Open Access Article
Open Access ArticleInsights into the production of extracellular polymeric substances of Cupriavidus pauculus 1490 under the stimulation of heavy metal ions
Weimin Zengabc,
Shishi Zhanga,
Mingchen Xiaa,
Xueling Wuab,
Guanzhou Qiuab and
Li Shen *ab
*ab
aSchool of Minerals Processing and Bioengineering, Central South University, Changsha 410083, China. E-mail: lishen@csu.edu.cn; Fax: +86-731-88879815; Tel: +86-731-88877472
bKey Laboratory of Biometallurgy, Ministry of Education, Changsha 410083, China
cCSIRO Process Science and Engineering, Clayton, Victoria 3168, Australia
First published on 27th May 2020
Abstract
Three different methods (a sulfuric acid method, sodium chloride method and vibration method) were used to extract extracellular polymeric substances (EPS) from Cupriavidus pauculus 1490 (C. pauculus 1490) in the present study. The sodium chloride method was able to extract the maximum amount of EPS (86.15 ± 1.50 mg g−1-DW), and could ensure minimum cell lysis by detecting glucose-6-phosphate dehydrogenase activity and using scanning electron microscopy. This method was therefore selected as the optimal extraction method and used in subsequent experiments. On this basis, the tolerance of C. pauculus 1490 and variations in EPS secretion after the addition of different metal ions was investigated. The tolerance levels of C. pauculus 1490 to Cd(II), Ni(II), Cu(II) and Co(II) were 300 mg L−1, 400 mg L−1, 400 mg L−1 and 400 mg L−1, respectively. Low concentrations of these heavy metal ions could promote bacterial growth, while increased concentrations were found to inhibit it. The results show that metal ions, especially Cd(II), stimulate the secretion of EPS, with an EPS yield reaching 956.12 ± 10.59 mg g−1-DW at 100 mg L−1. Real-time polymerase chain reaction (PCR) analysis showed that the key EPS synthetic genes, epsB, epsP and Wzz, were up-regulated. Fourier transform infrared spectroscopy analysis suggested that abundant functional groups in EPS play an important role in heavy metal ion complexation. These results will contribute to our understanding of the tolerance mechanism of microorganisms in the presence of different types and concentrations of metal ions.
1 Introduction
Heavy metals exist widely in nature at natural background concentrations.1 Due to the mining, smelting and processing of metals, heavy metals such as zinc (Zn), cadmium (Cd), copper (Cu), cobalt (Co) and nickel (Ni) have entered the atmosphere, water and soil, causing serious environmental pollution.2,3 Microorganisms are always at the start of the whole food chain, and thus metal ions will first affect their growth and activity, and subsequently pose a threat to other advanced living entities. Therefore, investigation of the behavior of microorganisms under the stimulation of metal ions is very important.4–6Cupriavidus is a Gram-negative bacterium of the class beta-proteus, which exists widely in the environment and clinic. So far, 17 species of bacteria from the genus Cupriavidus have been identified, most of which exhibit strong resistance to heavy metals (http://www.bacterio.net/cupriavidus.html). Cupriavidus pauculus (C. pauculus) shows tolerance to a variety of antibacterial agents, such as ampicillin, meropenem, aztreonam, gentamycin, tobramycin and fosfomycin.7 It has also been widely reported that C. pauculus could degrade and metabolize multiple harmful substances including benzene, toluene,8 biphenyl, phenol, 3-nitrophenol,9 herbicide 2,4-dichlorooxyacetic acid,10 pesticide chlorpyrifos and 3,5,6-trichloro-2-pyridol.11,12 Moreover, the tolerance of C. pauculus to Ni(II) and Cu(II) is much higher than that of other reported heavy metal-resistant strains.13 Pal and Paul studied changes in the yield of soluble extracellular polymeric substances (EPS) from C. pauculus KPS 201 under different growth conditions, including carbon sources, pH value and temperature.14 However, few studies have focused on variations in EPS component binding with the cell surface of C. pauculus under the stress of different heavy metal ions.
Because some microorganisms have evolved tolerance mechanisms that respond to heavy metals, they are able can survive in high heavy metal concentrations. Such tolerance mechanisms include the intracellular efflux pump mechanism and extracellular heavy metal complexation and precipitation, for example.6,15 Many studies have reported on the intracellular efflux pump at the gene level. The two-component signaling system (TCS) is the most common membrane conduction system in bacteria, and plays an important regulatory role in cell differentiation and metabolism, osmotic regulation, pathogenicity, chemotaxis, metal tolerance and other processes.16,17 In addition, microorganisms can also form EPS, an extracellular barrier, to prevent harmful exogenous substances entering the cells. For example, a large number of functional anionic groups in EPS can form complexation with metal ions to ensure that cells maintain normal cellular metabolism under heavy metal stress.18,19 On the other hand, in the field of wastewater treatment, EPS or biofilm is an important indicator for the ability of microorganisms to remove heavy metal. Ding observed that the EPS production of Bacillus vallismortis was very similar to its adsorption rate of heavy metals, within the corresponding range of heavy metal concentrations.20 EPS is a key factor of heavy metal biosorption.21,22 Therefore, EPS research can provide theoretical support for microorganism isolation and the adaptation process of heavy metal wastewater treatment.
EPS are macromolecule mixtures secreted by microorganisms.23 According to their location on cells, they can be divided into bound EPS or soluble EPS.24 The main components of EPS are polysaccharides, proteins and a small amount of nucleic acid and humic acid. The yield of EPS greatly depends upon the extraction methods, and varies in different species of microorganism or different growth phases.25 A suitable extraction method should achieve the maximum extracting yield of EPS and avoid contamination from cell lysis. Xia et al. studied the extraction methods of EPS from mixed fungi. The results showed that physical methods (heating, ultrasound and centrifugation) were less efficient than chemical methods, and resulted in higher cell lysis.23 However, Comte et al. found that the molecular weight of EPS extracted from activated sludge flocs by physical method was higher, and that the extraction reagents used in chemical methods might cause EPS pollution and affect the subsequent characterization of EPS.26 At present, there is no publication systematically discussing the best method of extracting EPS from the Cupriavidus species, so the work reported in this paper is necessary. In the relevant literature, three indicators have most commonly been used to evaluate the degree of cell damage in the process of extracting EPS. Many scholars believe that glucose-6-phosphate dehydrogenase (G6PDH) activity can be considered an indicator of cell lysis because it is only present inside the cells.27–29 Second, the content of 2-keto-3-deoxyoctonate acid (KOD) has been utilized as a similar indicator in some publications.30–32 Moreover, the proportion of nucleic acid in EPS and the ratio of exoprotein and exopolysaccharides are also regarded as key criteria to determine whether the cells were dissolved during the extraction process23,27,33 It is generally believed that the main reason for extracellular DNA (eDNA) accumulation is cell lysis and release.34–36
Many studies have shown that bacteria can be induced to produce more EPS under heavy metal stress. Research by Zeng et al. showed that Bacillus sp. S3 significantly increased EPS production under the stress of Cd(II), Cr(VI) and Cu(II), and that the surface functional groups of EPS complexed with heavy metal ions to indirectly reduce the toxicity of heavy metals to cells.37 In addition, Priester et al. found the same conclusion when they studied microbial cells in activated sludge and biofilm in the presence of heavy metals.18 The eps gene cluster has an obvious homology among different strains.38,39 It encodes a series of glycosyltransferases with different substrate specificities to guide the synthesis and secretion of exopolysaccharides. EpsB gene and epsP gene encoding tyrosine protein kinase and low-molecular-weight protein tyrosine phosphatase belong to these glycosyltransferases.40–43 In addition, the expression product of the Wzz gene is tyrosine protein kinase, which plays an important role in controlling the direction of extracellular polysaccharide chain length through transmembrane domains.44,45 Here, the gene expression of epsB, epsP and Wzz was analyzed to explore the influence of heavy metal ions on the extracellular polymer of C. pauculus 1490.
Different methods, including physical and chemical methods, were compared in extracting bound EPS from C. pauculus 1490. Thereafter the optimum method was applied in EPS extraction from C. pauculus 1490 under the stimulation of Cd(II), Co(II), Ni(II) and Cu(II). The expression of EPS synthesis key genes (epsB, epsP and Wzz) under heavy metal stress was studied. Fourier transform infrared spectroscopy (FTIR) detection was carried out to analyze the functional group changes of EPS before and after heavy metal stimulation. Investigations of the secretion and composition of EPS will be helpful in understanding the mechanism of the metal tolerance of bacteria and provide theoretical support for the treatment of heavy metal wastewater.
2 Materials and methods
2.1 Bacterial strain and culture conditions
The Cupriavidus pauculus 1490 used in this study was provided by China General Microbiological Culture Collection Center (CGMCC). The strain was inoculated into tris-buffered minimal medium, composed of (g L−1) tris-base (6.06), NaCl (4.68), KCl (1.49), NH4Cl (1.07), NaSO4 (0.43), MgSO4·6H2O (0.2), CaCl2·2H2O (0.03), Na2HPO4·12H2O (0.02) and 0.4% Na-gluconate, with initial pH of 7.0 ± 0.2 and incubated in the rotary shaker at 30 °C and 150 rpm.2.2 Optimization and evaluation of EPS extraction methods
2.3 Growth of bacteria under the stimulation of heavy metals
Analytically pure reagents, CdCl2, CoCl2, NiCl2 and CuCl2 (Shanghai Pharmaceutical Co. Ltd., China), provided the heavy metal ions Cd(II), Co(II), Ni(II) and Cu(II), respectively. The sterile mother liquor was prepared by dissolving heavy metals in distilled water and filtering with a 0.22 μm filter membrane. The heavy metals solution was diluted to the target concentration and the bacterial seed liquid was added to the culture medium simultaneously and incubated in a rotary shaker at 30 °C and 150 rpm. The optical density of C. pauculus 1490 suspension was measured at 600 nm (OD600) by ultraviolet (UV) spectrophotometer (Shimadzu UV-2550, Japan) at the same time intervals to determine the bacterial growth.2.4 EPS extraction from C. pauculus 1490 under heavy metal stimulation
Using the culture method described in section 2.2, samples were taken at regular intervals and the EPS of C. pauculus 1490 were extracted using the optimum method identified in section 2.3. The extracts were stored at 4 °C for the analysis of EPS components.2.5 Fourier transform infrared spectroscopy (FTIR) analysis
Changes in surface functional groups of bacterial cells under the stimulation of heavy metals were identified using Nicolet Nexus 670 FTIR Spectrometer (Thermo Nicolet Corporation, Madison, WI, USA) Spectrometer. Freeze-dried samples and spectroscopically pure potassium bromide (KBr) were prepared into tablets with a mass ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 for detection. The detection range was 4000 to 400 cm−1.
100 for detection. The detection range was 4000 to 400 cm−1.
2.6 Chemical analysis of the components and contents of EPS
The content of EPS components was expressed as mg g−1 of dry weight (mg g−1-DW). The content of polysaccharides in EPS was determined using the phenol–sulfuric acid method, with glucose as the standard.46 Using bovine serum albumin as standard, the protein content was determined using a BCA protein detection kit.47 DNA content was determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, USA). All extraction experiments were conducted in triplicate.2.7 Real-time polymerase chain reaction (PCR) analysis
TRIzol reagent was used to extract the total RNA of C. pauculus 1490 cultured with 200 mg L−1 Cd(II) for 24 h according to its operating instructions. Primers were designed by Primer Premier 5 software, and the sequences of epsB, epsP and Wzz were amplified with 16 s as the internal reference. The primers were synthesized by GenScript (Nanjing) Co. Ltd. The primer characteristics of the target gene are shown in Table 1. Gene expression was calculated by 2−ΔΔCT method48 as follows:| 2−ΔΔCT = 2−[(CT,TG−CT,CG)treated−(CT,TG−CT,CG)tested] |
| Gene name | Primer sequence (5′ to 3′) | Amplification length (bp) |
|---|---|---|
| epsB | F TGACTTCCCATCAGATCGACCCA | 199 |
| R GACGAACTGGAGGCTGATT | ||
| epsP | F GGCACGCACAACTATCACGTT | 125 |
| R AAGCGGTCGAAATCGGTCAC | ||
| Wzz | F AGGAATATGTCGAGCAGAACG | 190 |
| R ACCGACTGCGAGAGGATCAGC |
3 Results and discussion
3.1 Evaluation of EPS extraction from C. pauculus 1490 by different methods
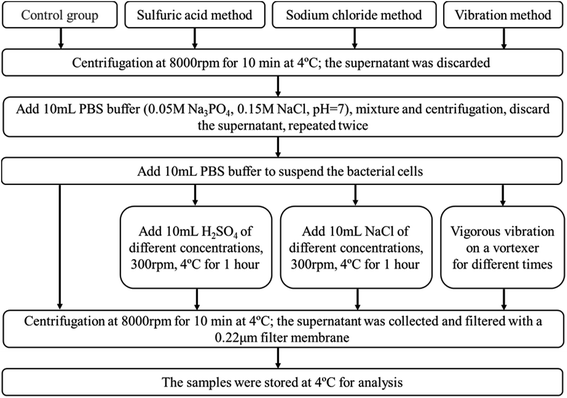
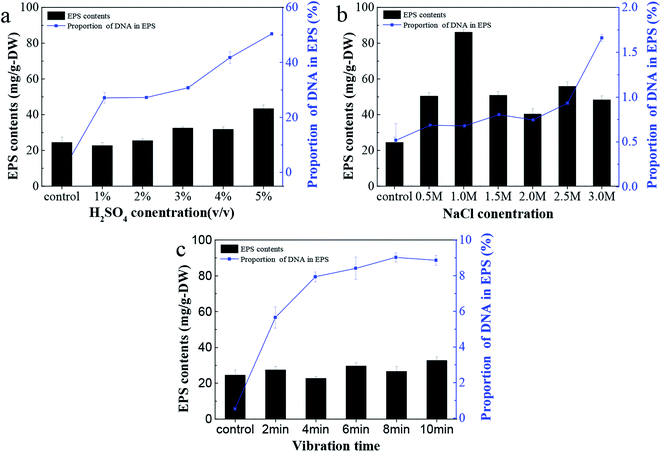
The EPS content from C. pauculus 1490 extracted by three different methods is shown in Fig. 2. It can be seen that as sulfuric acid concentration increases, the yield of EPS also increases (Fig. 2a). However, the proportion of DNA in EPS also increased substantially. Therefore, we concluded that sulfuric acid treatment could easily damage the cell wall and lead to the contamination of the EPS extracted.Treatment by NaCl and vibration caused less damage to the cell wall, and thus the release of DNA was obviously lower than previously (Fig. 2a and b). On the other hand, NaCl treatment could achieve higher EPS extraction. This is particularly the case when the concentration of NaCl added was 1 M, leading to a maximum EPS extraction of 86.15 ± 0.15 mg g−1-DW. Due to its outstanding advantages, such as mild reaction, high extraction efficiency and economy in terms of materials, many studies have reported the application of this method in extracting extracellular polymers.6,25,26,49,50 The components of EPS extracted with 1 M NaCl were analyzed, and it was shown that the content of polysaccharide and protein was 78.37 ± 0.65 mg g−1-DW and 7.18 ± 0.41 mg g−1-DW, respectively, while only a small amount of DNA was released from cell lysis.
Many authors have associated high activities of the cytosolic enzyme G6PDH in EPS extracts with a high extent of cell lysis.27 When the content of G6PDH in the EPS sample is relatively low, it can be considered that the degree of cell rupture is low.29 In order to further confirm the feasibility of NaCl treatment for EPS extraction in this study, G6PDH activity in EPS extracts was analyzed. The results show that when the concentration of NaCl was 1 M, the activity value of G6PDH (0.0718) was similar to that of the control experiment (0.0665). This further confirms that the cell lysis did not occur when NaCl solution was used. Therefore, in subsequent experiments, the NaCl treatment method was used to extract EPS from C. pauculus 1490.
In addition, the morphology of bacteria before and after EPS extraction was observed by SEM. As shown in Fig. 3a, the whole cell surface of C. pauculus 1490 is smooth and has relatively small striations. After NaCl treatment, the cell surface was rough with irregular pits, indicating that the NaCl method had a high efficiency (Fig. 3c). The cell morphology of the sample treated by the H2SO4 method (Fig. 3b) and the vibration method (Fig. 3d) are not different from that of the control sample, which further confirms that the NaCl method is more suitable for the EPS extraction from C. pauculus 1490.
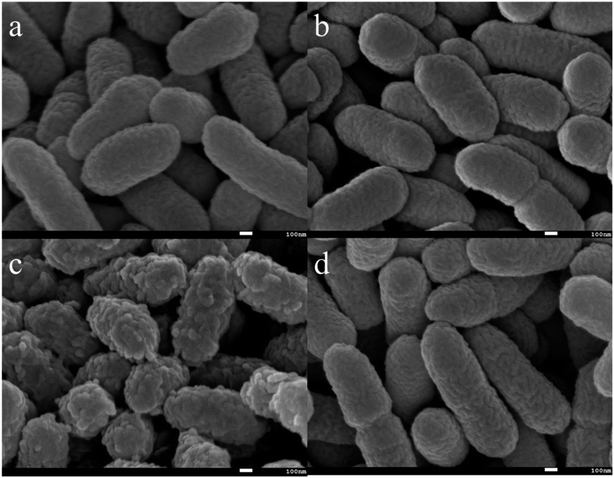 | ||
| Fig. 3 SEM images of C. pauculus 1490 after extracting EPS with different methods: (a) the control, (b) the sulfuric acid method, (c) the sodium chloride method, and (d) the vibration method. | ||
3.2 Tolerance of C. pauculus 1490 to various heavy metals
C. pauculus is short, rod-shaped and obtuse at both ends under optical microscope and can utilize multiple carbon sources for growth, such as glutamic acid, acetic acid, pyruvate, fumaric acid, malic acid, malonic acid, formic acid, citric acid and succinic acid.11In tris-buffered minimal medium with no heavy metal, C. pauculus 1490 could adapt well and enter into the logarithmic growth phase within 12 h. The maximum bacterial density of C. pauculus 1490 reached 8.2 × 108 cells per mL, with OD of 0.47 ± 0.01, when the carbon source was sodium gluconate (Fig. 4). In the medium with 100, 200 and 300 mg L−1 Cd(II), the OD600 value was 0.56 ± 0.02, 0.61 ± 0.01 and 0.64 ± 0.02, respectively. When the concentration of Cd(II) reached 400 mg L−1, the growth of the strain was completely inhibited (Fig. 4a). Under the stress of the heavy metals Co(II), Cu(II) and Ni(II), the growth of C. pauculus 1490 also showed similar phenomena (Fig. 4b and c). It can be concluded that heavy metals can promote the growth of C. pauculus 1490 within a certain concentration range, while at higher concentration, bacterial growth is severely restrained. Makkar and Casida also noted that C. necator strains grew faster in medium containing 0.4, 0.6 or 0.8 mM Cu(II) than the control strains.51
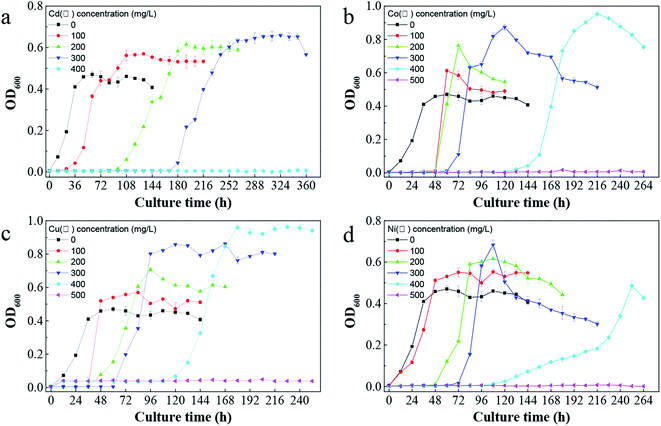 | ||
| Fig. 4 The impact of different heavy metals on the growth of C. pauculus 1490: (a) Cd(II), (b) Co(II), (c) Cu(II), and (d) Ni(II). | ||
In addition, Fig. 4 shows that the bacterial lag growth phase was clearly prolonged after adding heavy metals, and the delayed period increased with the increase of heavy metal concentration. In addition, the tolerance of C. pauculus 1490 to various heavy metals differed. Under the stress of Co(II), Cu(II) and Ni(II) at the same concentration (400 mg L−1), the OD600 values were 0.95 ± 0.02, 0.96 ± 0.91 and 0.48 ± 0.02, respectively, while cell growth was inhibited completely under 400 mg L−1 Cd(II). The tolerance of C. pauculus 1490 for Co(II), Cu(II), Ni(II) and Cd(II) was about 400 mg L−1, 400 mg L−1, 400 mg L−1 and 300 mg L−1, respectively, in the present study. It can be seen that Cd(II) was more toxic to C. pauculus 1490 than the other three metal ions. Due to their different degradation modes, easy accumulation and high toxicity, Cd(II) is one of the most frequently studied harmful heavy metals.52 Mohammad and Almas also concluded that cadmium is more toxic than other heavy metals such as copper, lead, zinc and nickel.53
SEM was carried out to analyze the cell surface under stimulation from different heavy metal ions. The cell surface was smooth in a pure culture (Fig. 5a). Meanwhile, cells surfaces were attached to the surface with sticky substances under the stress of Cd(II), which connects cells to each other (Fig. 5b). Comparing Fig. 5a and b, it can be seen that C. pauculus 1490 cells produced more extracellular polymers under heavy metal stimulation.
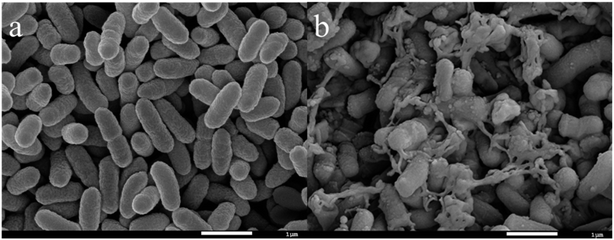 | ||
| Fig. 5 SEM images of C. pauculus 1490 before and after the stimulation of 200 mg L−1 Cd(II): (a) pure culture, and (b) Cd(II) stimulus. | ||
3.3 Effects of heavy metals on the production of EPS components
The change of EPS content from C. pauculus 1490 under the stress of different heavy metals is shown in Fig. 6. When the bacteria were exposed into the heavy metal solution, EPS was secreted at a very high level. As the incubation time was extended, the amount of EPS extracted decreased rapidly, to a very low value. For example, when 0.8 mg L−1 Cd(II) was added, the amount of EPS extracted reached 713.55 ± 24.03 mg g−1-DW at 12 h, which was reduced to 20.45 ± 1.34 mg g−1-DW at 24 h. It is reported that under heavy metal stress, bacteria tend to produce more EPS54–56 and form an immobile micro-precipitation by binding heavy metals with the functional groups on EPS, so as to alleviate the toxicity of heavy metals to cells.57 Once the bacteria adapted to the heavy metal environment or the intracellular defensive mechanism was initiated, EPS secretion decreased.37,58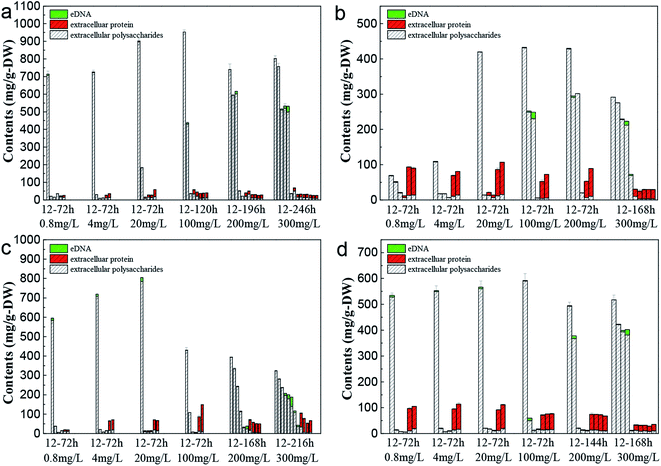 | ||
| Fig. 6 Changes in the EPS components of C. pauculus 1490 with time under different concentrations of heavy metals: (a) Cd(II), (b) Co(II), (c) Cu(II), and (d) Ni(II). | ||
Fig. 6 also shows that as the metal ion concentration increased, the amount of EPS first increased and then decreased. This phenomenon is consistent with the results reported by Sheng et al. and Ding et al., who found that a low concentration of metal ions would benefit the cell growth and EPS excretion, but that an excessive concentration of heavy metal would cause toxicity to the cell at physiological and molecular levels, resulting in enzyme inactivation and impeding EPS synthesis.20,59 On the other hand, the increase in metal ion concentration extended the lag growth phase of the cell (Fig. 2), and further led to a slow decrease of EPS extracted as a function of incubation time (Fig. 6).
The main components of EPS, extracellular polysaccharide and protein, showed different variation trends during the period of incubation. At the early stage, the percentage of polysaccharides in the EPS was very high. As the incubation time increased, the percentage of extracellular protein increased slowly, eventually becoming the main EPS components in the final stages. Particularly in the Ni(II), Co(II) and Cu(II) systems, the proportion of extracellular protein was higher, even exceeding 90%. Sheng and Yu found this same threshold when they studied the EPS secretion of R. acidophila.60 Ha et al. and Zhang et al. also reported that extracellular polysaccharides played an important role in the removal of heavy metals.19,61 This function is mainly because polysaccharides possess many functional anion groups, such as carboxyl, phosphate, mercapto, phenol and hydroxyl groups.18,62,63
During the whole growth cycle, the appearance of eDNA only occurred at the final incubation stage or at high metal ion concentrations. Lin and Wang's study also showed that bacteria secreted more eDNA under the stress of high Cu concentration.64 Sponz's study of activated sludge suggested that low levels of DNA were due to low mortality and cell lysis.65 A harsh environment, including a lack of nutrition and a high concentration of heavy metal, would lead to cell death and the release of large amounts of DNA.
To further understand the synthetic mechanism of EPS under the stimulation of typical metal ions, real-time PCR was carried out to investigate the key EPS synthetic gene (epsB, epsP and Wzz). It was found that the expression of the epsB gene and epsP gene increased approximately 3.31-times and 4.19-times during sampling, respectively, and that expression was significantly up-regulated. Another Wzz gene was also up-regulated, by about 2.93-times compared with the control group. It can be seen that the extracellular polysaccharide secretion of C. pauculus 1490 is increased under Cd2+ stimulation, which is consistent with the above results. Research by Elsholz et al. and Peize et al. showed that the epsB gene in the Bacillus subtilis and Bacillus licheniformis strains plays a significant role in the synthesis of extracellular polysaccharides, and overexpression can promote the increase of extracellular polysaccharides.40,41 The absence of the epsP gene mutant leads to the formation of heterocyst with a damaged polysaccharide layer in Streptococcus pneumoniae.42,43 Furthermore, the expression of the Wzz gene has also been studied by other researchers. Wzz gene expression products play a key role in the production of exopolysaccharides.44,45 The study of related genes can help us deepen the biosynthetic pathway of C. pauculus 1490 EPS under heavy metal-ion stress.
3.4 FTIR analysis of bacterial surface functional groups under heavy metal ion stress
Fig. 7 shows the IR absorption peaks of C. pauculus 1490 before and after the addition of heavy metal ions. Overall, the changing trends in the FTIR spectra of different kinds of heavy metal samples (Cd(II), Ni(II), Cu(II) and Co(II)) are similar, and phenomena such as peak shift and peak intensity change are noted. There is a wide band at 3422.77 cm−1 in the control group assigned to the –OH stretching vibration, mainly originating from exopolysaccharides.58,66 After the stimulation of 20 mg L−1 metal ions, this group shifted slightly to 3421.21 cm−1 (Cd), 3387.38 cm−1 (Ni), 3372.05 cm−1 (Cu) and 3390.26 cm−1 (Co), and further shifted to 3386.56 cm−1 (Cd), 3309.60 cm−1 (Ni), 3305.76 cm−1 (Cu) and 3192.52 cm−1 (Co), respectively, even overlapping with the –NH stretching vibration.58,66,67 Similarly, the peak at 1059.36 cm−1 representing the “fingerprint zone of polysaccharides” also shifted, and the degree of migration increased with increasing heavy metal concentration. Furthermore, the peaks at 2927.01 cm−1, 1649.39 cm−1, 1536.54 cm−1, 1400.53 cm−1 and 1239.38 cm−1, attributed to the –CH stretching vibration, amide I band (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and –CN stretching vibration), amide II band (–NH bending vibration and –CN stretching vibration) and P
O and –CN stretching vibration), amide II band (–NH bending vibration and –CN stretching vibration) and P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, respectively, showed a sharp shape and increased strength after exposure to the metal. This indicates that these groups were involved in the complexation of heavy metals.68–70
O, respectively, showed a sharp shape and increased strength after exposure to the metal. This indicates that these groups were involved in the complexation of heavy metals.68–70
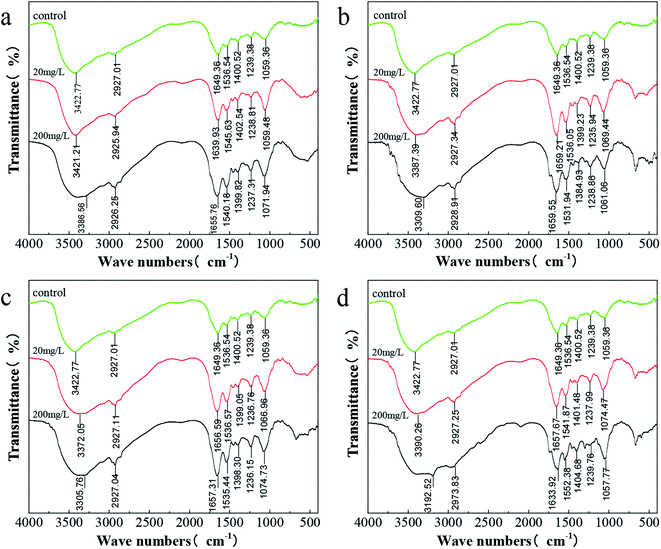 | ||
| Fig. 7 FTIR spectra of C. pauculus 1490 before and after the stimulation of different concentrations of heavy metals: (a) Cd(II), (b) Co(II), (c) Cu(II), and (d) Ni(II). | ||
4 Conclusions
In this study, EPS secretion from C. pauculus 1490 under the stimulation of Cd(II), Ni(II), Cu(II) and Co(II), and its tolerance to these metals, were investigated systematically for the first time. The optimum EPS extraction method was selected, and the results indicated that the NaCl method achieved the highest EPS yield and lowest cell lysis compared with the H2SO4 method and vibration method. It was found that the addition of heavy metal ions could promote cell growth at a relatively high concentration. Of the four heavy metal ions tested, Cd(II) displayed the strongest toxicity to C. pauculus 1490. Analysis of the EPS components of C. pauculus 1490 under the stimulation of heavy metals suggested that EPS secretion accelerated initially, but then decreased gradually. In addition, different kinds of heavy metal ions also have various effects on the amount of EPS secreted, among which Cd(II) more notably promoted the secretion of EPS. This effect was confirmed based on the expression of EPS-related genes. FTIR analysis showed that hydroxyl, carbonyl, carboxyl, and amino groups presented on the cell surface and their intensity and position changed at different levels under the simulation of different metal ions. In our next study, we will focus on the molecular mechanism of EPS secretion stimulated by heavy metals, in order to further clarify the tolerance of microorganisms to heavy metals and the removal mechanism.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31470230, 51320105006, 51604308), the Youth Talent Foundation of Hunan Province of China (No. 2017RS3003), the Natural Science Foundation of Hunan Province of China (No. 2018JJ2486), Key Research and Development Projects in Hunan Province (2018WK2012) and GDAS’ Special Project of Science and Technology Development (2018GDASCX-0917).References
- T. T. T. Dung, V. Cappuyns, R. Swennen and N. K. Phung, Rev. Environ. Sci. Biotechnol., 2013, 12, 335–353 CrossRef CAS.
- K. Kadirvelu, M. Palanival, R. Kalpana and S. Rajeswari, Bioresour. Technol., 2000, 74, 263–265 CrossRef CAS.
- P. Giovanella, L. Cabral, A. Costa, F. Camargo and F. Bento, Ecotoxicol. Environ. Saf., 2017, 140, 162–169 CrossRef CAS PubMed.
- W. Peng, X. Li and S. Xiao, J. Soils Sediments, 2018, 18 Search PubMed.
- A. Mishra and A. Malik, Crit. Rev. Environ. Sci. Technol., 2013, 43, 1162–1222 CrossRef CAS.
- T. T. More, J. S. S. Yadav, S. Yan, R. D. Tyagi and R. Y. Surampalli, J. Environ. Manage., 2014, 144, 1–25 CrossRef CAS PubMed.
- D. Q. Fu, J. F. Zhang, S. J. Ding, X. P. Ge and Y. X. Sun, Chin. J. Health Lab. Technol., 2018, 28, 166–168 Search PubMed.
- E. H. Lee, H. Park and K. S. Cho, J. Environ. Sci. Health - Part A Toxic/Hazard. Subst. Environ. Eng., 2013, 48, 273–278 CrossRef CAS PubMed.
- X. Q. Chi, J. J. Zhang, S. Zhao and N. Y. Zhou, Environ. Pollut., 2013, 172, 33–41 CrossRef CAS PubMed.
- T. Kraiser, M. Stuardo, M. Manzano, T. Ledger and B. Gonzalez, Plant Soil, 2013, 366, 377–388 CrossRef CAS.
- L. Cao, H. M. Liu, H. Zhang, K. Huang, T. Gu, H. Y. Ni, Q. Hong and S. P. Li, Curr. Microbiol., 2012, 65, 231–236 CrossRef CAS PubMed.
- P. Lu, Q. Li, H. Liu, Z. Feng, X. Yan, Q. Hong and S. Li, Bioresour. Technol., 2013, 127, 337–342 CrossRef CAS PubMed.
- M. Ramirez, J. Obrzydowski, M. Ayers, S. Virparia, M. Wang, K. Stefan, R. Linchangco and D. Castignetti, TheScientificWorldJournal, 2014, 2014, 901702 Search PubMed.
- A. Pal and A. K. Paul, J. Polym., 2013, 2013, 7 Search PubMed.
- m.-l. Ledrich, S. Stemmler, P. Laval-Gilly, L. Foucaud and J. Falla, Biometals, 2006, 18, 643–650 CrossRef PubMed.
- D. Beier and R. Gross, Curr. Opin. Microbiol., 2006, 9, 143–152 CrossRef CAS PubMed.
- M. Galperin, BMC Microbiol., 2005, 5, 35 CrossRef PubMed.
- J. Priester, S. Olson, S. Webb, M. Neu, L. Hersman and P. Holden, Appl. Environ. Microbiol., 2006, 72, 1988–1996 CrossRef CAS PubMed.
- J. Ha, A. Gelabert, A. Spormann and G. Brown, Geochim. Cosmochim. Acta, 2010, 74, 1–15 CrossRef CAS.
- P. Ding, S. Weifeng, Z. Yang and J. Jian, Bioprocess Biosyst. Eng., 2018, 41 Search PubMed.
- S. Comte, G. Guibaud and M. Baudu, Process Biochem., 2006, 41, 815–823 CrossRef CAS.
- M. Solís, A. Solís, H. I. Pérez, N. Manjarrez and M. Flores, Process Biochem., 2012, 47, 1723–1748 CrossRef.
- M. C. Xia, P. Bao, S. S. Zhang, A. J. Liu, L. Shen, R. L. Yu, Y. D. Liu, M. Chen, J. K. Li, X. L. Wu, G. Z. Qiu and W. M. Zeng, Environ. Sci. Pollut. Res., 2019, 26, 22137–22146 CrossRef CAS PubMed.
- J. Tourney and B. Ngwenya, Chem. Geol., 2014, 386, 115–132 CrossRef CAS.
- J. Wingender, T. Neu and H.-C. Flemming, Microbial Extracellular Polymeric Substances: characterization, structure and function, 1999, DOI:10.1007/978-3-642-60147-7_1.
- S. Comte, G. Guibaud and M. Baudu, J. Hazard. Mater., 2007, 140, 129–137 CrossRef CAS PubMed.
- B. Frølund, R. Palmgren, K. Keiding and P. Nielsen, Water Res., 1996, 30, 1749–1758 CrossRef.
- J. F. Wu and C. W. Xi, Appl. Environ. Microbiol., 2009, 75, 5390–5395 CrossRef CAS PubMed.
- C. Pellicer-Nàcher, C. Domingo-Félez, A. G. Mutlu and B. Smets, Water Res., 2013, 47, 5564–5574 CrossRef PubMed.
- W. Zeng, G. Qiu, H.-b. Zhou, X. Liu, M. Chen, W. Chao, Z. Chenggui and J. Peng, Hydrometallurgy, 2010, 100, 177–180 CrossRef CAS.
- Y. D. Karkhanis, J. Y. Zeltner, J. J. Jackson and D. J. Carlo, Anal. Biochem., 1978, 85, 595–601 CrossRef CAS PubMed.
- T. Gehrke, J. Telegdi, D. Thierry and W. Sand, Appl. Environ. Microbiol., 1998, 64, 2743–2747 CrossRef CAS PubMed.
- B. Q. Liao, D. G. Allen, I. G. Droppo, G. G. Leppard and S. N. Liss, Water Res., 2001, 35, 339–350 CrossRef CAS PubMed.
- H. Hamilton, N. Domínguez, K. Schwartz, K. Hackett and J. Dillard, Mol. Microbiol., 2005, 55, 1704–1721 CrossRef CAS PubMed.
- Z. Qin, Y. Ou, L. Yang, Y. Zhu, T. Tolker-Nielsen, S. Molin and Q. Di, Microbiology, 2007, 153, 2083–2092 CrossRef CAS PubMed.
- H. Steinmoen, E. Knutsen and L. Håvarstein, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 7681–7686 CrossRef CAS PubMed.
- W. Zeng, F. Li, C. Wu, R. Yu, X. Wu, L. Shen, Y. Liu, G. Qiu and J. Li, Bioprocess Biosyst. Eng., 2019, 43, 153–167 CrossRef PubMed.
- B. Veith, C. Herzberg, S. Steckel, J. Feesche, K. H. Maurer, P. Ehrenreich, S. Baumer, A. Henne, H. Liesegang, R. Merkl, A. Ehrenreich and G. Gottschalk, J. Mol. Microbiol. Biotechnol., 2004, 7, 204–211 CrossRef CAS PubMed.
- S. Yan, N. Wang, Z. Chen, Y. Wang, N. He, Y. Peng, Q. Li and X. Deng, Funct. Integr. Genomics, 2013, 13, 425–434 CrossRef CAS PubMed.
- A. Elsholz, S. Wacker and R. Losick, Genes Dev., 2014, 28, 1710–1720 CrossRef CAS PubMed.
- P. Liu, Z. Chen, L. Yang, Q. Li and N. He, Microb. Cell Factories, 2017, 16, 163 CrossRef PubMed.
- H. Tan, S. Wan, P.-Q. Liu, W. Li, C.-C. Zhang and W.-L. Chen, Res. Microbiol., 2013, 164 Search PubMed.
- Z. Ahmad, R. Morona and A. Standish, Microbiology, 2018, 164 Search PubMed.
- R. Collins, V. Kargas, B. Clarke, C. Siebert, D. Clare, P. Bond, C. Whitfield and R. Ford, Structure, 2017, 25, 806–815 CrossRef CAS PubMed.
- S. M. Huszczynski, C. Coumoundouros, P. Pham, J. S. Lam and C. M. Khursigara, J. Bacteriol., 2019, 201, e00165-19 CrossRef PubMed.
- A. Gong, C. Bolster, M. Benavides and S. Walker, Environ. Eng. Sci., 2009, 26, 1523–1532 CrossRef CAS.
- R. Yu, C. Hou, A. Liu, T. Peng, X. Mingchen, X. Wu, L. Shen, Y. Liu, J. Li, F. Yang, G. Qiu, M. Chen and W. Zeng, Hydrometallurgy, 2018, 176 Search PubMed.
- H. VanGuilder, K. Vrana and W. Freeman, BioTechniques, 2008, 44, 619–626 CrossRef CAS PubMed.
- M. Sesay, G. Ozcengiz and D. Sanin, Water Res., 2006, 40, 1359–1366 CrossRef CAS PubMed.
- G.-P. Sheng, H.-Q. Yu and X.-Y. Li, Biotechnol. Adv., 2010, 28, 882–894 CrossRef CAS PubMed.
- S. Monchy, R. Benotmane, P. Janssen, T. Vallaeys, S. Taghavi, D. Lelie and M. Mergeay, J. Bacteriol., 2007, 189, 7417–7425 CrossRef CAS PubMed.
- Y. Wang, J. Shi, H. Wang, Q. Lin, X. Chen and Y. Chen, Ecotoxicol. Environ. Saf., 2007, 67, 75–81 CrossRef CAS PubMed.
- M. Oves, M. Khan and A. Zaidi, Saudi J. Biol. Sci., 2013, 20, 121–129 CrossRef CAS PubMed.
- B. Mohite, S. Koli and S. Patil, Appl. Biochem. Biotechnol., 2018, 186 Search PubMed.
- G.-P. Sheng, H.-Q. Yu and Z.-B. Yue, Appl. Microbiol. Biotechnol., 2005, 69, 216–222 CrossRef CAS PubMed.
- X. Wei, L. Fang, P. Cai, Q. Huang, H. Chen, W. Liang and X. Rong, Environ. Pollut., 2013, 159, 1369–1374 CrossRef PubMed.
- M. L. Fisher, R. Allen, Y. Q. Luo and R. Curtiss, PLoS One, 2013, 8, 10 Search PubMed.
- L. Shen, Z. F. Li, J. J. Wang, A. J. Liu, Z. H. Li, R. L. Yu, X. L. Wu, Y. D. Liu, J. K. Li and W. M. Zeng, Environ. Sci. Pollut. Res., 2018, 25, 20713–20722 CrossRef CAS PubMed.
- G.-P. Sheng, H.-Q. Yu and Z. Yu, Appl. Microbiol. Biotechnol., 2005, 67, 125–130 CrossRef CAS PubMed.
- G.-P. Sheng and H.-Q. Yu, Appl. Microbiol. Biotechnol., 2006, 72, 126–131 CrossRef CAS PubMed.
- D. Y. Zhang, J. L. Wang and X. L. Pan, J. Hazard. Mater., 2006, 138, 589–593 CrossRef CAS PubMed.
- P. M. Joshi and A. A. Juwarkar, Environ. Sci. Technol., 2009, 43, 5884–5889 CrossRef CAS PubMed.
- H. Liu and H. H. P. Fang, Biotechnol. Bioeng., 2002, 80, 806–811 CrossRef CAS PubMed.
- H. Lin, C. Wang, H. Zhao, G. Chen and X. Chen, Environ. Sci. Pollut. Res., 2018, 25, 24172–24180 CrossRef CAS PubMed.
- D. T. Sponza, Process Biochem., 2002, 37, 983–998 CrossRef CAS.
- S. Ozturk, B. Aslim and Z. Suludere, Bioresour. Technol., 2010, 101, 9742–9748 CrossRef CAS PubMed.
- S. Singh, S. Pradhan and L. C. Rai, Process Biochem., 2000, 36, 175–182 CrossRef CAS.
- W. J. Yuan, J. Cheng, H. X. Huang, S. L. Xiong, J. Q. Gao, J. Zhang and S. Feng, Ecotoxicol. Environ. Saf., 2019, 175, 138–147 CrossRef CAS PubMed.
- M. Kl, C. Kirschner, L.-P. i. Choo-Smith, N. van den Braak, H. Endtz, D. Naumann and G. Puppels, J. Microbiol. Methods, 2002, 51, 255–271 CrossRef.
- S. Comte, G. Guibaud and M. Baudu, Enzyme Microb. Technol., 2006, 38, 237–245 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2020 |