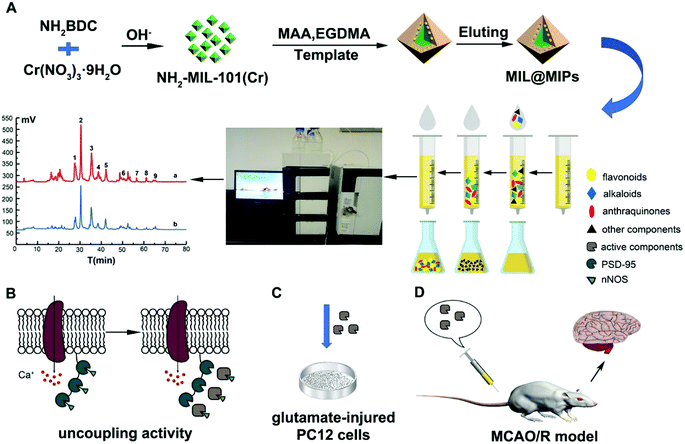
Open Access Article
Open Access ArticleModeling rapid and selective capture of nNOS–PSD-95 uncouplers from Sanhuang Xiexin decoction by novel molecularly imprinted polymers based on metal–organic frameworks†
Linli Pana,
Yingying Dinga,
Xiaoting Nia,
Chong-Zhi Wangb,
Bo Jianga,
Yu Zhanga,
Nan Jianga,
Yulin Tanga,
Lina Chen *a and
Chun-Su Yuanb
*a and
Chun-Su Yuanb
aSchool of Pharmacy, Nanjing Medical University, Nanjing, Jiangsu 211166, China. E-mail: chenlina@njmu.edu.cn; Fax: +86 25 8686 8467; Tel: +86 25 8686 8478
bTang Center for Herbal Medicine Research, Department of Anesthesia & Critical Care, University of Chicago, Chicago, IL 60637, USA
First published on 21st February 2020
Abstract
Novel and highly selective molecularly imprinted polymers based on the surface of metal–organic frameworks, NH2-MIL-101(Cr) (MIL@MIPS), were successfully fabricated to capture neuronal nitric oxide synthase–postsynaptic density protein-95 (nNOS–PSD-95) uncouplers from Sanhuang Xiexin Decoction (SXD) for stroke treatment. The resultant polymers were characterized by Fourier transform infrared spectroscopy, scanning electron microscopy, thermogravimetric analysis, and X-ray diffraction. The performance tests revealed that MIL@MIPs had a large binding capacity, fast kinetics, and excellent selectivity. Then the obtained polymers were satisfactorily applied to solid-phase extraction coupled with high-performance liquid chromatography to selectively capture nNOS–PSD-95 uncouplers from SXD. Furthermore, the biological activities of components obtained from SXD were evaluated in vivo and in vitro. As a consequence, the components showed a potent neuroprotective effect from the MTS assay and uncoupling activity from the co-immunoprecipitation experiment. In addition, the anti-ischemic stroke assay in vivo was further investigated to determine the effect of reducing infarct size and ameliorating neurological deficit by the active components. Therefore, this present study contributes a valuable new method and new tendency to selectively capture active components for stroke treatment from SXD and other natural medicines.
1. Introduction
Ischemic stroke, characterized by high mortality, a high rate of disability, and high recurrence rate, seriously endangers human health.1,2 Under the condition of cerebral ischemia, excitatory amino acids are released excessively, causing the over-activation of N-methyl-D-aspartate receptor (NMDAR) and the excitation of neuronal nitric oxide synthase (nNOS), which finally results in the over-release of nitric oxide (NO), apoptosis, and nerve damage.3,4 It was found that blocking the coupling of postsynaptic density protein-95 (PSD-95) and nNOS, downstream of the NMDAR–PSD-95–nNOS signaling pathway, can inhibit the pathological release of NO caused by cerebral ischemia and then cure ischemic stroke without affecting the physiological functions of NMDAR and nNOS.5–7 IC87201 (ref. 8) and ZL006,9 small molecule inhibitors of the nNOS–PSD-95 interaction, were developed and reported to have potent neuroprotective activity both in vivo and in vitro. However, considering the strong hydrophilicity and unsatisfactory brain distribution of those small molecules, new nNOS–PSD-95 uncouplers need to be developed urgently.10Natural medicine appears to be an important source to find novel nNOS–PSD-95 uncouplers due to its complex components and excellent neuroprotection, and we had captured potential nNOS–PSD-95 uncouplers such as baicalein (BAI), berberine (BER), and emodin (EMO) from natural medicine11–13 in our previous work. Sanhuang Xiexin Decoction (SXD), a classic prescription of natural medicine, is composed of Scutellariae Radix (SR), Coptidis Rhizome (CR), and Rhei Rhizome (RR) at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.14,15 Modern pharmacological studies have found that SXD has a lot of bioactivities including anti-inflammatory,16 anti-oxidative,17 antidiabetics,18 anti-hypertension and anti-hyperlipidemia,19 anti-atherogenic20 and anti-cerebral ischemia reperfusion injury activities.21 According to our preliminary experiment, SXD also had obvious uncoupling activity of nNOS–PSD-95. However, it is a time-consuming and laborious task to discover and capture powerful uncouplers from SXD due to its complicated chemical composition. Therefore, a highly sensitive, specific and rapid separation detection method is urgently needed to quickly find nNOS–PSD-95 uncouplers from SXD for anti-stroke applications.
2.14,15 Modern pharmacological studies have found that SXD has a lot of bioactivities including anti-inflammatory,16 anti-oxidative,17 antidiabetics,18 anti-hypertension and anti-hyperlipidemia,19 anti-atherogenic20 and anti-cerebral ischemia reperfusion injury activities.21 According to our preliminary experiment, SXD also had obvious uncoupling activity of nNOS–PSD-95. However, it is a time-consuming and laborious task to discover and capture powerful uncouplers from SXD due to its complicated chemical composition. Therefore, a highly sensitive, specific and rapid separation detection method is urgently needed to quickly find nNOS–PSD-95 uncouplers from SXD for anti-stroke applications.
Recently, the developments of molecularly imprinted polymers (MIPs) most likely benefit the study of active components.22–24 Molecular imprinting technology (MIT), imprinting the template molecule onto a substrate and leaving it with specific recognition sites that match the imprinted molecule in three dimensions,25 has shown great advantages such as strong resistance to harsh environments, good stability, low cost and easy synthesis, and superior adsorption capacity, and has been applied in natural medicine metabolism analysis,26 proteins separation,27 environmental surveillance,28 immunoassay,29 biosensors,30 and simulated enzyme catalysis.31 Therefore, MIPs can be used as artificial receptors to capture new nNOS–PSD-95 uncouplers.
However, the applications of traditional MIPs are limited by their low adsorption and selectivity, irregular shape, and nonuniform distribution of binding sites.32 To overcome the above disadvantages, metal–organic frameworks (MOFs) were introduced into the field of MIPs. MOFs are a relatively new type of porous materials with a three-dimensional pore structure that have developed rapidly in recent years, whose surface area is large, porosity is high, structure is regular, and quality is controllable.33,34 Among which, MIL-101(Cr) has aroused scientific interest for its high chemical and hydrothermal stability, tremendous functional multiplicity, and high specific surface area, which make it an ideal support material.35,36 Moreover, MIL-101(Cr) was employed as adsorbents for solid phase extraction (SPE) for the determination of sulphonamides in environmental water samples coupling with UPLC-MS/MS.37 A core–shell MIPs coated onto the surface of MIL-101(Cr) was prepared, which was used as absorbents for the detection of pyrraline in milk and milk powder.38 NH2-MIL-101(Cr) was synthesized via a direct hydrothermal method and was coated with polydimethylsiloxane and stir bar for the determination of organophosphorus pesticides in environmental water samples.39
Herein, we have prepared highly selective surface molecularly imprinted polymers based on NH2-MIL-101(Cr) (MIL@MIPs) by molecular imprinting technique (Fig. 1). The characterization, adsorption performance and selectivity capacity were systematically investigated to evaluate the obtained polymers. Then the prepared MIL@MIPs were successfully used as sorbents to quickly discover and efficiently capture potential nNOS–PSD-95 uncouplers from SXD coupled with SPE and HPLC (MIL@MIPs-SPE-HPLC). Finally, SXD and the potential uncouplers captured from SXD were tested with neuroprotective effects on PC12 cells and uncoupling activity by coimmunoprecipitation experiments in vitro. Furthermore, anti-ischemic stroke efficacy was investigated using a reperfusion (MCAO/R) model in vivo.
2. Experimental
2.1 Materials
2-Aminoterephthalic acid (NH2BDC) and edaravone (EDA) were purchased from Macklin (Shanghai, China). Chromium(III) nitrate nonahydrate (Cr(NO3)3·9H2O) was obtained from Xilong Scientific Co., Ltd. (Beijing, China). Ultrapure water was produced by the Millipore water purification system (Darmstadt, Germany). Ethylene glycol dimethacrylate (EGDMA), 2,2-azobisisobutyronitrile (AIBN), methacrylic acid (MAA), methyl methacrylate (MMA), 4-vinylpyridine (4-VP), acetonitrile (ACN), and formic acid of HPLC grade were acquired from Aladdin (Shanghai, China). 2,3,5-Triphenyltetrazolium chloride (TTC) and dimethyl sulfoxide (DMSO) were obtained from Sigma-Aldrich (Steinheim, Germany). 3-(4,5-Dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) was provided by Promega (Madison, America). ZL006 was provided by Prof. F Li from school of Pharmacy, Nanjing Medical University (Nanjing, China). All the herbal medicines and standards were purchased from Nanjing Yuanbaofeng Pharm-Tech Co., Ltd. (Nanjing, China). All other chemicals used in this paper were of analytical reagent grade.2.2 Apparatus
A heating magnetic stirrer of C-MAG HS 7 (Staufen, Germany) and an ultrasonic cleaner of KQ-3200B (Jiangsu, China) were applied during the preparation of MIPs. A shake culture box of ZHLY-180 (Shanghai, China) and a centrifugal machine of TG16-WS (Shanghai, China) were used during the elution and adsorption experiment. The optimizing of preparation conditions and evaluation of adsorptive property were conducted by an ultraviolet spectrophotometer of Shimadzu UV-2450 (Kyoto, Japan). The characteristics were carried out with a scanning electron microscopy of FEI Nova Nano SEM 450 (SEM, Hillsboro, USA), a thermogravimetric analyzer of STA449C (TGA, Netzsch, Germany), a Fourier Transform Infrared spectrometer of TENSOR27 (FT-IR, Bruker, Germany), a N2 adsorption–desorption analyzer of Autosorb-iQ EVO (Quantachrome, USA), and an elemental analyzer of Vario EL III (Elementar, Germany). A HPLC system of LC-20 (Shimadzu, Japan) was used for HPLC analysis and a solid phase extraction apparatus of ASE-12 (Auto Science, China) was used for extraction of target objects.2.3 Synthesis of NH2-MIL-101(Cr)
NH2-MIL-101(Cr) was synthesized according to a previous report with some modifications.40 Cr(NO3)3·9H2O (2 mmol), NH2BDC (2 mmol), and NaOH (4 mmol) were added to 15 mL deionized water and stirred for 12 min, and then the reaction solution was transferred to a 30 mL Teflon-lined stainless steel autoclave and kept at 150 °C for 12 h. Afterwards, the autoclave was left to naturally cool down to room temperature. Then it was taken out and the green suspension was centrifuged and washed with DMF at room temperature to remove unreacted NH2BDC for several times and ethanol for 2–3 times. The products were then dispersed into a 50 mL autoclave with 25 mL hot ethanol and kept at 100 °C for 24 h to remove unreacted NH2BDC in the pores of the product. Finally, the products were taken out and dried at 50 °C for 24 h in a vacuum.2.4 Preparation of MIL@MIPs
In our previous work, BAI, BER, and EMO were found to have potential uncoupling ability,11–13 thus they were chosen as templates to synthesize different imprinted polymers (BAI@MIPs, BER@MIPs and EMO@MIPs) respectively, using a surface imprinted polymerization method. Firstly, the template and the functional monomer were dispersed in 40 mL solvent and left to incubate for 4 h at room temperature for self-assembly. Then NH2-MIL-101(Cr), the cross-linker (EGDMA) and the initiator (AIBN) were added and the prepared solution was stirred for 24 h at 60 °C under the protection of nitrogen. After polymerization, the products were repeatedly eluted by methanol–acetic acid (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) to absolutely remove templates, and then they were washed with methanol to remove traces of acetic acid until the solution was neutral. Finally, the obtained MIL@MIPs were dried at 50 °C for 24 h in a vacuum. MIL@NIPs were prepared in the same condition just in the absence of templates.
1, v/v) to absolutely remove templates, and then they were washed with methanol to remove traces of acetic acid until the solution was neutral. Finally, the obtained MIL@MIPs were dried at 50 °C for 24 h in a vacuum. MIL@NIPs were prepared in the same condition just in the absence of templates.
2.5 Evaluation of adsorption performance of MIL@MIPs
To evaluate the adsorption capacity of obtained MIL@MIPs, the static, kinetic, selective, regenerate and reusable adsorption experiments were carried out. All the experiments were conducted for three times in parallel.For static adsorption experiments, 20 mg MIL@MIPs and MIL@NIPs were respectively suspended in 5 mL EMO, BAI, or BER acetonitrile/ethyl alcohol solution (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v) with different initial concentrations ranging from 0.5 to 10.0 mmol L−1. The solution was dispersed by being shaken for 2 h and then detected by UV-Vis spectrophotometer. The adsorption amount (Qe) of polymers was calculated using the following eqn (1):
2, v/v) with different initial concentrations ranging from 0.5 to 10.0 mmol L−1. The solution was dispersed by being shaken for 2 h and then detected by UV-Vis spectrophotometer. The adsorption amount (Qe) of polymers was calculated using the following eqn (1):
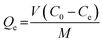 | (1) |
Similarly, for kinetic experiments, 20 mg MIL@MIPs were suspended in 5 mL EMO, BAI, or BER acetonitrile/ethyl alcohol solution with a concentration of 7, 7, or 9, respectively. After that, the solution was shaken under room temperature for a certain interval (5, 10, 15, 20, 25, 30, 40, 50 min).
For selective adsorption experiments, EMO, BAI, BER, and their three structural-liked compounds were assessed, respectively. To investigate the reusability of MIL@MIPs, adsorption–desorption experiments were conducted. The adsorption procedure was the same as described above, and then the MIL@MIPs were eluted with methanol/acetic acid (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). After that, the regenerated MIL@MIPs were reused for the next adsorption.
1, v/v). After that, the regenerated MIL@MIPs were reused for the next adsorption.
2.6 Purification and capturing nNOS–PSD-95 uncouplers from SXD by MIL@MIPs-SPE-HPLC system
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) to obtain active components. Both the washing and elution solutions were evaporated to dryness by a nitrogen stream at room temperature and the obtained residues were dissolved in 1 mL methanol for further HPLC analysis.
1, v/v) to obtain active components. Both the washing and elution solutions were evaporated to dryness by a nitrogen stream at room temperature and the obtained residues were dissolved in 1 mL methanol for further HPLC analysis.2.7 Activity evaluation of active components obtained from SXD
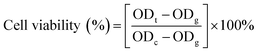 | (2) |
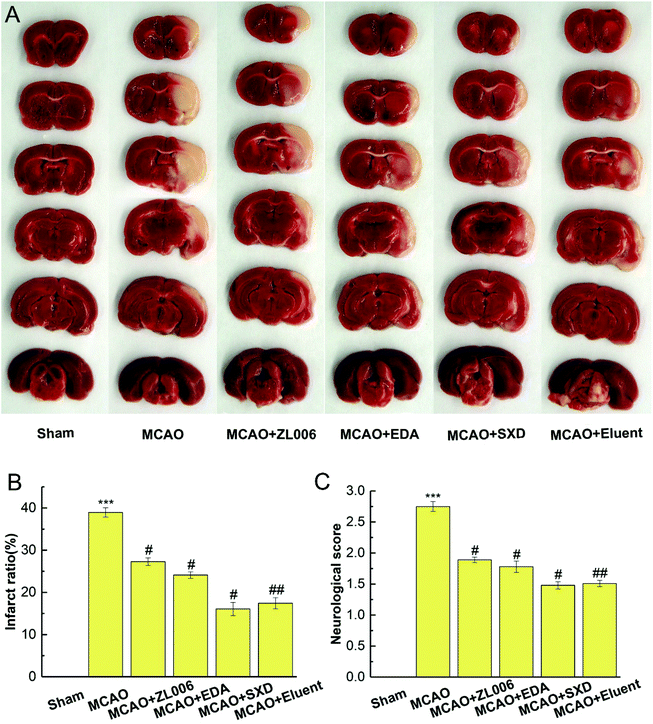
The right middle cerebral artery occlusion was operated to induce focal cerebral ischemia with MCAO occlusion line. Briefly, the rats were anesthetized with 10% chloral hydrate (4 mL kg−1) by intraperitoneal injection. Then their right common carotid artery was exposed, and the coated filament was introduced about 20 mm. After 2 h of occlusion, the line was lightly withdrawn and the rats were treated with drugs or 0.9% physiological saline via tail vein injection right away. After reperfusion for 24 h, a 5-point neurological function score was employed to evaluate neurological deficit by an observer blinded to the trial. Subsequently, the rats were decapitated and the brains were dissected coronally, and then the slices were immediately stained with 2% TTC solution at 40 °C for 60 min. The stained slices were finally photographed and quantified for ischemic injury by an image assay system. The percentage of infarction was calculated using the following eqn (3):
 | (3) |
3. Results and discussion
3.1 Optimization the preparation condition of MIL@MIPs
The adsorption ability and selectivity of MIL@MIPs mainly depended on the amount of template, solvent, functional monomer, and the ratio of template/functional monomer/cross-linker. Thus, we prepared different polymers considering of these factors, and the results were shown in Table S2–S4.†EMO, BAI, and BER were chosen as templates because they were the main components of RR, SR, and CR, respectively. Above all, EMO@MIPs-1, EMO@MIPs-2, and EMO@MIPs-3 were prepared to investigate the effect of the amounts of templates. As shown in Table S2,† EMO@MIPs-2 performed the best adsorption ability when the quantities of NH2-MIL-101(Cr) used were 0.05 g and 0.5 mmol EMO was chosen as the optimal amount of template finally. Solvent plays an important role in preparing EMO@MIPs as well because it is not only the agent to induce holes but also the solvent to distribute the drug evenly. It could be found that EMO@MIPs-2 (acetonitrile/ethanol, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v) exhibited the highest adsorption capacity compared with EMO@MIPs-4 and EMO@MIPs-5. EMO@MIPs-4 was prepared with tetrahydrofuran/ethanol. The polymer was hard and difficult to disperse completely, thus its application was limited.
2, v/v) exhibited the highest adsorption capacity compared with EMO@MIPs-4 and EMO@MIPs-5. EMO@MIPs-4 was prepared with tetrahydrofuran/ethanol. The polymer was hard and difficult to disperse completely, thus its application was limited.
In addition, a functional monomer was used to self-assemble with template, and three kinds of functional monomers (MAA, MMA and 4-VP) were investigated to obtain the best one. It was shown that the polymers synthesized using MAA had the best adsorption ability. Moreover, we synthesized different polymers with a different ratio of template/functional monomer/cross-linker because it had a great influence on the adsorption properties, and the results showed that the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 was superior to the others. In conclusion, we found that 0.5 mmol EMO, acetonitrile/ethanol (1
30 was superior to the others. In conclusion, we found that 0.5 mmol EMO, acetonitrile/ethanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v), MAA, and a ratio of 1
2, v/v), MAA, and a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 were the optimal conditions to synthesize EMO@MIPs. The optimal synthesis conditions and the optimization process of BAI@MIPs and BER@MIPs were exhibited in Table S3 and S4,† respectively.
30 were the optimal conditions to synthesize EMO@MIPs. The optimal synthesis conditions and the optimization process of BAI@MIPs and BER@MIPs were exhibited in Table S3 and S4,† respectively.
3.2 Characterization of MIL@MIPs
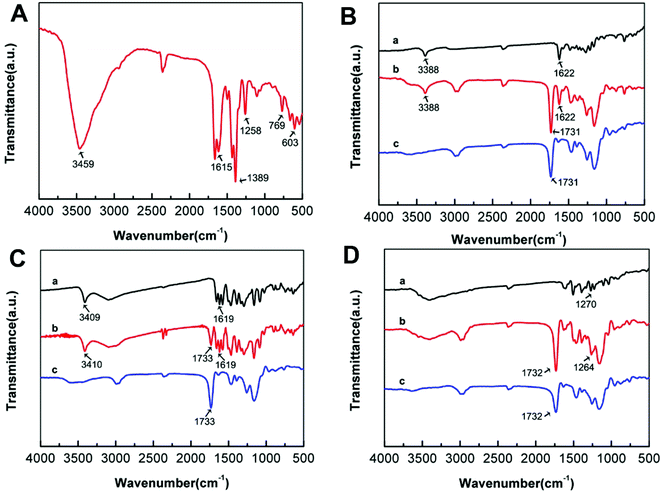
The FT-IR spectra was detected to demonstrate the successful synthesis of NH2-MIL-101(Cr) and MIL@MIPs. As shown in Fig. 2A, the 3459 cm−1 peak was ascribed to the asymmetrical stretching vibration of the amine groups, and the N–H bending vibration was located at 1615 cm−1. Moreover, the peaks at 1389 cm−1, 1258 cm−1, and 769 cm−1 corresponded to the C–N stretching vibrations of aromatic amines. Meanwhile, the 603 cm−1 was observed, which was attributed to the symmetric stretching of Cr–O. All of the above proved that NH2-MIL-101(Cr) was successfully prepared. EMO@MIPs, BAI@MIPs, and BER@MIPs before eluting templates apparently showed the same characteristic peaks as the templates at 1622 cm−1, 1619 cm−1, and 1270 cm−1 as shown in Fig. 2B, C, and D respectively, while they were not found in MIL@NIPs. What's more, the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching bands at 1731 cm−1, 1733 cm−1, and 1732 cm−1 appearing in EMO@MIPs, BAI@MIPs, and BER@MIPs indicated the successful reacting between MAA and EGDMA. All the results demonstrated that MIL@MIPs were successfully made. The TEM and SEM investigations were carried out to exhibit the morphology of NH2-MIL-101(Cr) and MIL@MIPs. From Fig. 3A, we can see that NH2-MIL-101(Cr) showed a uniform size distribution with an average of about 50 nm. The SEM images (Fig. 3B–D) showed that the size of MIL@MIPs increased after the process of encapsulation and the surface was rough, which had various amounts of imprinting cavities.
O stretching bands at 1731 cm−1, 1733 cm−1, and 1732 cm−1 appearing in EMO@MIPs, BAI@MIPs, and BER@MIPs indicated the successful reacting between MAA and EGDMA. All the results demonstrated that MIL@MIPs were successfully made. The TEM and SEM investigations were carried out to exhibit the morphology of NH2-MIL-101(Cr) and MIL@MIPs. From Fig. 3A, we can see that NH2-MIL-101(Cr) showed a uniform size distribution with an average of about 50 nm. The SEM images (Fig. 3B–D) showed that the size of MIL@MIPs increased after the process of encapsulation and the surface was rough, which had various amounts of imprinting cavities.
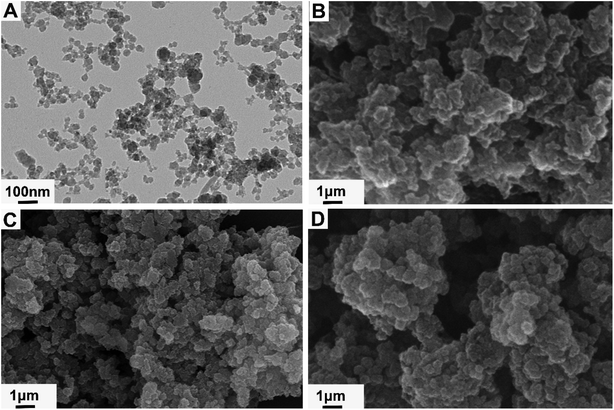 | ||
| Fig. 3 TEM images of (A) NH2-MIL-101(Cr); SEM images of (B) EMO@MIPs, (C) BAI@MIPs, and (D) BER@MIPs. | ||
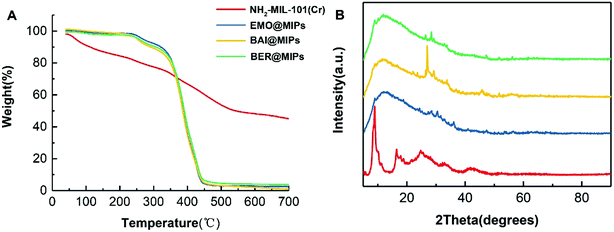
To evaluate the thermostability of NH2-MIL-101(Cr) and MIL@MIPs, TGA was performed under a temperature of 40 °C to 700 °C. As seen in Fig. 4A (red), there were two obvious weight loss times during the whole process, and the first loss below 180 °C can be mainly attributed to the volatilization of residual solvent, and moisture was about 13%, whereas the other weight loss in a temperature ranging from 200 °C to 600 °C was about 37%, which principally resulted from the structural collapse. From the other three lines in Fig. 4A, we can see that there was a slight decline of about 6% below 300 °C, which was assigned to the elimination of water and residual solvent, and the loss of MIL@MIPs below 700 °C was mainly contributed to the decomposition of the imprinted layer. To further demonstrate the successful coating of the imprinted layer, the XRD patterns were employed. As shown in Fig. 4B, the pattern of NH2-MIL-101(Cr) was similar to the report in literature39 and the decline of the patterns of MIL@MIPs also indentified the successful grafting of the imprinted layer.
The elemental analysis of NH2-MIL-101(Cr) and MIL@MIPs was carried out using XPS and the results were shown in Table 1. As we can see, the amount of chromium in MIL@MIPs decreased considerably compared with that of NH2-MIL-101(Cr), and the amount of carbon and protium increased significantly, which demonstrated the successful encapsulation greatly.
| Cr | C | N | H | |
|---|---|---|---|---|
| NH2-MIL-101(Cr) | 9.41 | 29.19 | 4.81 | 4.85 |
| EMO@MIPs | 0.16 | 55.28 | <0.3 | 7.23 |
| BAI@MIPs | 0.19 | 57.25 | <0.3 | 7.32 |
| BER@MIPs | 0.28 | 57.07 | <0.3 | 7.24 |
The N2 adsorption–desorption measurement was performed to determine the porosity features of NH2-MIL-101(Cr) and MIL@MIPs, and the results were shown in Table 2. The obtained NH2-MIL-101(Cr) showed obvious large surface area (918.3 m2 g−1) and pore volume (0.721 cm3 g−1), which greatly improved the efficiency of MIL@MIPs. With the occurrence of imprinting, the BET surface area and pore volume of MIL@MIPs decreased, which could also identify the successful encapsulation of MIL@MIPs on the surface of NH2-MIL-101(Cr).
| SBET (m2 g−1) | Vt (cm3 g−1) | |
|---|---|---|
| NH2-MIL-101(Cr) | 918.3 | 0.721 |
| EMO@MIPs | 144.18 | 0.092 |
| BAI@MIPs | 175.7 | 0.214 |
| BER@MIPs | 294.6 | 0.273 |
3.3 Adsorption performance of MIL@MIPs
Static adsorption experiments were conducted to evaluate the adsorption ability of MIL@MIPs and MIL@NIPs using the initial concentration of 0.5–10 mmol L−1. From Fig. S1,† we can see that MIL@MIPs showed a rising trend of binding capacity until the initial template concentration was about 7, 7, and 9 mmol L−1 for EMO, BAI, and BER respectively, and the saturated adsorption capacity was 108.54 μmol g−1, 81.52 μmol g−1, and 114.46 μmol g−1 severally. However, MIL@NIPs showed much lower adsorption amounts, nearly less than half of MIL@MIPs, which was mainly attributed to its nonspecific binding.Kinetic adsorption curves were used to illustrate the mass transfer property of MIL@MIPs at different time intervals of 5–50 min, and the results were shown in Fig. S2.† The adsorption ability reached equilibrium at the time of 40, 30 and 30 min respectively for EMO@MIPs, BAI@MIPs, and BER@MIPs, and the rapid adsorption property was primarily ascribed to the specific imprinting sites of templates, because of which, the templates could reach the imprinting cavities easily and quickly.
Selective adsorption experiments were performed to estimate the specificity recognition capacity of MIL@MIPs and MIL@NIPs. Fig. S3† clearly indicated that MIL@MIPs showed better binding capacity to the template than its structural analogues, which was owed to the specific recognition sites encapsulated on the surface of MIL@MIPs previously. Nevertheless, MIL@NIPs showed no obvious difference in binding capacity to both the template and its structural analogues, which suggested that MIL@NIPs had no specificity recognition capacity but physical adsorption.
Subsequently, desorption experiments and reusability were detected because regeneration and reusability play an important role in the application of MIL@MIPs. As shown in Fig. S4,† the desorption efficiency reached equilibrium quickly at the time of 10, 20, and 10 min for EMO@MIPs, BAI@MIPs, and BER@MIPs respectively, which was mainly due to the specific binding site and fast mass transfer property. After that, the regenerated polymers were reused for adsorption and the consecutive adsorption–desorption process was repeated with the same MIL@MIPs for six times. Fig. S5† showed that the binding capacity of MIL@MIPs decreased slowly, which highly indicated that the MIL@MIPs were stable and reusable and could be used for the following adsorption experiments.
3.4 Application
| Analyte | Linear regression date | RSD (%) | LOD (μg mL−1) | LOQ (μg mL−1) | |||
|---|---|---|---|---|---|---|---|
| Regression equation | R2 | Test range (mg mL−1) | Intra-day | Inter-day | |||
| Coptisine | y = 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 849 849![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 668x − 173 668x − 173![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 567 567 |
0.9999 | 0.001–1.0 | 1.7 | 2.2 | 0.02 | 0.36 |
| Baicalin | y = 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 920 920![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 753x − 34 753x − 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 619 619 |
0.9998 | 0.001–0.5 | 1.1 | 1.8 | 0.06 | 0.33 |
| Berberine | y = 53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 235 235![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 032x − 31 032x − 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 678 678 |
0.9995 | 0.01–1.0 | 1.3 | 1.0 | 0.09 | 0.32 |
| Emodin-8-O-β-D-glucoside | y = 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 475 475![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 821x − 31 821x − 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 351 351 |
0.9998 | 0.001–0.5 | 2.1 | 1.6 | 0.08 | 0.48 |
| Wogonoside | y = 56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 404 404![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 870x + 27 870x + 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 287 287 |
0.9999 | 0.001–1.0 | 2.3 | 1.2 | 0.08 | 0.33 |
| Baicalein | y = 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 389 389![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 185x + 390 185x + 390![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 926 926 |
0.9996 | 0.01–1.0 | 3.1 | 2.4 | 0.13 | 0.28 |
| Emodin | y = 48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 624 624![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 763x + 118 763x + 118![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 102 102 |
0.9997 | 0.01–1.0 | 1.9 | 2.0 | 0.06 | 0.12 |
| Chrysophanol | y = 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 029 029![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 676x + 56 676x + 56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 580 580 |
0.9998 | 0.001–0.3 | 1.2 | 1.8 | 0.02 | 0.54 |
| Physcion | y = 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 807 807![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 416x − 5875 416x − 5875 |
0.9999 | 0.001–0.2 | 3.2 | 2.9 | 0.07 | 0.45 |
3.4.2 Purification and capturing nNOS–PSD-95 uncouplers from SXD by MIL@MIPs-SPE-HPLC
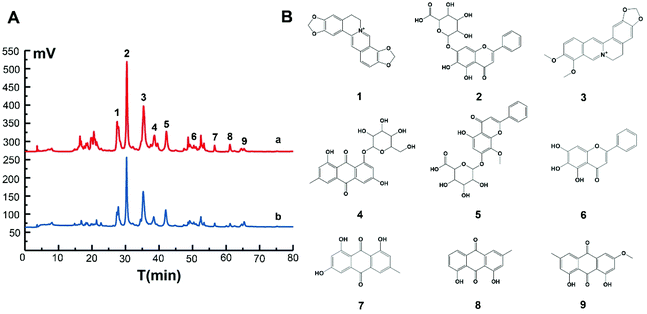
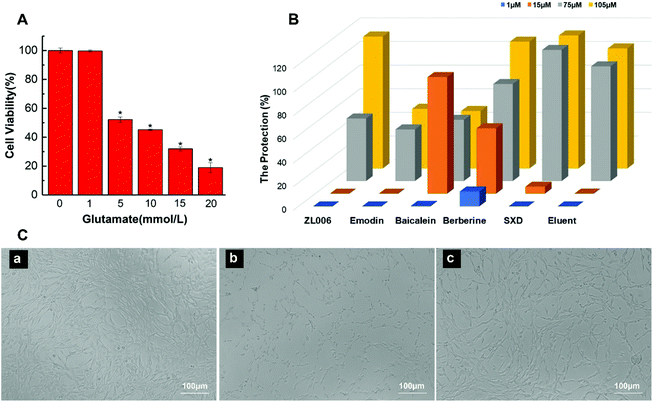
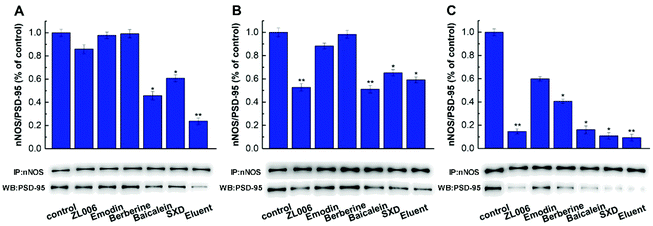
MIL@MIPs were used as sorbents for SPE column to capture nNOS–PSD-95 uncouplers from SXD. The initial extract and active components treated with MIL@MIPs were detected by HPLC and the results were shown in Fig. 5. It could be seen that active components (the blue curve of Fig. 5A) were much simpler than the initial extract (the red curve of Fig. 5A), which was complicated and diverse. From the chromatograms, we can find that (1) coptisine, (2) baicalin, (3) berberine, (4) emodin-8-O-β-D-glucopyranoside, (5) wogonoside, (6) baicalein, (7) emodin, (8) chrysophanol, and (9) physcion (Fig. 5B) were obtained in the eluent by HPLC-MS and all the nine compounds were then further confirmed by comparing the retention time and molecular weight with standards (Table S5†). As a consequence, we can see that the method established by combining MIL@MIPs-SPE with HPLC has highly selective capacity and is feasible, simple, and time-saving.3.5 Activity evaluation of active components obtained from SXD
4. Conclusion
Novel molecularly imprinted polymers based on NH2-MIL-101 (MIL@MIPs) were successfully designed, optimized, and characterized, which exhibited large binding capacity, fast kinetics, excellent selectivity, rapid desorption ability, and satisfactory reusability by adsorption performance experiments. Then a model of MIL@MIPs-SPE-HPLC was applied to capture nNOS–PSD-95 uncouplers from SXD. MTS assay and coimmunoprecipitation experiment demonstrated that the active components obtained from SXD showed excellent neuroprotective effect and uncoupling activity in vitro. At the same time, it could also reduce the infarct size and ameliorate neurological deficits significantly estimated in anti-ischemic stroke assay in vivo. Furthermore, this study can provide a scientific basis for the application of SXD and its optimized prescriptions. Meanwhile, it can also provide new ideas and new methods for efficiently capturing nNOS–PSD-95 uncouplers from other nature medicines.Live subject statement
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Nanjing Medical University and experiments were approved by the Animal Ethics Committee of Nanjing Medical University.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (81373895, 81173538), Natural Science Foundation of Jiangsu Province in China (BK20191352), Jiangsu Salt Industry Group Co., Ltd. (NMU-SY201805) and Innovative Team of Jiangsu Province (201910312037Z).References
- B. Chen, G. X. Wang, W. W. Li, W. L. Liu, R. H. Lin, J. Tao, M. Jiang, L. D. Chen and Y. Wang, Exp. Cell Res., 2017, 351, 163–172 CrossRef CAS PubMed.
- A. S. Ernst, L. I. Böhler, A. M. Hagenston, A. Hoffmann, S. Heiland, C. Sticht, M. Bendszus, M. Hecker, H. Bading, H. H. Marti, T. Korff and R. Kunze, Acta Neuropathol. Commun., 2019, 7, 15 CrossRef PubMed.
- Q. J. Wu and M. Tymianski, Mol. Brain, 2018, 11, 15 CrossRef PubMed.
- Y. J. Kaneko, J. P. Tuazon, X. M. Ji and C. V. Borlongan, Cell. Physiol. Biochem., 2018, 51, 1982–1995 CrossRef CAS PubMed.
- A. Bach, S. W. Pedersen, L. A. Dorr, G. Vallon, I. Ripoche, S. Ducki and L. Y. Lian, Sci. Rep., 2015, 5, 12157 CrossRef CAS PubMed.
- S. F. Mo, G. Y. Liao, J. Yang, M. Y. Wang, Y. Hu, G. N. Lian, L. D. Kong and Y. Zhao, Brain Res., 2016, 1648, 250–256 CrossRef CAS PubMed.
- S. C. Wu, Y. Yue, H. Tian, L. Tao, Y. T. Wang, J. Xiang, S. Wang and H. Ding, Neuropharmacology, 2014, 83, 107–117 CrossRef CAS PubMed.
- S. K. Florio, C. Loh, S. M. Huang, A. E. Iwamaye, K. F. Kitto, K. W. Fowler, J. A. Treiberg, J. S. Hayflick, J. M. Walker, C. A. Fairbanks and Y. Lai, Br. J. Pharmacol., 2009, 158, 494–506 CrossRef CAS PubMed.
- L. Zhou, F. Li, H. B. Xu, C. X. Luo, H. Y. Wu, M. M. Zhu, W. Lu, X. Ji, Q. G. Zhou and D. Y. Zhu, Nat. Med., 2010, 16, 1439–1443 CrossRef CAS PubMed.
- D. Y. Chen, T. Zhao, K. D. Ni, P. Dai, L. Yang, Y. Xu and F. Li, Bioorg. Med. Chem. Lett., 2016, 26, 2152–2155 CrossRef CAS PubMed.
- X. L. Gu, J. J. Huang, L. Zhang, Y. Zhang, C. Z. Wang, C. H. Sun, D. D. Yao, F. Li, L. N. Chen and C. S. Yuan, J. Sep. Sci., 2017, 40, 3522–3534 CrossRef CAS PubMed.
- D. D. Yao, L. Zhang, J. J. Huang, C. H. Sun, Y. Zhang, X. L. Gu, C. Z. Wang, F. Li, L. N. Chen and C. S. Yuan, J. Pharm. Biomed. Anal., 2018, 154, 180–190 CrossRef CAS PubMed.
- J. J. Huang, C. H. Sun, D. D. Yao, C. Z. Wang, L. Zhang, Y. Zhang, L. N. Chen and C. S. Yuan, J. Mater. Chem. B, 2018, 6, 1531–1542 RSC.
- X. Y. Wei, J. H. Tao, X. Cui, S. Jiang, D. W. Qian and J. A. Duan, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2017, 1061, 248–255 CrossRef PubMed.
- T. Y. Wu, F. R. Chang, J. R. Liou, I. W. Lo, T. C. Chung, L. Y. Lee, C. C. Chi, Y. C. Du, M. H. Wong, S. H. H Juo, C. C. Lee and Y. C. Wu, Front. Pharmacol., 2016, 7, 374 CAS.
- Y. T. Shih, D. C. Wu, C. M. Liu, Y. C. Yang, I. J. Chen and Y. C. Lo, J. Ethnopharmacol., 2007, 112, 537–544 CrossRef PubMed.
- S. F. Liou, J. H. Hsu, J. C. Liang, H. J. Ke, I. J. Chen, J. R. Wu and J. L. Yeh, J. Nat. Med., 2012, 66, 311–320 CrossRef CAS PubMed.
- X. Y. Wei, J. H. Tao, Y. M. Shen, S. W. Xiao, S. Jiang, E. X. Shang, Z. H. Zhu, D. W. Qian and J. N. Duan, Front. Pharmacol., 2018, 9, 955 CrossRef PubMed.
- K. Lee, B. Kim, H. Hur, K. S. Chinannai, I. Ham and H. Y. Choi, Chin. J. Integr. Med., 2017 DOI:10.1007/s11655-017-2771-7.
- Y. S. Wang, R. T. Lin, H. Y. Cheng, S. F. Yang, W. W. Chou and S. H. H. Juo, J. Ethnopharmacol., 2011, 133, 442–447 CrossRef PubMed.
- J. X. Song, X. Chen, Y. Lyu, W. Zhuang, J. Zhang, L. Gao and X. L. Tong, Brain and Behavior, 2019, 9, e01185 CrossRef PubMed.
- L. Uzun and A. P. F. Turner, Biosens. Bioelectron., 2016, 76, 131–144 CrossRef CAS.
- O. S. Ahmad, T. S. Bedwell, C. Esen, A. Garcia-Cruz and S. A. Piletsky, Trends Biotechnol., 2019, 37, 294–309 CrossRef CAS PubMed.
- R. I. Boysen, J. Sep. Sci., 2019, 42, 51–71 CrossRef CAS PubMed.
- Z. Z. Wu, D. Y. He, B. Cui and Z. Y. Jin, Food Chem., 2019, 283, 517–521 CrossRef CAS PubMed.
- C. Li, M. H. Ngai, K. K. Reddy, S. C. Y. Leong, Y. W. Tong and C. L. L. Chai, Anal. Chim. Acta, 2019, 1066, 121–130 CrossRef CAS PubMed.
- W. Shi, S. Q. Zhang, K. B. Li, W. P. Jia and D. M. Han, Sep. Purif. Technol., 2018, 202, 165–173 CrossRef CAS.
- Y. C. Lu, M. H. Guo, J. H. Mao, X. H. Xiong, Y. J. Liu and Y. Li, Ecotoxicol. Environ. Saf., 2019, 177, 66–76 CrossRef CAS PubMed.
- R. Sedghi, B. Heidari and M. Yassari, J. Colloid Interface Sci., 2017, 503, 47–56 CrossRef CAS PubMed.
- J. Ashley, M. A. Shahbazi, K. Kant, V. A. Chidambara, A. Wolff, D. D. Bang and Y. Sun, Biosens. Bioelectron., 2017, 91, 606–615 CrossRef CAS PubMed.
- M. D. Attieh, Y. Zhao, A. Elkak, A. Falcimaigne-Cordin and K. Haupt, Angew. Chem., Int. Ed., 2017, 56, 3339–3343 CrossRef PubMed.
- M. Tatemichi, M. Sakamoto, M. Mizuhata, S. Deki and T. Takeuchi, J. Am. Chem. Soc., 2007, 129, 10906–10910 CrossRef CAS.
- T. Guo, Q. L. Deng, G. Z. Fang, D. H. Gu, Y. K. Yang and S. Wang, Biosens. Bioelectron., 2016, 79, 341–346 CrossRef CAS PubMed.
- M. Khdhayyer, A. F. Bushell, P. M. Budd, M. P. Attfield, D. M. Jiang, A. D. Burrows, E. Esposito, P. Bernardo, M. Monteleone, A. Fuoco, G. Clarizia, F. Bazzarelli, A. Gordano and J. C. Jansen, Sep. Purif. Technol., 2019, 212, 545–554 CrossRef CAS.
- K. Qian, Q. L. Deng, G. Z. Fang, J. P. Wang, M. F. Pan, S. Wang and Y. H. Pu, Biosens. Bioelectron., 2016, 79, 359–363 CrossRef CAS PubMed.
- S. Clauzier, L. N. Ho, M. Pera-Titus, B. Coasne and D. Farrusseng, J. Am. Chem. Soc., 2012, 134, 17369–17371 CrossRef CAS.
- X. P. Dai, X. N. Jia, P. Zhao, T. Wang, J. Wang, P. T. Huang, L. He and X. H. Hou, Talanta, 2016, 154, 581–588 CrossRef CAS PubMed.
- H. L. Liu, L. Mu, X. M. Chen, J. Wang, S. Wang and B. G. Sun, J. Agric. Food Chem., 2017, 65, 986–992 CrossRef CAS PubMed.
- Z. W. Xiao, M. He, B. B. Chen and B. Hu, Talanta, 2016, 156, 126–133 CrossRef PubMed.
- Y. Lin, C. Kong and L. Chen, RSC Adv., 2012, 2, 6417–6419 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra10537a |
| This journal is © The Royal Society of Chemistry 2020 |