Open Access Article
Open Access ArticleA strategy for preparing non-fluorescent graphene oxide quantum dots as fluorescence quenchers in quantitative real-time PCR
Chenyan Hu†
a,
Zhongzhu Yang†a,
Zhen Songa,
Linghui Xiao*b and
Yang He *a
*a
aCollege of Medical Technology, State Key Laboratory of Characteristic Chinese Medicine Resources in Southwest China, Chengdu University of Traditional Chinese Medicine, Chengdu, 611137, China. E-mail: heyang@cdutcm.edu.cn
bHospital of Chengdu University of Traditional Chinese Medicine, Chengdu, 610075, China. E-mail: xiaolh2010@hotmail.com
First published on 16th April 2020
Abstract
In recent years, graphene oxide quantum dots (GOQDs) have emerged as novel nanomaterials for optical sensing, bioimaging, clinical testing, and environmental testing. However, GOQDs demonstrate unique photoluminescence properties, with GOQDs having quantum limitations and edge effects that often affect the accuracy of the test results in the sensory field. Herein, GOQDs with a large content of hydroxyl groups and low fluorescence intensity were first prepared via an improved Fenton reaction in this study, which introduces a large amount of epoxy groups to break the C–C bonds. The synthesized GOQDs show no significant variation in the fluorescence intensity upon ultraviolet and visible light excitations. We further utilized the GOQDs as fluorescence quenchers for different fluorescent dyes in real-time fluorescence quantitative polymerase chain reaction (qRT-PCR), and verified that the addition of GOQDs (5.3 μg ml−1) into a qRT-PCR system could reduce the background fluorescence intensity of the reaction by fluorescence resonance energy transfer (FRET) during its initial stage and its non-specific amplification, and improve its specificity. In addition, the qRT-PCR method could detect two different lengths of DNA sequences with a high specificity in the 104 to 1010 copies per μl range. It is of paramount importance to carry out further investigations to establish an efficient, sensitive, and specific RT-PCR method based on the use of GOQD nanomaterials as fluorescence quenchers.
Introduction
Recently, biosensors have been widely studied to be employed in rapid detection methods for pathogens or pathogenic genes in the sensory field.1,2 In particular, fluorescence sensors with modified fluorophores, which are based on deoxyribonucleic acid (DNA), appear to be promising for application due to their stability, high specificity, and sensitivity.3,4 Fluorescence sensors to detect the presence of a target molecule are typically based on the fluorescence resonance energy transfer (FRET) process,5 thus the choice of the receptor pair usually determines the analytical performance of the fluorescence sensor.6 To reduce the limitations that characterize the receptor pairs, studies have shown that novel nanomaterials, which exhibit an effective quenching ability in a broad range of emission wavelengths, can be used as broad-spectrum quenching molecules.7–9 For instance, fluorescence sensors based on graphene oxide quantum dots (GOQDs) have been applied in a wide range of fields for cancer research, clinical trials, and medical practice.10–12GOQDs are zero-dimensional materials, which are nanoscale fragments of graphene oxide (GO).13,14 Due to the quantum confinement and the edge effect, GOQDs have ignited tremendous research interest for their outstanding properties in terms of physical and chemical properties when compared to layered GO. For instance, the quantum limitations and the edge effects of GOQDs can be greatly enhanced to achieve distinctive photoluminescence properties by adjusting the size of the GOQDs and the number of oxygen-containing functional groups.15 In addition, due to their good biocompatibility, fluorescence stability, and low toxicity, GOQDs are suitable for optical sensing, bioimaging, clinical testing, and environmental testing.16,17 Nevertheless, the high background fluorescence intensity, which characterizes GOQDs, often affects the accuracy of the test results. Fluorescence sensors usually present a low concentration of GOQDs to improve the accuracy of the detection results.18 Therefore, it is of pivotal importance to reduce the influence of the background fluorescence of the GOQDs to enable their use in a broader variety of sensing and detection techniques.
Polymerase chain reaction (PCR) is a process that the mimics DNA replication in vitro and that can detect a target DNA sequence from the final reaction product.19 In recent years, a real-time fluorescent quantitative PCR technology (qRT-PCR) has been developed based on the traditional PCR. This technique monitors the PCR amplification via the variations in the fluorescence intensity.20 Compared to the traditional PCR, qRT-PCR exhibits a significant advantage to detect the evolution of the amplification reaction by detecting the fluorescence signal in real-time. However, regardless of the dye method or the probe method used, a high background fluorescence intensity is present in qRT-PCR. Moreover, the detection accuracy is lower in the dye method when the primers with poor specificity to the templates.21,22 Therefore, it is necessary to solve the issue generated by the presence of such high original background fluorescence intensity to improve the specificity of the qRT-PCR method.
Based on the improved Fenton reaction, a graphite layer was initially separated by using MnO3+ as the intercalating agent. Successively, the graphene was oxidized into GO by H2O2, which contains a large quantity of epoxy functional groups. At this point, the Fenton reaction occurs between the zero-valent iron and H2O2. In this fashion, non-fluorescent GOQDs with a large number of hydroxyl groups were obtained. So far, there are no studies about non-fluorescent GOQDs. In this work, we report a new qRT-PCR method that employs easily prepared and non-fluorescent GOQDs to efficaciously enhance the specificity of qRT-PCR reactions. At first, the fluorescence quenching ability of non-fluorescent GOQDs was investigated by combining together various fluorescent dyes. Furthermore, the non-fluorescent GOQDs were applied to qRT-PCR via the formation of a π–π stacking and hydrogen bonds between the TaqMan probe and the GOQDs to reduce the original background fluorescence intensity. The π–π stacking and the hydrogen bond between the primers and the GOQDs significantly improved the specificity of qRT-PCR. Moreover, two different lengths DNA fragments (106 bp and 65 bp) were detected via qRT-PCR based on the GOQDs. The superior performance of the GOQDs in qRT-PCR indicates that this enables new applications in the biological and medical fields.
Experimental
Reagents and materials
Graphite (crystalline powder, 100 mesh) was purchased from Qingdao Laixi Colloidal Graphite Factory (Qingdao, China). Potassium nitrate (KNO3), potassium permanganate (KMnO4), sulphuric acid (H2SO4), and 30% hydrogen peroxide (H2O2) were supplied by Chengdu Kelong Chemical Reagent Factory (Chengdu, China). The 1000 Da dialysis bag was provided by Shanghai Yuanye Biological Science & Technology Company (Shanghai, China). The solutions were prepared by using ultrapure water (>18 MΩ) from the ELGA water purification system (ELGA, London, UK). All the reagents used in this study were of analytical reagent grade.Improved Fenton reaction
Graphite (0.5 g) was mixed with sulphuric acid (95–98%) and the solution was stirred at room temperature prior to the addition of NaNO3 (0.5 g). Meanwhile, KMnO4 (3 g) was added to the sulphuric acid (95–98%) before adding NaNO3 (0.5 g) to form MnO3+. The manganese-embedded graphite was formed by the slow addition of MnO3+ into graphite. Successively, H2O2 (30%) and zero-valent iron were added into 10 ml of manganese-embedded graphite. The zero-valent iron in the liquid was removed by boiling the solution. The liquid required a continuous addition of H2O2 (30%) and was left boiling for 6 min to obtain the GOQDs solution. The experiment was carried out by using a magnetic stirrer. The GOQDs solution was dialyzed (pH 2.0, 1000 Da) for 7 days to remove the excess of Fe2+, Fe3+, the traces of H2O2, and of other low-molecular mass reaction products. Finally, a pale yellow transparent GOQDs solution was obtained.Characterization
Atomic force microscopy (AFM) of the GOQDs was conducted on a Multimode Nanoscope V scanning probe microscopy system (Bruker, USA). In the test, a commercial AFM cantilever tip (Bruker, USA) with a force constant of ∼50 N m−1 and a resonance vibration frequency of ∼350 kHz was used. The X-ray photoelectron spectroscopy (XPS) pattern of the samples was measured by using a Thermo ESCALAB 250XI scanning XPS microscope (a monochromated Al Kα radiant energy of 1486.6 eV). Fourier transform infrared spectroscopy (FT-IR) spectra were obtained on a NICOLET 5700 FT-IR spectrometer (Waltham, USA). The samples to perform the FT-IR measurements were prepared by grinding a dry powder of GOQDs with KBr, which was then compressed into a thin slice. UV-visible absorption spectra were obtained by using a MAPADA UV-6300 ultraviolet-visible spectrophotometer (Shanghai, China). The photoluminescence spectrum (PL) of GO, 6-FAM, JOE, and Cy3 was recorded on a Lengguang F97PRO Fluorescence Spectrophotometer (Shanghai, China). The emission spectrum of GOQDs was obtained by using a detection wavelength of 440 nm and an excitation wavelength of 380 nm. The emitted fluorescence intensity of 6-FAM, JOE and Cy3 were detected by applying excitation wavelengths of 485 nm, 520 nm and 535 nm and emission wavelengths of 525 nm, 548 nm and 556 nm, respectively.Single-stranded DNA templates and primers
The primers and the probes were designed by employing the Primer Express 3.0.1 software, which was developed based on the related works. All the single-stranded DNA templates and primers were synthesized by Shanghai Sangon Biological Science & Technology Company (Shanghai, China). The specific sequences are listed in Table 1. The single-stranded DNA, primers, probes, and templates were dissolved in a solution of ultrapure water and stored at −20 °C.| Name | Sequence (5′ → 3′) |
|---|---|
| 6-FAM-ssDNA | 6-FAM-CCATGCTAATCTTCTCTGTATCGTTCCA |
| JOE-ssDNA | JOE-CCATGCTAATCTTCTCTGTATCGTTCCA |
| Cy3-ssDNA | Cy3-CCATGCTAATCTTCTCTGTATCGTTCCA |
| Primer 1 | CTGCTTCGGCAGCACA |
| Primer 2 | AACGCTTCACGAATTTGCGT |
| Primer 3 | GCGCGTAGCAGCACAGAAAT |
| Primer 4 | AGTGCAGGGTCCGAGGTATT |
| Probe 1 | 6-FAM-CCATGCTAATCTTCTCTGTATCGTTCCA-BHQ1 |
| Probe 2 | HEX-CACTGGATACGACGCCAATATTTCT-BHQ1 |
| 65 bp DNA | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCCAATATTTCTGTGCTGCTA |
| 106 bp DNA | GTGCTCGCTTCGGCAGCACATATACTAAAATTGGAACGATACAGAGAAGATTAGCATGGCCCCTGCGCAAGGATGACACGCAAATTCGTGAAGCGTTCCATATTTT |
Construction of 106 bp plasmid DNA
MicroRNA was extracted from human breast epithelial cell line (MCF-10A). The final reverse transcription reaction system was containing 200 ng of microRNA, 0.4 μl of 10 μM Primer 2 (U6 RT-Primer), 1 μl of TransScript RT/RI Enzyme SuperMix (Transgen, China), and 10 μl of 2× TS Reaction Mix (Transgen, China) for a total volume of 25 μl. The reverse transcription cDNA was stored at −20 °C. Next, a direct PCR was carried out, containing 2 μl of reverse transcription cDNA, 0.5 μl of 10 μM Primer 1, 0.5 μl of 10 μM Primer 2, and 12.5 μl of 2× EasyTaq PCR SuperMix (Transgen, China) for a total volume of 25 μl. The direct PCR reaction process consisted in a pre-denaturation at 94 °C for 5 min, a denaturation at 94 °C for 5 s, an annealing at 60 °C for 30 s, an extension at 72 °C for 30 s for a total of 30 cycles, and a final extension at 72 °C for 5 min. Subsequently, a 2% agarose gel electrophoresis of 5 μl PCR products was performed, and a DiaSpin DNA Gel Extraction Kit (Diamond, China) was used to extract and purify the PCR products. The target fragments were ligated with pEASY-T3 Cloning Vector (Transgen, China), and then Trans1-T1 Phage Resistant Chemically Competent Cell (Transgen, China) was used for transformation of linkage products. The constructed plasmid was sent to Beijing TSINGKE Biological & Technology Company for identification and sequencing.Fluorescence quenching of 6-FAM, JOE, and Cy3 by GOQDs
Initially, 1500 μl of 6-FAM-ssDNA (200 nM), JOE-ssDNA (200 nM), and Cy3-ssDNA (200 nM) solutions were separately inserted into three plastic tubes without nuclease, then 1500 μl of GOQDs working solution (26 μg ml−1) were added. Successively, each tube was incubated at 37 °C for 30 min in the dark. Finally, the fluorescence intensity of the mixture was measured by a Lengguang F97PRO Fluorescence Spectrophotometer (Shanghai, China). The emission wavelengths and the excitation wavelengths of 6-FAM, JOE, and Cy3 were 520 nm and 494 nm, 548 nm and 520 nm, and 570 nm and 550 nm, respectively. Each measurement was repeated three times.qRT-PCR method based on the GOQDs
A qRT-PCR experiment was carried out in an analytikJena qTOWER 2.2 Fluorescence Quantitative Gradient Polymerase Chain Reactor (Jena, Germany). When the TaqMan probe is cleaved by the Taq DNA polymerase during the qRT-PCR reaction, the fluorescent energy of the fluorophore cannot be adsorbed by the quenching group, thus a fluorescence signal is generated and can be used to monitor the amplification of the PCR.23 The experiment showed that the final reaction systems contained 1 μl of template (106 bp or 65 bp DNA), 0.5 μl of 10 μM forward primer (Primer 1 or Primer 3), 0.5 μl of 10 μM reverse primer (Primer 2 or Primer 4), 12.5 μl of 2 × TransStart Probe qRT-PCR SuperMix (Transgen, China), and 0.5 μl of GOQDs solution for a total volume of 25 μl. The final concentration of the GOQDs in the reaction measured 0.00014 μg μl−1. Moreover, the qRT-PCR reaction mixture without either 106 bp or 65 bp DNA but containing GOQDs was used as an internal control solution. The 106 bp plasmid DNA, the 65 bp DNA, and the GOQDs were then replaced by deionized water in a negative control solution. To define the optimal concentration of GOQDs, a quantity of 26 μg ml−1 was diluted into ultrapure water to prepare a series of working solutions with concentrations of 11 μg ml−1, 5.3 μg ml−1, 3.3 μg ml−1, 2.2 μg ml−1, and 1.5 μg ml−1 calculated via the concentration-absorbance standard curve. The 106 bp and 65 bp DNA was diluted into a working solution (1010 copies per μl) with ultrapure water. This solution was then continuously diluted to 109 to 101 copies per μl to generate a standard curve. The qRT-PCR reaction process consisted in a pre-denaturation of the sample at 94 °C for 30 s, which was followed by a denaturation at 94 °C for 5 s and annealing at 60 °C for 30 s for a total of 40 cycles. During each cycle and after the annealing process, the fluorescence emission intensity of the specimen at 520 nm or 580 nm was probed by using an excitation wavelength of 490 nm or 535 nm. 2% agarose gel electrophoresis of qRT-PCR products were performed and the measurements were repeated three times.Statistical analyses
All statistical analyses were performed using the SPSS statistical package (Version 23.0; IBM Company, Chicago, IL, USA), as mean ± standard deviation (SD). T test were used to compare the treatment means with controls. The statistically significance level was set at P < 0.01.Results and discussion
Improved Fenton reaction to prepare the GOQDs
To obtain high-quality GOQDs from graphite, three consecutive chemical reactions (intercalation, oxidation, and Fenton reaction) are required. First, the intercalation reaction is dominated by the transformation of planar triangular MnO3+ into tetrahedral potassium permanganate under the action of sulphuric acid (95–98%). Graphite is oxidized and forms hydroxyl groups on the surface of the sample upon addition of the sulphuric acid. Furthermore, MnO3+, which is formed onto the GO surface, penetrates between the graphite carbon layers by exploiting the action of the hydroxyl group as intercalation agent.24 After the intercalation process is completed, the planar triangular MnO3+ structure spontaneously transforms into a –MnO4 regular tetrahedral structure to support the graphite sheets.25 Finally, a purple manganese-embedded graphite sample with a large MnO4− content is formed. Subsequently, upon the addition of H2O2 into the solution, the carbon atoms, which are present in the manganese-embedded graphite, react with the sulphuric acid to produce carbon dioxide, which dissolves in the water and forms HCO3−. Simultaneously, MnO4− can react directly with H2O and graphite to form carbonate, bicarbonate, and manganese dioxide. Manganese dioxide reacts with the sulphuric acid and produces Mn2+. The HCO3− ions, which are formed via these two mechanisms, react with H2O2 to produce percarbonate. Then, they interact with Mn2+ to form [MnO]2+, which reacts with graphite to generate GO which contains a host of epoxy groups.26,27Successively, the Fenton reaction occurs. The epoxy groups are attacked via hydroxyl radicals on the surface of GO and the GOQDs are obtained. The mixed solution undergoes an exothermic reaction and eventually boils upon the addition of zero-valent iron and H2O2 (30%), due to the presence of a large amount of hydroxyl radicals produced during the Fenton reaction. The carbon atoms, which are bound to the hydroxyl and epoxy groups on the surface of the graphene lamellae, are attacked by the large hydroxyl radicals, breaking the C–C bonds. Simultaneously, the newly formed oxygen-containing functional groups, such as the quinone groups, are attacked by the hydroxyl radicals.28–30 In this fashion, the Fenton reaction induced by the presence of zero-valent iron and H2O2 (30%) can restart. An excessive presence of zero-valent iron reduces the –COOH on the surface of the GOQDs into –OH under acidic conditions. In this way, GOQDs sample rich in hydroxyl groups can be formed.
Characterization of GOQDs
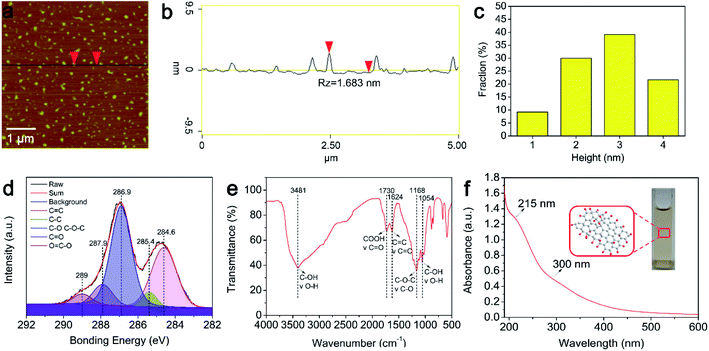
The surface structure and size of the GOQDs were characterized via AFM (Fig. 1a–c). A statistical analysis of 120 samples shows that about 80% of the GOQDs are less than 3 nm thickness (Fig. 1c), indicating that most of the GOQDs are 1–3 layers, which is very close to that is reported in the previous papers.31 In particular, the GOQDs prepared by the improved Fenton reaction can be freely dispersed in water with transparent appearance. In order to analyze the composition of the GOQDs, XPS experiments were carried out. Fig. 1d depicts the XPS spectrum: the C 1s peak consists of five features, which are located at 284.6 eV, 285.4 eV, 286.9 eV, 287.9 eV, and 289 eV and they correspond to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C–C, C–O, C
C, C–C, C–O, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, and –COOH characteristic peaks of the carbon skeleton, respectively.32 It indicates that the sp3 and sp2 hybridization carbon atoms coexist in GOQDs. Moreover, the XPS results also suggest that a large amount of hydroxyl groups and epoxy groups are present on the surface of the GOQDs. The content of carbon and oxygen measures 53.77% and 39.27%, respectively and the ratio of carbon to oxygen is 1.37. The FT-IR spectrum of the GOQDs was also investigated (Fig. 1e). The peak at 1168 cm−1 corresponds to the characteristic absorption fingerprint of the epoxy (–O–) group,33 whereas the feature at 1730 cm−1 is produced by the stretching vibration of the C
O, and –COOH characteristic peaks of the carbon skeleton, respectively.32 It indicates that the sp3 and sp2 hybridization carbon atoms coexist in GOQDs. Moreover, the XPS results also suggest that a large amount of hydroxyl groups and epoxy groups are present on the surface of the GOQDs. The content of carbon and oxygen measures 53.77% and 39.27%, respectively and the ratio of carbon to oxygen is 1.37. The FT-IR spectrum of the GOQDs was also investigated (Fig. 1e). The peak at 1168 cm−1 corresponds to the characteristic absorption fingerprint of the epoxy (–O–) group,33 whereas the feature at 1730 cm−1 is produced by the stretching vibration of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group of the GOQDs.34 Moreover, a wide and strong absorption peak appears in the 3250–3750 cm−1 range. This feature is attributed to the stretching vibration of the –OH group.35 FT-IR spectroscopy implies that these functional groups induce excellent hydrophilic properties in the GOQDs sample. Based on the UV-vis results (Fig. 1f), the absorption peak at 215 nm is attributed to the π → π* transition of the sp2 C–C bonds.14 Furthermore, the n → π* transition of the C
O group of the GOQDs.34 Moreover, a wide and strong absorption peak appears in the 3250–3750 cm−1 range. This feature is attributed to the stretching vibration of the –OH group.35 FT-IR spectroscopy implies that these functional groups induce excellent hydrophilic properties in the GOQDs sample. Based on the UV-vis results (Fig. 1f), the absorption peak at 215 nm is attributed to the π → π* transition of the sp2 C–C bonds.14 Furthermore, the n → π* transition of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds or other functional groups on the surface of the GOQDs present a shoulder peak in approximately 300 nm.36 In order to better understand luminescence characteristics of the GOQDs, the luminescence spectrum and photoluminescence excitation spectrum measurements were carried out. Fig. 2 illustrated the luminescence spectrum of the GOQDs. One can see that the synthesized GOQDs show no significant variation in the fluorescence intensity upon ultraviolet and visible light excitations.
O bonds or other functional groups on the surface of the GOQDs present a shoulder peak in approximately 300 nm.36 In order to better understand luminescence characteristics of the GOQDs, the luminescence spectrum and photoluminescence excitation spectrum measurements were carried out. Fig. 2 illustrated the luminescence spectrum of the GOQDs. One can see that the synthesized GOQDs show no significant variation in the fluorescence intensity upon ultraviolet and visible light excitations.
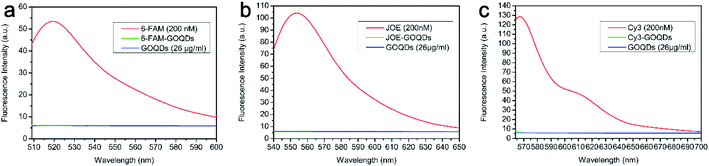 | ||
| Fig. 2 Fluorescence quenching ability of GOQDs. (a) Fluorescence quenching of 6-FAM by GOQDs. (b) Fluorescence quenching of JOE by GOQDs. (c) Fluorescence quenching of Cy3 by GOQDs. | ||
Fluorescence quenching of 6-FAM, JOE, and Cy3
In order to explore the properties of the GOQDs and illustrate their fluorescence quenching effect, they were incubated with three different fluorescent dyes. The emission wavelengths of the fluorescent dyes 6-FAM, JOE, and Cy3 are 520 nm, 548 nm, and 570 nm, respectively. As shown in Fig. 2, the fluorescence intensity of these three dyes decreases after the addition of the GOQDs, indicating that their effective quenching effect on the fluorescence of the group. The sp2 aromatic structure of GO can adsorb the ssDNA via its π–π stacking cloud.18,37–39 For this reason, even a very small layer of GO can bond with the base of the fluorescent probe via such π–π cloud. In addition, the hydroxyl groups on the surface of GOQDs can also form hydrogen bonds with the base of the fluorescent probe, promoting the adsorption of the fluorescent probe equally. Ultimately, FRET occurs between the fluorescent probe and the GOQDs. The fluorescent group on the probe acts as a donor to transfer the energy to the GOQDs, which results in a fluorescence quenching effect.10 Additionally, the combination of the characterization data and the literature shows that the background fluorescence intensity of GOQDs prepared via the improved Fenton reaction is much lower than for traditional GOQDs (Fig. 2).40 According to the literature, the higher the –COOH content on the surface of the GOQDs is, the stronger is the fluorescence intensity of the GOQDs. However, when –COOH is reduced into –OH a reduction of its fluorescence intensity can be detected.28 The reasons for low background fluorescence intensity of GOQDs were strongly supported by the results of XPS, FT-IR and UV-vis. The results also suggest that the GOQDs may have potential applications in the clinical disease detection field and may be used in qRT-PCR as an effective fluorescence quencher to detect target genes.Screening the optimal concentration of GOQDs in qRT-PCR
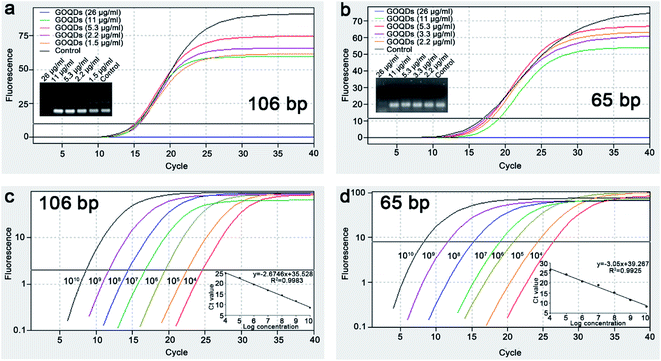
The qRT-PCR is a highly sensitive method for DNA quantitative analysis. In this experiment, the effects of the GOQDs on the qRT-PCR process were investigated by adding GOQDs in different concentrations into a traditional qRT-PCR system. To avoid the contributions of the different templates into the estimate of the qRT-PCR amplification efficiency, 106 bp plasmid DNA and synthesized 65 bp DNA was used as the templates.During the qRT-PCR process, the amplification curve changes as the concentrations of GOQDs decreases (Fig. 3a and b). Moreover, the curve resembles a typical amplification control curve with an optimal concentration of GOQDs of 5.3 μg ml−1 (106 bp and 65 bp). The agarose gel electrophoresis results are consistent with the trend of the amplification curve and indicate that the excess of GOQDs can inhibit the qRT-PCR amplification reaction by adsorbing a high quantity of the probe, primer, or template through its sp2 conjugated region. Moreover, when the concentration of the GOQDs is below the optimal concentration (5.3 μg ml−1), a host in the non-specific amplification occurs in the qRT-PCR system of 106 bp plasmid DNA. Contrarily, the specificity of the qRT-PCR process increases upon the increase of the GOQDs concentrations. The results suggested that GOQDs could improve qRT-PCR specificity in the concentration range of 5.3–11 μg ml−1. Additionally, a concentration of GOQDs of 5.3 μg ml−1 is an optimal concentration to maintain high specificity in our qRT-PCR system (106 bp and 65 bp).
Specificity of qRT-PCR based on GOQDs
To further understand the effect of the GOQDs on the TaqMan qRT-PCR process, agarose gel electrophoresis was performed after qRT-PCR. As shown in Fig. 3a, the non-specific amplification bands disappear upon the addition of GOQDs (>5.3 μg ml−1). During the qRT-PCR process, when the dNTPs, the cDNA polymerase, and the template are mixed, the background signals generated by the non-specific amplification appear even if the enzyme is not heat-activated.41 Due to the lack of a biofeedback mechanism in vitro, the effect of the GOQDs on the specificity of qRT-PCR may be the result of the interaction of multiple factors. Therefore, we assume that the enhancement effects of GOQDs can be explained as follows. First, after the optimal concentration of GOQDs is achieved, the large number of hydroxyl groups on the surface of GOQDs may interact with the primers to form hydrogen bonds and the π–π stacking can form together, thereby inhibiting the mismatches between the primers and the templates at the non-target sites. When the primers are fully paired with the templates, a qRT-PCR reaction can occur due to detachment of the primers from the surface of GOQDs. This may reduce the non-specific amplification products (Fig. 4). Second, the GOQDs, which are added into the system, can interact with excessive probes, primers, or templates, reducing the binding of primers to the templates to inhibit the occurrence of non-specific reactions. Herein, the addition of GOQDs in the qRT-PCR reaction can reduce the non-specific amplification process and improve the specificity of the qRT-PCR method. In addition, an increase in the concentration of the GOQDs resulted in an enhancement in the adsorption of the GOQDs on the primers. At the same time, due to the existence of the above two mechanisms, the specificity of qRT-PCR can be scarcely improved by the increase of the GOQDs concentrations. However, it can be largely inhibited by GOQDs upon addition of >11 μg ml−1. The possible reason could be that the adsorption capacity of GOQDs for the primers are far stronger than the complementary pairing ability of the primers and the templates.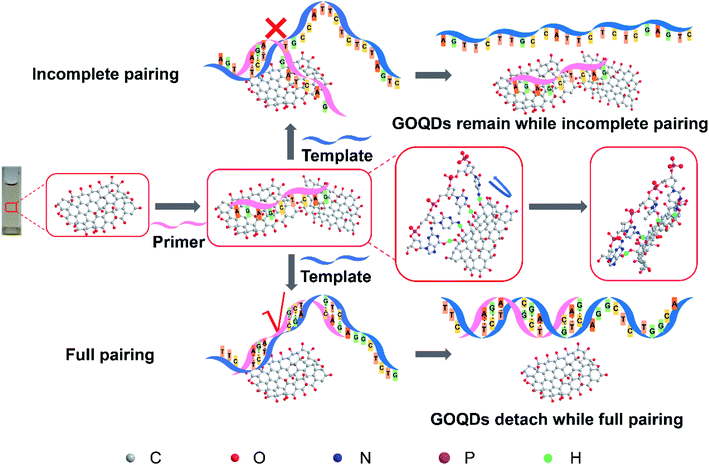 | ||
| Fig. 4 Scheme of enhancing qRT-PCR specificity by GOQDs. The whole picture shows the schematic diagram of the hypothesis of enhancing qRT-PCR specificity by GOQDs. | ||
Sensitivity of qRT-PCR based on GOQDs
Under the optimal concentration of GOQDs (5.3 μg ml−1), the detection performance of this qRT-PCR method was evaluated by adding varying concentrations of the DNA templates into the qRT-PCR system. As shown in Fig. 3c and d, the Ct value decreases upon the decrease of the DNA copy number. The standard curve shows that the Ct value is linear as a function of the logarithm of the initial 106 bp plasmid DNA and 65 bp DNA templates copy number in the 104 to 1010 copies per μl range with a determining coefficient of 0.9983 and 0.9925. According to the theory,42 a quantitative relationship can be established between the initial copy number of the DNA templates and the amount of PCR product at any cycle. Therefore, the qRT-PCR method based on the GOQDs allows on to detect the DNA in a wide range of DNA copy number and the detection range is higher than 7 orders of magnitude with a detection limit of 104 copies per μl. This is the first time to achieve sensitive analysis of DNA using the qRT-PCR method based on the GOQDs.Detection of 106 bp and 65 bp DNA by qRT-PCR based on GOQDs
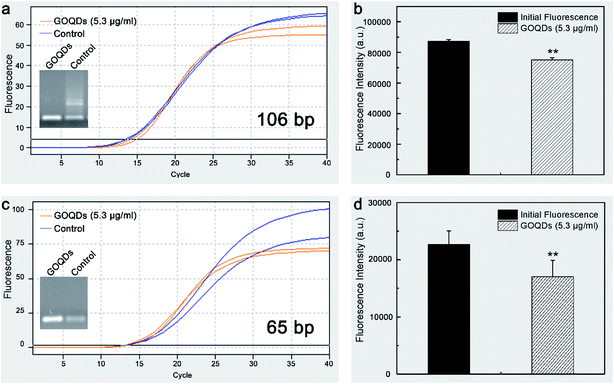
The effect of the GOQDs on the qRT-PCR amplification was further investigated by detecting two different lengths of DNA sequences (106 bp and 65 bp). Typical qRT-PCR results obtained by using the GOQDs are shown in Fig. 5. Upon the addition of the GOQDs (5.3 μg ml−1) into the two qRT-PCR systems, a significant reduction in the background fluorescence intensity of the TaqMan probes of 20% (P = 0.0006 < 0.01) and 25% (P = 0.0014 < 0.01) can be observed (Fig. 5b and d). However, the plateau of the amplification curves is lower than that of the control groups (Fig. 5a and c), probably due to the formation of the π–π stacking between the fluorophores and the GOQDs to trigger FRET after the TaqMan probe was hydrolyzed by the Taq DNA polymerase. This, in fact, generates a quenching mechanism on the fluorescence intensity of several fluorescent groups under the action of the GOQDs. The improved specificity of qRT-PCR by GOQDs indicates that the GOQDs-based qRT-PCR method can be used to detect DNA specifically and selectively.Conclusions
In summary, GOQDs, rich in hydroxyl groups and with a low background fluorescence intensity, were prepared via the improved Fenton reaction to efficiently quench the fluorescence of three fluorescent dyes (6-FAM, JOE, and Cy3). In this study, a simple and effective qRT-PCR method was established to evaluate the effect of the GOQDs. The results show that the GOQDs effectively decrease the background fluorescence intensity of the TaqMan probe, reduce the non-specific amplification, and improve the specificity of the qRT-PCR reaction. Furthermore, the qRT-PCR method based on the GOQDs can detect the 106 bp and 65 bp DNA in a concentration of 104 copies per μl, in a linear range of 104 to 1010 copies per μl and with a detection range spanning 7 orders of magnitude. This technique can also detect two different lengths of DNA sequences with a high specificity. These results indicate that the GOQDs prepared by using the improved Fenton reaction are promising materials to develop an efficient qRT-PCR method for a high-specific and selective detection of DNA.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was financially supported by the Outstanding Youth Science Foundation of Sichuan Province (Grant No. 2017JQ0015), by project first-class disciplines development supported by Chengdu University of Traditional Chinese Medicine (Grant No. CZYJC1904), by scientific research fund of Sichuan provincial education department (Grant No. 18CZ0010). We thank the Innovative Institute of Chinese Medicine and Pharmacy at Chengdu University of Traditional Chinese Medicine for its instrumental support (PL, FT-IR, and vacuum freeze drier).Notes and references
- S. Ellairaja, N. Krithiga, S. Ponmariappan and V. S. Vasantha, J. Agric. Food Chem., 2017, 65, 1802–1812 CrossRef CAS PubMed.
- Y. T. Yew, A. H. Loo, Z. Sofer, K. Klímová and M. Pumera, Appl. Mater. Today, 2017, 7, 138–143 CrossRef.
- C. Lungu, S. Pinter, J. Broche, P. Rathert and A. Jeltsch, Nat. Commun., 2017, 8, 649 CrossRef PubMed.
- L. Andreeva, B. Hiller, D. Kostrewa, C. Lässig, C. C. de Oliveira Mann, D. J. Drexler, A. Maiser, M. Gaidt, H. Leonhardt, V. Hornung and K. P. Hopfner, Nature, 2017, 549, 394–398 CrossRef CAS PubMed.
- A. H. Loo, Z. Sofer, D. Bouša, P. Ulbrich, A. Bonanni and M. Pumera, ACS Appl. Mater. Interfaces, 2016, 8, 1951–1957 CrossRef CAS PubMed.
- Z. S. Qian, X. Y. Shan, L. J. Chai, J. J. Ma, J. R. Chen and H. Feng, Biosens. Bioelectron., 2014, 60, 64–70 CrossRef CAS PubMed.
- R. M. Kong, X. B. Zhang, Z. Chen and W. H. Tan, Small, 2011, 7, 2428–2436 CAS.
- T. Sun, N. Xia and L. Liu, Nanomaterials, 2016, 6, 20 CrossRef PubMed.
- Y. W. Lin, C. C. Huang and H. T. Chang, Analyst, 2011, 136, 863 RSC.
- D. M. Kim, D. H. Kim, W. Jung, K. Y. Lee and D. E. Kim, Analyst, 2018, 143, 1797–1804 RSC.
- R. V. Goreham, K. L. Schroeder, A. Holmes, S. J. Bradley and T. Nann, Mikrochim. Acta, 2018, 185, 128 CrossRef PubMed.
- S. Y. Choi, S. H. Baek, S. J. Chang, Y. Song, R. Rafique, K. T. Lee and T. J. Park, Biosens. Bioelectron., 2017, 93, 267 CrossRef CAS PubMed.
- R. Gao, Z. Zhong, X. Gao and L. Jia, J. Agric. Food Chem., 2018, 66, 10898–10905 CrossRef CAS PubMed.
- S. Ahirwar, S. Mallick and D. Bahadur, ACS Omega, 2017, 2, 8343–8353 CrossRef CAS PubMed.
- F. Zhang, F. Liu, C. Wang, X. Xin, J. Liu, S. Guo and J. Zhang, ACS Appl. Mater. Interfaces, 2016, 8, 2104–2110 CrossRef CAS PubMed.
- Y. Liu, Y. Xu, X. Geng, Y. Huo, D. Chen, K. Sun, G. Zhou, B. Chen and K. Tao, Small, 2018, 14, 1800293 CrossRef PubMed.
- E. Lai, Anal. Biochem., 2016, 5, e165 Search PubMed.
- Y. He, B. Zhang and Z. Fan, Mikrochim. Acta, 2018, 185, 163 CrossRef PubMed.
- L. M. Zhang, T. Jiang, X. F. Li, Y. Wang, C. Zhao, S. Zhao, L. Xi, S. J. Zhang, X. Z. Liu, Y. J. Jia, H. Yang, J. P. Shi, C. X. Su, S. X. Ren and C. C. Zhou, Cancer, 2017, 123, 2927–2935 CrossRef CAS PubMed.
- T. S. Kang, Trends Food Sci. Technol., 2019, 91, 574–585 CrossRef CAS.
- A. Nakashima, N. Ihara, M. Shigeta, H. Kiyonari, Y. Ikegaya and H. Takeuchi, Science, 2019, 365, eaaw5030 CrossRef CAS PubMed.
- I. Baris, O. Etlik, V. Koksal, Z. Ocak and S. T. Baris, Anal. Biochem., 2013, 441, 225–231 CrossRef CAS PubMed.
- S. Mocellin, C. R. Rossi, P. Pilati, D. Nitti and F. M. Marincola, Trends Mol. Med., 2003, 9, 189–195 CrossRef CAS PubMed.
- D. J. Royer, J. Inorg. Nucl. Chem., 1961, 17, 159–167 CrossRef CAS.
- X. Hu, L. Shi, D. Zhang, X. Zhao and L. Huang, RSC Adv., 2016, 6, 14192–14198 RSC.
- B. S. Lane, M. Vogt, V. J. DeRose and K. Burgess, J. Am. Chem. Soc., 2002, 124, 11946–11954 CrossRef CAS PubMed.
- M. M. Najafpour, F. Rahimi, M. Amini, S. Nayeri and M. Bagherzadeh, Dalton Trans., 2012, 41, 11026–11031 RSC.
- X. Zhou, Y. Zhang, C. Wang, X. Wu, Y. Yang, B. Zheng, H. Wu, S. Guo and J. Zhang, ACS Nano, 2012, 6, 6592–6599 CrossRef CAS PubMed.
- H. Bai, W. Jiang, G. P. Kotchey, W. A. Saidi, B. J. Bythell, J. M. Jarvis, J. M. Jarvis, A. G. Marshall, R. A. Robinson and A. Star, J. Phys. Chem. C, 2014, 118, 10519 CrossRef CAS PubMed.
- X. Zhou and L. Xu, Chem. Res. Chin. Univ., 2017, 33, 1–6 CrossRef CAS.
- F. Shi, Y. Zhang, W. Na, X. Zhang, Y. Li and X. Su, J. Mater. Chem. B, 2016, 4, 3278–3285 RSC.
- L. Tang, R. Ji, X. Cao, J. Lin, H. Jiang, X. Li, K. S. Teng, C. M. Luk, S. Zeng, J. Hao and S. P. Lau, ACS Nano, 2012, 6, 5102–5110 CrossRef CAS PubMed.
- M. Balaji, S. Jegatheeswaran, P. Nithya, P. Boomi, S. Selvam and M. Sundrarajan, J. Photochem. Photobiol., B, 2018, 178, 371–379 CrossRef PubMed.
- P. Routh, S. Das, A. Shit, P. Bairi, P. Das and A. K. Nandi, ACS Appl. Mater. Interfaces, 2013, 5, 12672–12680 CrossRef CAS PubMed.
- D. W. Lee, V. L. De Los Santos, J. W. Seo, L. L. Felix, D. A. Bustamante, J. M. Cole and C. H. W. Barnes, J. Phys. Chem. B, 2010, 114, 5723–5728 CrossRef CAS PubMed.
- Q. J. Lu, C. Y. Wu, D. Liu, H. Y. Wang, W. Su, H. T. Li, Y. Y. Zhang and S. Z. Yao, Green Chem., 2017, 19, 900–904 RSC.
- N. Duan, S. Wu, S. Dai, T. Miao, J. Chen and Z. Wang, Microchim. Acta, 2014, 182, 917–923 CrossRef.
- X. Cui, L. Zhu, J. Wu, Y. Hou, P. Wang, Z. Wang and M. Yang, Biosens. Bioelectron., 2015, 63, 506–512 CrossRef CAS PubMed.
- Z. Liu and X. Su, Biosens. Bioelectron., 2017, 87, 66–72 CrossRef CAS PubMed.
- F. Liu, M. H. Jang, H. D. Ha, J. H. Kim, Y. H. Cho and T. S. Seo, Adv. Mater., 2013, 25, 3657–3662 CrossRef CAS PubMed.
- E. Tan, B. Erwin, S. Dames, T. Ferguson, M. Buechel, B. Irvine, K. Voelkerding and A. Niemz, Biochemistry, 2008, 47, 9987–9999 CrossRef CAS PubMed.
- K. Roh, D. M. Kim, E. H. Lee, H. Kim, H. S. Park, J. H. Jang, S. H. Hwang and D. E. Kim, Chem. Commun., 2015, 51, 6960–6963 RSC.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |