrane fractionation. The permeate derived from the 10 kDa membrane was then passed via the 5 kDa membrane, recycling the retentate into a second 5 kDa MWCO membrane fractionation. This sequence was then repeated with the 3 kDa MWCO membrane, thus lowering the potential for smaller molecular weight fractions to contaminate the larger fractions.
900 × g, 30 min, 4 °C), they were then passed through the 10 kDa MWCO membrane first, recycling the retentate into a second 10 kDa MWCO membrane fractionation. The permeate derived from the 10 kDa membrane was then passed via the 5 kDa membrane, recycling the retentate into a second 5 kDa MWCO membrane fractionation. This sequence was then repeated with the 3 kDa MWCO membrane, thus lowering the potential for smaller molecular weight fractions to contaminate the larger fractions.
2.6 Assay of ACEI activity
The technique described by Yodjun et al.23 was used to measure the ACEI activity, whereby 50 μL of the crude protein derived from longan seeds was combined with an equal amount of ACE (25 mU mL−1) and underwent pre-incubation for 10 min at 37 °C. The next step then involved re-incubation for 30 min using 150 μL of substrate (10 mM HHL in PBS) at 37 °C. The reaction was stopped by the addition of 250 mL of 1 M HCl. The resulting hippuric acid was extracted in 500 μL of ethylacetate with centrifugation at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 × g, and 4 °C for 15 min, whereupon 200 μL of the upper layer was placed in a new test tube at room temperature and evaporated in a vacuum. Once the hippuric acid had subsequently been dissolved in 500 μL of double deionized water, the absorbance was measured at 250 nm (A250) using a UV-spectrophotometer. The amount of hippuric acid released in the assay mixture was then determined from the A250 value by comparison to a standard curve derived from known hippuric acid concentrations. Captopril was used as the positive control. The equation 100 × [(C − Bi) − (S − Bs)/(C − Bi)] was employed to assess the percentage ACEI activity, where C represents the A250 of the control, S indicates the A250 of the sample, Bs denotes the A250 of the peptide only sample, and Bi indicates the A250 of the blank inhibitor. The IC50 value, as the peptide concentration of hydrolysate required to obtain a 50% ACEI activity, was calculated by non-linear regression using the GraphPad Prism Version 6 software (GraphPad Software Inc., La Jolla, CA, USA).
900 × g, and 4 °C for 15 min, whereupon 200 μL of the upper layer was placed in a new test tube at room temperature and evaporated in a vacuum. Once the hippuric acid had subsequently been dissolved in 500 μL of double deionized water, the absorbance was measured at 250 nm (A250) using a UV-spectrophotometer. The amount of hippuric acid released in the assay mixture was then determined from the A250 value by comparison to a standard curve derived from known hippuric acid concentrations. Captopril was used as the positive control. The equation 100 × [(C − Bi) − (S − Bs)/(C − Bi)] was employed to assess the percentage ACEI activity, where C represents the A250 of the control, S indicates the A250 of the sample, Bs denotes the A250 of the peptide only sample, and Bi indicates the A250 of the blank inhibitor. The IC50 value, as the peptide concentration of hydrolysate required to obtain a 50% ACEI activity, was calculated by non-linear regression using the GraphPad Prism Version 6 software (GraphPad Software Inc., La Jolla, CA, USA).
2.7 Assessment of protein content
Bradford's procedure24,25 was used to determine the protein content, using BSA as the standard at 5, 10, 15, and 20 μg mL−1 in order to plot the calibration curve. A 50 μL aliquot of each two-fold sample dilution in deionized water was added to each of three wells in a microtiter plate, followed by the addition of 50 μL of Bradford's reagent to each of the wells, shaken to mix for 5 min and left to stand for 10 min. The optical density at 595 nm was then measured using an ELISA plate reader and converted to the protein concentration using the linear equation derived directly from the standard curve.2.8 RP-HPLC
The peptide solution offering the best ACEI activity after ultrafiltration underwent further separation by RP-HPLC (Spectra System, Thermo Fisher Scientific) using a Luna 5U C18 100 A° column (4.6 × 250 mm, Luna 5 μM; Phenomenex, Torrance, CA). Samples were filtered through a nylon membrane of 0.45 μm (Whatman, GE, Buckinghamshire, UK) and loaded into the column at 20 μL (protein concentration of 1.0–1.50 mg protein per mL). Elution was performed at 25 °C with a three-phase linear gradient of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0% (v/v) A
0% (v/v) A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B declining to 90
B declining to 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 over 15 min, to 65
10 over 15 min, to 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 over 30 min, and to 55
35 over 30 min, and to 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 over 40 min with a flow rate of 0.7 mL min−1. Mobile phase A was 0.1% (v/v) TFA, while B was 70% (v/v) ACN in 0.05% (v/v) TFA. Chromatographic analysis was performed using ChromQuest Software (Thermo Fisher Scientific). Assessment of the peptide peaks at 280 nm was based upon from leaving the baseline until returning to the baseline.
45 over 40 min with a flow rate of 0.7 mL min−1. Mobile phase A was 0.1% (v/v) TFA, while B was 70% (v/v) ACN in 0.05% (v/v) TFA. Chromatographic analysis was performed using ChromQuest Software (Thermo Fisher Scientific). Assessment of the peptide peaks at 280 nm was based upon from leaving the baseline until returning to the baseline.
2.9 Liquid chromatography-quadrupole time-of-flight electrospray-ionization tandem mass spectrometry (LC-Q-TOF-ESI-MS/MS) and de novo peptide sequencing
The RP-HPLC fraction (F5) with a strong ACEI activity and sufficient yield was subject to LC-Q-TOF-ESI-MS/MS analysis to identify the peptides after calibrating the tool to identify peptide chains in the mass range of m/z 50–25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (Model Amazon SL, Bruker, Germany). The evaluation was performed with de novo sequencing, while Mascot was used in the analysis of all of the MS data. The sequences which were obtained were then compared with findings available through the NCBI database using BLASTp.
000 (Model Amazon SL, Bruker, Germany). The evaluation was performed with de novo sequencing, while Mascot was used in the analysis of all of the MS data. The sequences which were obtained were then compared with findings available through the NCBI database using BLASTp.
2.10 Synthesis of peptides
For the two peptides whose sequences were obtained via the MS analysis (Section 2.9) their synthesis was performed using a 433A Synergy solid phase peptide synthesizer (Applied Biosystems, CA). The synthetic peptides Glu–Thr–Ser–Gly–Met–Lys–Pro–Thr–Glu–Leu (ETSGMKPTEL) and Ile–Ser–Ser–Met–Gly–Ile–Leu–Val–Cys–Leu (ISSMGILVCL) were first verified through analytical MS (Model Finnigan™ LXQ™, Thermo Fisher Scientific Inc., MA) along with the use of a Surveyor HPLC. These pure synthesized peptides were then screened to confirm and characterize their ACEI activity (Section 2.6) for comparison with that of the peptide fraction obtained from RP-HPLC fractionation (Sections 2.8 and 2.9).2.11 Peptide properties
For the design of future research, it is important to understand the ACEI peptide properties. It is possible to locate useful parameter scans in the online database. Toxicity analysis of the peptides were performed using the ToxinPred server (http://crdd.osdd.net/raghava/toxinpred/), while the online Innovagen server was used to assess peptide solubility (http://www.innovagen.com/proteomics-tools).2.12 Temperature influence on ACE inhibitory activity
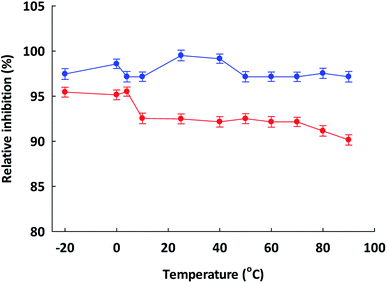
Temperature can also affect the ACEI activity, and was evaluated by incubating TSGMKPTEL and ISSMGILVCL in 20 mM phosphate buffer at pH 7.2 at various temperatures (−20 to 90 °C using 10 °C increments) for 60 min. The samples were then cooled to 4 °C before examination of the residual ACEI activity (Section 2.6). The uninhibited mixture, set as 100% inhibition, was used to normalize the attained activities for the positive control, while the negative control was a maximum inhibited mixture (attained how?). Inhibitory activities are reported as relative to the uninhibited mixture activity. All assays had ACE.2.13 Influence of pH upon ACE inhibitory activity
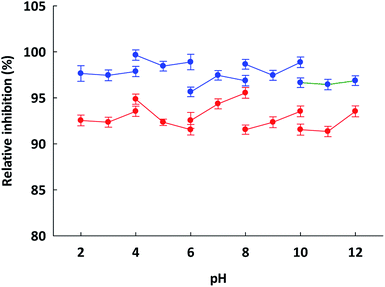
The pH stability and pH optima for the ACEI activity were assessed through the incubation of ETSGMKPTEL and ISSMGILVCL in buffers at similar levels of salinity but with pH values varying in the range of 2–14 using 20 mm glycine–HCl (pH 2–4), sodium acetate (pH 4–6), potassium phosphate (pH 6–8), Tris–HCl (pH 8–10), and glycine–NaOH (pH 10–12). The respective peptide–buffer mixture was left to stand at room temperature for 60 min before the ACEI activity was ascertained (Section 2.6) and compared to the negative or positive controls and reported as the relative percentage ACEI activity. All assays had ACE.2.14 Kinetic parameter estimation
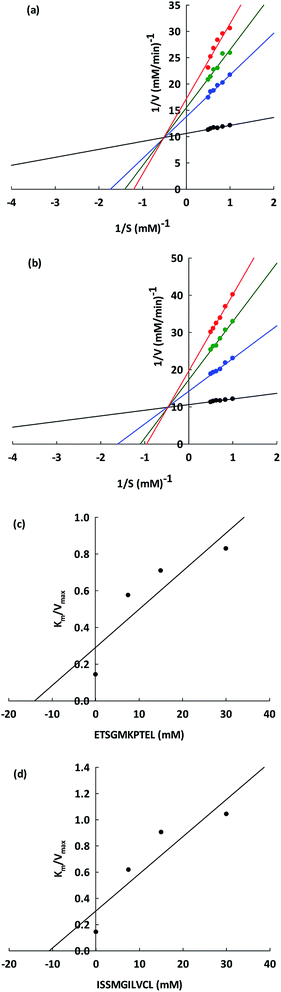
The hydrolysis rate for HHL by ACE over a range of HHL concentrations of 0–5.0 mM was measured while varying the concentrations of ETSGMKPTEL and ISSMGILVCL. The data underwent double-reciprocal analysis using the Lineweaver–Burk plot in order to establish the Michaelis–Menten constant (Km), the maximum velocity (Vmax), and the mode of ACE inhibition. The Dixon plot was used to assist in determining the inhibitor constant (Ki) by serving as a representation of the Lineweaver–Burk plot slope.2.15 Molecular docking of ETSGMKPTEL and ISSMGILVCL at the ACE binding site
Molecular docking was performed using the Hermes 1.10.1 software. The Protein Data Bank (http://www.rcsb.org/pdb/home/home.do) provided details of the three-dimensional (3D) crystal structure for a human ACE–lisinopril complex (1O8A.pdb), which had a binding site with a radius of 6 Å capable of serving as the receptor for this research study. Discovery Studio 2.0 software was used to prepare the molecular structures for each of the ETSGMKPTEL and ISSMGILVCL peptides.26 Prior to docking, preparation of the model receptor ACE molecule was performed by removing all of the water molecules, and adding the hydrogen atoms to the receptor model. Molecular docking evaluation was performed by assessing the Goldscore, ChemScore, ChemPLP, and ASP. The binding energy and fitness scores provided the detail to determine the most suitable pose for each of the residues. The hydrogen bonds (H-bonds) were identified with the Discovery Studio 20 software (specify the cut-off criteria used to accept a H-bond), along with the nature of the various interactions between the ACE molecules and the peptide residues (hydrophobic, hydrophilic, electrostatic, van der Waals, or coordination interactions).2.16 Statistical analysis
Each experiment was performed in triplicate, and the findings are presented as the mean ± one standard error of the mean (SEM). Analysis was performed using SPSS 11.5 statistical software. Hypotheses were tested using one-way ANOVA and the least significant difference (LSD) test accepting significance at the P < 0.05 level.3. Results and discussion
3.1 Longan seeds' amino acid composition
Food proteins that offer suitable levels of amino acids can also potentially be used for antihypertensive purposes. For each specific peptide, the particular function is established by the type of the constituent amino acids along with the R-group differences. In general, longan seeds offer aromatic amino acids in addition to hydrophobic aliphatic amino acids (Table 1), including methionine, isoleucine, alanine, leucine, and proline, which are able to support the ACEI ability of food proteins. Daskaya-Dikmen et al.27 stated that a strong ACEI activity is clearly linked to hydrophobic amino acid residues in peptides, especially at the C-terminal position. Moreover, those peptides that contain proline are understood to resist enzyme digestion in the intestines, and can easily reach the blood circulatory system. Manoharan et al.28 suggested that the proline found in the C-terminal position of the peptides could bind effectively to ACE molecules. Furthermore, the ACEI drugs that are commercially available, such as captopril and enalapril, have proline as a part of their structure.29 From analysis of the amino acid composition it was apparent that longan seeds might contain peptides which offer strong ACEI activity.| Amino acid type | Amino acid | % (w/w) |
|---|---|---|
| a Findings appear in the form of mean ± SEM (n = 3). | ||
| Hydrophilic | Arginine (Arg) | 0.54 ± 0.022 |
| Aspartic acid (Asp) | 0.49 ± 0.014 | |
| Glutamic acid (Gln) | 0.55 ± 0.071 | |
| Histidine (His) | 0.11 ± 0.006 | |
| Lysine (Lys) | 0.35 ± 0.011 | |
| Serine (Ser) | 0.30 ± 0.031 | |
| Threonine (Thr) | 0.29 ± 0.048 | |
| Hydrophobic | Alanine (Ala) | 0.80 ± 0.077 |
| Cysteine (Cys) | 0.73 ± 0.045 | |
| Glycine (Gly) | 0.44 ± 0.002 | |
| Isoleucine (Ile) | 0.76 ± 0.007 | |
| Leucine (Leu) | 0.36 ± 0.018 | |
| Methionine (Met) | 1.08 ± 0.086 | |
| Phenylalanine (Phe) | 0.28 ± 0.091 | |
| Proline (Pro) | 1.25 ± 0.074 | |
| Tryptophan (Trp) | 1.09 ± 0.031 | |
| Tyrosine (Tyr) | 0.14 ± 0.054 | |
| Valine (Val) | 0.29 ± 0.008 | |
| Total | 9.44 ± 0.161 | |
3.2 Hydrolysis of longan seed protein isolates using pepsin–pancreatin
Pepsin is a proteolytic enzyme released in the stomach, while the pancreas inside the small intestine produces pancreatin, which contains elastase, trypsin, and chymotrypsin proteases. The peptides produced by extensive pepsin–pancreatin digestion are, therefore, likely to be formed by and resistant to human gastro-intestinal digestion and may be absorbed by digestive epithelial cells in the small intestine. This indicates their potential bioavailability to perform their normal biological activities.30 The longan seed protein isolates underwent hydrolysis with pepsin–pancreatin and then were screened for ACEI activity. The longan seed protein isolates that had not been hydrolyzed did not demonstrate any ACEI activity, but rather ACEI activity was only detected after the enzymatic hydrolysis. The pepsin–pancreatin hydrolysate presented an ACEI activity with an IC50 value of 1.74 ± 0.006 mg mL−1 (Table 2). Previously, ACEIs were reported in the protein hydrolysates of peas and whey but at a much greater ACEI activity with IC50 values of 0.070 and 0.041 mg mL−1, respectively.31 Further ACEI inhibitory activity was reported from cowpea protein hydrolysates, but with IC50 values in the range of 0.04–170.6 (Flavourzyme hydrolysate), 24.3–123 (Alcalase hydrolysate), and 44.7–112 (pepsin–pancreatin hydrolysate) μg mL−1.32 Meanwhile, the IC50 values for chickpea desi and chickpea kabuli proteins, which had undergone hydrolysis by simulated in vitro gastrointestinal processes, were 140 ± 1 μg mL−1 and 229 ± 1 μg mL−1 respectively.33| Molecular weight (kDa) | ACE inhibitory activity (IC50; mg mL−1) |
|---|---|
| a The findings appear in the form of mean ± SEM (n = 3). Those mean values which are shown with a different superscript in each column exhibit significant differences (P < 0.05). | |
| Crude extract | >5.00 |
| Crude protein | >5.00 |
| Protein hydrolysate | 1.74 ± 0.006c |
| >10 | 1.37 ± 0.017b,c |
| 5–10 | 0.95 ± 0.006a,b |
| 3–5 | 0.43 ± 0.011a,b |
| <3 | 0.25 ± 0.004a |
3.3 Ultrafiltration
Peptides can be separated on the basis of their molecular weight (as 3D size) by ultrafiltration. This approach is a relatively simple, reliable, and efficient means of preparing samples for RP-HPLC. Using 10, 5 and 3 kDa MWCO ultrafiltration membranes sequentially, the longan seed protein hydrolysate (IC50 = 1.74 ± 0.006 mg mL−1) was separated into four size fractions of >10 kDa, 5–10 kDa, 3–5 kDa, and <3 kDa, and then each was screened for ACEI activity. The <3 kDa fraction showed the greatest ACEI activity with an IC50 value of 250 ± 4 μg mL−1 (Table 2). It is possible for smaller peptides to pass through the wall of the intestines to enter the bloodstream if they survive the process of gastrointestinal digestion. Because these peptides in this study were obtained after extensive (assumed complete) pepsin–pancreatin digestion they are likely to be stable to human digestion and so able to exert their inhibitory influence even after they have reached the bloodstream.That these small peptides display ACEI activity in part may reflect that there is a tiny groove in ACE which prevents large polypeptides from accessing the active site. The MW of the hydrolysate is lower than that of the strongest ACEI peptides derived from the tuna frame hydrolysate generated by pepsin (MW of 2482 Da and IC50 value of 11.3 μM). The findings reported by Arise et al.34 were similar, in that the smallest groundnut protein hydrolysate (MW < 1 kDa) revealed the greatest ACEI activity, where a five-fold improvement in the ACEI purity was achieved following sequential ultrafiltration. Wan Mohtar et al.35 described the winged bean (Psophocarpus tetragonolobus) protein hydrolysate in the context of the four proteolytic enzymes (flavourzyme, alcalase, bromelain, and papain) after ultrafiltration at a MWCO of 10, 5 and 2 kDa, where the <2 kDa fraction provided the greatest level of ACEI activity (IC50 values of 0.003 and 0.130 mg mL−1). Accordingly, the MW < 3 kDa fraction of the longan seed hydrolysate was further fractionated by RP-HPLC.
3.4 RP-HPLC fractionation of the <3 kDa MWCO fraction of logan seed hydrolysate
The <3 kDa MWCO peptide fraction was further fractionated by RP-HPLC using a RP-C18 column with gradient elution. Fig. 1 presents the chromatographic profile, where five main fractions were obtained on the basis of their hydrophobicity. The fractions (F1–5) were collected and lyophilized, whereupon the peptide concentration and ACEI activity of each fraction was assessed. The F2 and F3 subfraction yields were not high enough to accurately measure the ACEI IC50 value, while those for fractions F1, F4, and F5 were 24.57 ± 0.015, 37.36 ± 0.007, and 18.22 ± 0.029 μg mL−1, respectively. The F5 sub-fraction was then subjected to peptide amino acid sequencing through the use of Q-TOF-ESI-MS/MS analysis.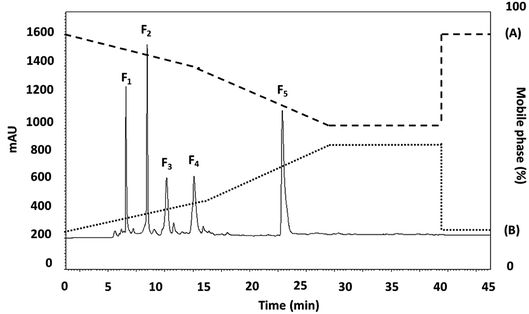 | ||
| Fig. 1 The RP-HPLC profile of the active fraction (<3 kDa) obtained from longan seeds protein hydrolysate. | ||
It was previously reported for chickpea protein hydrolysates that the peptides that were the last to be eluted from the RP-HPLC had the highest ACEI activity.36 One argument is that ACEI peptides have a high level of hydrophobic amino acids, and so have a greater retention time within the hydrophobic chromatography column. For example, it is very important that ACEI peptides have hydrophobic qualities, often seen at the peptide N-terminal, and hydrophilic peptides are incompatible with the active ACE site, and so have almost no ACEI activity as a consequence. Accordingly, if hydrophobic amino acids are removed, the inhibition capability of ACE might be adversely affected because of the heightened affinity for active ACE sub-sites of the hydrophobic peptides.37
3.5 Identification of peptide sequences in fraction F5
The amino acid sequences for identification of the peptides in the RP-HPLC F5 sub-fraction, which had ACEI activity was determined using LC-MS/MS ESI Q-TOF de novo sequencing aided by searching the MASCOT database. The de novo sequencing process resulted in 100% identification on the basis of the Sapindaceae family as the peptides Glu–Thr–Ser–Gly–Met–Lys–Pro–Thr–Glu–Leu (ETSGMKPTEL; 1092.22 Da) and Ile–Ser–Ser–Met–Gly–Ile–Leu–Val–Cys–Leu (ISSMGILVCL; 1035.33 Da) (see Fig. S1†).These two peptide sequences were then commercially synthesized and their ACEI activity was evaluated. The findings confirmed that both synthesized peptides had an ACEI activity with an IC50 value of 2.15 ± 0.016 μM and 3.88 ± 0.004 μM for ETSGMKPTEL and ISSMGILVCL, respectively. In the case of ETSGMKPTEL, a 100% (10/10) amino acid sequence identity to 6-phosphogluconate dehydrogenase (partial – all from D. longan) was found, with 88% similarity to the kelch repeat-containing protein of D. longan. The ISSMGILVCL peptide showed similarity to a ferredoxin (partial), and the NBS-LRR disease-resistant protein NBS11 (100%), with 80% similarity to the glutathione transferase from D. longan. These peptides can be investigated for specific conflict sites, and might be able to replace polymorphic regions with proteins possibly derived from divergent sub-units.
The BIOPEP database was used to search for possible ACEI activity from the peptides identified in the study through comparison with previously identified sequences (Table 3). A majority of the peptides identified had sequences which contained peptides with potential ACEI characteristics. In the case of those peptides derived from longan seed protein hydrolysate, the relevant sequences were quite short, with just 10 amino acids. These findings matched those reported by Natesh et al.,38 who showed by crystallography that large peptide molecules cannot access the active ACE sites. It is not yet, however, fully understood how peptide structures affects the relationship with ACEI activity, as the structure–activity relationship of ACEI peptides has yet to be fully established. When considering different peptides and their ACEI activity, there is some correlation between structure and activity. For example, the C-terminal tripeptide sequence has a powerful influence on the binding to ACE, while branched aliphatic amino acids at the N-terminal end are the most suitable to increase the peptide binding activity to ACE.25
3.6 Peptide properties
To prepare functional peptides it is first necessary to understand their properties and stabilities. Data provided by the Innovagen server indicated that ISSMGILVCL exhibits poor solubility in water (Table 3). However, in this research we noted that ISSMGILVCL presented a good solubility in water at concentrations up to 5.0 mg mL−1 (4.8 mM, above the IC50 value of 3.88 μM), and thereafter the solubility decreased as the concentration rose. The hydrophobicity of ISSMGILVCL is due to the existence of hydrophobic amino acids, including Ile, Met, Gly, Leu, and Val. Hydrophobicity can support the peptide binding to the hydrophobic active ACE center, thereby leading to improved inhibitory activity.In assessing stability, the pH of the peptides was measured along with the thermal stability. The ACEI activity of both peptides was also stable to heat, at up to 90 °C for 60 min (Fig. 2), and a broad pH range of 2–12 for 60 min (Fig. 3). The findings, therefore, support that these peptides offer gastrointestinal stability, and might also, in vivo, serve to generate antihypertensive effects.
3.7 Kinetics of enzyme inhibition
The ACEI kinetic properties must be carefully examined in order to discover the underlying mechanisms that allow peptides to serve as potential antihypertensive agents. From the Lineweaver–Burk plots (Fig. 4a and b), it is apparent that ETSGMKPTEL and ISSMGILVCL can both act as non-competitive inhibitors of ACE. The kinetic parameters of the ACEI by these two peptides are presented in Table 4. From these data, a constant value for Km was observed (0.14 ± 0.06 mM), indicating the existence of non-competitive inhibition. In non-competitive inhibition, the binding of the substrate and the inhibitor occurs at different areas of the enzyme,39 and so there is a constant affinity between the enzyme and the substrate. The Vmax (10.94 ± 0.03 mM min−1) value decreased as the inhibitory peptide concentration increased because of a lowering of the ACE catalytic efficiency, which in turn reduces the efficiency of the enzymatic reaction. At identical concentrations, the Vmax for ETSGMKPTEL was lower than that of ISSMGILVCL, suggesting a superior ACEI activity for ETSGMKPTEL compared to ISSMGILVCL.| Kinetics parameters | Control | ETSGMKPTEL (mM) | ISSMGILVCL (mM) | ||||
|---|---|---|---|---|---|---|---|
| 30 | 15 | 7.5 | 30 | 15 | 7.5 | ||
| a Findings appear as mean ± SEM (n = 3). | |||||||
| Km (mM) | 0.14 ± 0.06 | 0.14 ± 0.06 | |||||
| Vmax (mM min−1) | 10.94 ± 0.03 | 17.21 ± 0.02 | 15.51 ± 0.05 | 13.80 ± 0.02 | 19.67 ± 0.06 | 17.32 ± 0.04 | 14.23 ± 0.01 |
| Ki (mM) | 14.02 ± 0.11 | 10.78 ± 0.02 | |||||
From Fig. 4c and d, the Ki values were constant (14.02 ± 0.11 and 10.78 ± 0.02 mM for ETSGMKPTEL and ISSMGILVCL, respectively), thereby supporting non-competitive inhibition, which is an important trait. The majority of peptide ACEIs are competitive, while only very few are non-competitive, including those from sunflowers30 and chick peas.36 Accordingly, there is an advantage to be obtained from finding non-competitive inhibitors, such as ETSGMKPTEL and ISSMGILVCL, especially when compared to a competitive inhibitor since the concentration of the cellular substrate, such as angiotensin I, is quite high in relation to the Km value. In that scenario, ETSGMKPTEL and ISSMGILVCL would not be competing with the substrate to bind with the free enzyme, and would, therefore, retain their effectiveness.
3.8 Molecular docking of ETSGMKPTEL and ISSMGILVCL at the ACE binding site
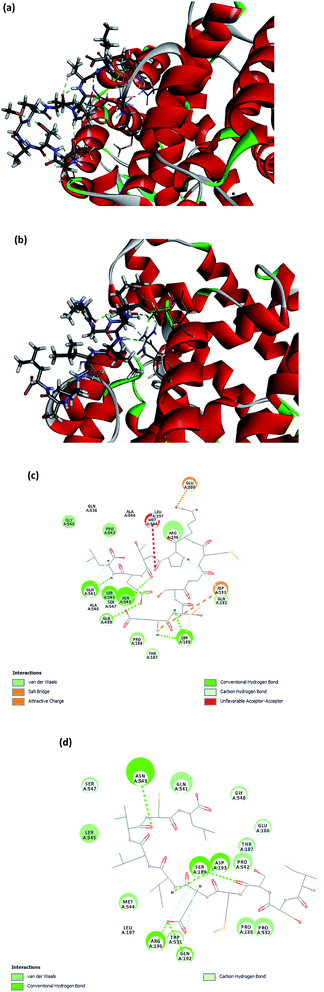
The molecular docking between ACE and the two small peptides was simulated using a computational approach that presents the ligand–receptor complex structures through the use of a scoring function.40 The Hermes 1.10.1 software was used to performed the calculations involved in docking between ETSGMKPTEL or ISSMGILVCL and ACE, which was established to be non-competitive. The molecular docking supported that the effectiveness of the peptides' ACEI activity was due to the creation of H-bonds with a non-active site of ACE, leading to a distortion of the catalysis configuration. Fig. 5a and b present the ideal docking position in 3D structures, while Fig. 5c and d show the bonds between ACE and the peptides. It is noteworthy that the greatest potential from ligands and peptides comes from position ligand A6, which offers a ChemPLP fitness score of 53.8349 for ETSGMKPTEL and 70.8909 for ISSMGILVCL. These ligand-receptor combination scores serve to show the potential stability involved.The ETSGMKPTEL peptide connected to ACE through six H-bonds linking the residues Gln541, Asn543, Gln489, Asn543, along with Ser189 with 2 H-bonds following docking. Moreover, ETSGMKPTEL exhibited five van der Waals interactions with ACE at Gln541, Asn543, Ser189, Gln192, and Glu200, creating the docked configuration for ACE with the ligand ISSMGILVCL. The most important interacting sites for ACE are those of the five H-bonds that involve residues Asn543, Ser189, Asp192, Gln192, and Arg196. Meanwhile, the van der Waals interactions in the bonding between ACE and ISSMGILVCL further served to stabilize residues Ser189 and Asp193. The docking results suggest multiple H-bonds linking ACE and the peptides, indicating that when peptides inhibit ACE activity they do so by stabilizing the enzyme–peptide complex through H-bonding.41,42
For ACE, the active site includes the residues Ala354, Glu384, Tyr523, Gln281, His353, Lys511, His513, Tyr520, and Glu162, which are the likely contact sites for competitive inhibitors. It is, therefore, the case that when binding arises between ETSGMKPTEL or ISSMGILVCL and ACE this does not use an active ACE site pocket. The findings support the potential use of ETSGMKPTEL and ISSMGILVCL as ACEIs and indicate that their development into functional foods to treat hypertension may be feasible.
4. Conclusions
The researchers believe that this study represents the first demonstration that stable ACEI activity from the enzymatic hydrolysate fractions of longan seeds by pepsin and pancreatin. Following fractionation via ultrafiltration and RP-HPLC, the peptides ETSGMKPTEL and ISSMGILVCL were shown to exhibit ACEI activity, which occurred non-competitively according to the Lineweaver–Burk plot. The results from molecular docking indicated that the links between ACE and the peptides were predominantly in the form of H-bonds, which were able to generate the inhibitory activity. It is broadly possible that such peptides produced from the pepsin–pancreatin hydrolysates of longan seeds might find applications in functional foods produced on an industrial scale. In the case of the antihypertensive properties, it will be necessary, however, to perform further studies, in particular assessing the effects observed during experiments involving mice.Funding
Sincere thanks are expressed to the Annual Government Statement of Expenditure (GRB_BSS_79_57_61_11), the Ratchadapisek Sompoch Endowment Fund, Chulalongkorn University (R_016_2556), and the Ratchadapisek Sompoch Endowment Fund (2019), Chulalongkorn University (762008) in recognition of the financial assistance offered which enabled this research study to be completed.Conflicts of interest
The authors have no conflict of interest. This publication has not been submitted earlier to any journal and is not being considered for publication elsewhere. All of the authors, including the corresponding authors, have read and approved the final submitted manuscript.Acknowledgements
The researchers are grateful to the Institute of Biotechnology and Genetic Engineering at Chulalongkorn University for the facilities they made available during the course of the study. We also thank Dr Robert Butcher (Publication Counseling Unit, Chulalongkorn University) for his constructive comments in preparing this manuscript.References
- N. Lionakis, D. Mendrinos, E. Sanidas, G. Favatas and M. Georgopoulou, World J. Cardiol., 2012, 26, 135–147 CrossRef PubMed.
- C. Y. Wu, H. Y. Hu, Y. J. Chou, N. Huang, Y. C. Chou and C. P. Li, Medicine, 2015, 94, e2160 CrossRef PubMed.
- J. R. Petrie, T. J. Guzik and R. M. Touyz, Can. J. Cardiol., 2018, 34, 575–584 CrossRef PubMed.
- D. Regoli and F. Gobeil Jr, Vasc. Pharmacol., 2015, 64, 1–10 CrossRef CAS PubMed.
- G. Saye and G. Bhat, Cardiol. Clin., 2014, 32, 21–32 CrossRef PubMed.
- C. A. Dézsi, Am. J. Cardiovasc. Drugs, 2014, 14, 167–173 CrossRef PubMed.
- F. H. Messerli, S. Bangalore, C. Bavishi and S. F. Rimoldi, J. Am. Coll. Cardiol., 2018, 71, 1474–1482 CrossRef CAS PubMed.
- C. Guang and R. D. Phillips, J. Agric. Food Chem., 2009, 57, 5113–5120 CrossRef CAS PubMed.
- I. Wijesekara and S. K. Kim, Mar. Drugs, 2010, 8, 1080–1093 CrossRef CAS PubMed.
- M. Martin and A. Deussen, Crit. Rev. Food Sci. Nutr., 2019, 59, 1264–1283 CrossRef CAS PubMed.
- Y. W. Sari, W. J. Mulder, J. P. Sanders and M. E. Bruins, Biotechnol. J., 2015, 10, 1138–1157 CrossRef CAS PubMed.
- C. S. K. Lin, L. A. Pfaltzgraff, L. Herrero-Davila, E. B. Mubofu, A. Solhy, P. J. Clark, A. Koutinas, N. Kopsahelis, K. Stamatelatou, F. Dickson, S. Thankappan, M. Zahouily, R. Brocklesby and R. Luque, Energy Environ. Sci., 2013, 6, 426–464 RSC.
- S. Panprom, K. Somtrakool and P. Thidpad, Asian Cult. Hist., 2014, 6, 43–51 Search PubMed.
- T. Varzakas, G. Zakynthinos and F. Verpoort, Foods, 2016, 5, E88 CrossRef PubMed.
- N. Rangkadilok, S. Tongchusak, R. Boonhok, S. C. Chaiyaroj, V. B. Junyaprasert, W. Buajeeb, J. Akanimanee, T. Raksasuk, T. Suddhasthira and J. Satayavivad, Fitoterapia, 2012, 83, 545–553 CrossRef CAS PubMed.
- H. C. Tseng, W. T. Wu, H. S. Huang and M. C. Wu, Int. J. Food Sci. Nutr., 2014, 65, 589–593 CrossRef CAS PubMed.
- J. Chen, Z. Z. Ge, W. Zhu, Z. Xu and C. M. Li, J. Agric. Food Chem., 2014, 62, 9744–9750 CrossRef CAS PubMed.
- T. Saisavoey, P. Sangtanoo, O. Reamtong and A. Karnchanatat, Food Biotechnol., 2018, 32, 79–94 CrossRef CAS.
- G. W. Latimer, Official Methods AOAC. 986.25, Gaithersburg, MD, USA, 2012 Search PubMed.
- G. W. Latimer, Official Method AOAC. 988.15, Gaithersburg, MD, USA, 2005 Search PubMed.
- K. Inthuwanarud, P. Sangvanich, S. Puthong and A. Karnchanatat, Pak. J. Pharm. Sci., 2016, 6, 1893–1900 Search PubMed.
- R. Suttisuwan, S. Phunpruch, T. Saisavoey, P. Sangtanoo, N. Thongchul and A. Karnchanatat, Food Biotechnol., 2019, 33, 303–324 CrossRef CAS.
- M. Yodjun, A. Karnchanatat and P. Sangvanich, Appl. Biochem. Biotechnol., 2012, 166, 2037–2050 CrossRef CAS PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- P. Wongtay, P. Sangtanoo, P. Sangvanich and A. Karnchanatat, Protein J., 2019, 38, 565–575 CrossRef CAS PubMed.
- C. Zhong, L. C. Sun, L. J. Yan, Y. C. Lin, G. M. Liu and M. J. Cao, Food Funct., 2018, 9, 594–603 RSC.
- C. Daskaya-Dikmen, A. Yucetepe, F. Karbancioglu-Guler, H. Daskaya and B. Ozcelik, Nutrients, 2017, 9, E316 CrossRef PubMed.
- S. Manoharan, A. S. Shuib and N. Abdullah, Afr. J. Tradit., Complementary Altern. Med., 2017, 14, 383–406 CrossRef CAS PubMed.
- B. J. Bhuyan and G. Mugesh, Org. Biomol. Chem., 2011, 9, 1356–1365 RSC.
- C. Megías, M. del Mar Yust, J. Pedroche, H. Lquari, J. Girón-Calle, M. Alaiz, F. Millán and J. Vioque, J. Agric. Food Chem., 2004, 52, 1928–1932 CrossRef PubMed.
- V. Vermeirssen, J. van Camp and W. Verstraete, J. Sci. Food Agric., 2005, 85, 399–405 CrossRef CAS.
- M. R. Segura Campos, L. A. Chel Guerrero and D. A. Betancur Ancona, J. Sci. Food Agric., 2010, 90, 2512–2518 CrossRef CAS PubMed.
- C. Barbana and J. I. Boye, Food Res. Int., 2010, 43, 1642–1649 CrossRef CAS.
- A. K. Arise, A. M. Alashi, I. D. Nwachukwu, S. A. Malomo, R. E. Aluko and E. O. Amonsou, J. Sci. Food Agric., 2017, 97, 2834–2841 CrossRef CAS PubMed.
- W. A. Wan Mohtar, A. A. Hamid, S. Abd-Aziz, S. K. Muhamad and N. Saari, J. Food Sci. Technol., 2014, 51, 3658–3668 CrossRef CAS PubMed.
- J. Pedroche, M. M. Yust, J. Girón-Calle, M. Alaiz, F. Millán and J. Vioque, J. Sci. Food Agric., 2002, 82, 960–965 CrossRef CAS.
- D. Y. Pujiastuti, M. N. Ghoyatul Amin, M. A. Alamsjah and J. L. Hsu, Molecules, 2019, 24, E2541 CrossRef PubMed.
- R. Natesh, S. L. U. Schwager, E. D. Sturrock and K. R. Acharya, Nature, 2003, 421, 1427–1429 CrossRef PubMed.
- Y. Blat, Chem. Biol. Drug Des., 2010, 75, 535–540 CrossRef CAS PubMed.
- X. Y. Meng, H. X. Zhang, M. Mezei and M. Cui, Curr. Comput.-Aided Drug Des., 2011, 7, 146–157 CrossRef CAS PubMed.
- D. Yu, C. Wang, Y. Song, J. Zhu and X. Zhang, Int. J. Mol. Sci., 2019, 20, E4159 CrossRef PubMed.
- M. Zarei, N. B. Z. Abidin, S. M. Auwal, S. Y. Chay, Z. A. Haiyee, A. M. Sikin and N. Saari, Biomolecules, 2019, 9, E569 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra00093k |
| This journal is © The Royal Society of Chemistry 2020 |