Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
N- and C-terminal regions of the small heat shock protein IbpA from Acholeplasma laidlawii competitively govern its oligomerization pattern and chaperone-like activity†
Liliya S. Chernovaab,
Mikhail I. Bogachev c,
Vitaly V. Chasov
c,
Vitaly V. Chasov a,
Innokentii E. Vishnyakov‡
a,
Innokentii E. Vishnyakov‡
 *bd and
Airat R. Kayumov‡
*bd and
Airat R. Kayumov‡ *ab
*ab
aKazan Federal University, 18 Kremlevskaya street, 420008, Kazan, Russia. E-mail: kairatr@yandex.ru; Tel: +7-843-233-78-02
bInstitute of Cytology, Russian Academy of Sciences, 4 Tikhoretsky Avenue, 194064 St-Petersburg, Russia. E-mail: innvish@gmail.com; Tel: +7-812-297-03-28
cSt. Petersburg Electrotechnical University, 5 Professor Popov street, 197376, St. Petersburg, Russia
dPeter the Great St.Petersburg Polytechnic University, 29 Polytechnicheskaya street, 195251, St-Petersburg, Russia
First published on 26th February 2020
Abstract
Small heat shock proteins (sHSPs) are ubiquitous molecular chaperones preventing the irreversible denaturation of proteins. While in Escherichia coli two sHSPs IbpA and IbpB work in strong cooperation, the sole Mollicute with free-living ability Acholeplasma laidlawii carries a single gene encoding the sHSP protein AlIbpA. In vitro, independently of the temperature, AlIbpA forms a heterogeneous mixture of approximately 24-mer globules, fibrils and huge protein aggregates. The removal of either 12 or 25 N-terminal amino acids led to the formation of fibrils and enhanced the protein ability to prevent the temperature-induced aggregation of insulin, assuming the fibrillar form as an active protein. In turn, the deletion of the C-terminus or substitution of C-terminal LEL motif by SEP decreased the temperature stability of AlIbpA and eliminated its chaperone function completely, although the protein remained predominantly in a globular state. This suggests that the C-terminal LEL motif is necessary for the chaperon-like activity of AlIbpA and fibril formation. Double N- and C-terminal truncations abolished both the chaperone-like activity and huge oligomer formation. Since the globular form of sHSPs is considered as their inactive form, our data suggest that the N-terminus of AlIbpA is responsible for the huge globule (low-active form) formation and behaves as an intramolecular inhibitor of the fibrils (active form) formation and substrates binding. Taken together these data demonstrate non-trivial properties of AlIbpA, in which the competitive action of N- and C-termini governs the equilibrium between either fibrillar or globular structures representing a possible molecular mechanism of the AlIbpA activity regulation.
Introduction
Protein aggregation induced by heat-shock, and oxidative or salt stress is a major threat for cell viability. Small heat shock proteins (sHSPs) represent an abundant and ubiquitous family of molecular chaperones that are believed to prevent irreversible aggregation of other cellular proteins under stress conditions.1–4In cells sHSPs form huge homo- and/or heterooligomeric complexes consisting of typically between 9 and 48 subunits.1,5,6 This assembly of sHSPs involves several parts of the subunits in a flexible manner (reviewed in ref. 7 and 8). The oligomerization state of sHSPs can be influenced by the changes in pH,9 temperature10 as well as by post-translational modifications, such as phosphorylation11 that affects their affinity to other proteins and chaperone-like activity.12 This relationship between changes in oligomerization and chaperone-like activity has been clearly shown for the human HSPB1 and HSPB5 as well as for the yeast HSP42.13–15
While the dynamics of sHSPs oligomerization and oligomers structure were analyzed in detail mainly for archaeal and eukaryotic proteins, the common features of sHSPs seem to be universal in all kingdoms of life (for an extensive review, we refer to ref. 2). The molecules of sHSPs are composed of three structural domains. The central α-crystalline domain (ACD) consisting of about 90-aa is extremely conserved in structure and comprises seven or eight anti-parallel β-strands that form a β-sandwich.16 The ACD is flanked by variable N-terminal domain (NTD) and a short C-terminal domain (CTD).5,14,17–19 The importance of N- and C-terminal domains for the modulation of sHSP chaperone-like activity by the formation of stress-induced macromolecular assemblies has been previously multiply reported. Being in an oligomeric state, sHSPs recruit the misfolded proteins and prevent their further aggregation.5,13–15,17,20,21
The intrinsically disordered N-terminal domain is generally more or less conserved and contains either one or two (W/F)(D/F)PF-like motifs, which are shown to participate in the oligomerization of sHSPs and are required for their chaperone-like activity.16 Thus, the removal of the NTD in sHSP26 from Saccharomyces cerevisiae led to the formation of di- and tetra-mers which were unable to protect the substrates from thermal denaturation.22,23 The removal of 11 N-terminal amino acids of the Escherichia coli sHSP also led to the loss of the chaperone-like activity.24
The C-terminal domain of many sHSPs also plays an important role in their oligomerization and chaperone-like activity. Generally it contains the conserved V/IXI/V motif necessary for the subunit assembly through interaction with the α-crystallin domain during the oligomerization process.14,25 The truncation of V/IXI/V motif or substitution of the isoleucine residues by glycine results in the dissociation of the subunits and thereby in the loss of the chaperone-like activity.18,25 In sHSP IbpA from E. coli, in addition to the conserved V/IXI/V motif, the arginine 133 in the C-terminus was shown to interact with glutamic acid at position 62 in the α-crystalline domain thereby providing higher order structure formation and playing an important role in the IbpA chaperone function.24
In E. coli, two sHSP proteins IbpA and IbpB (inclusion body associated proteins), the best characterized bacterial sHSPs, work in strong cooperation24,26,27 and belong to the energy-independent part of the proteins quality control network.28 These proteins bind partially denatured proteins and prevent their further aggregation thereby facilitating their subsequent refolding by the ATP-dependent DnaK–DnaJ–GrpE and GroEL/GroES chaperone systems in cooperation with the AAA+ proteases ClpP and Lon.10,29–31 In vitro studies revealed that both IbpA and IbpB are required to efficiently stabilize denatured proteins in a folding-competent state; however, they play different roles in this process.10,32 IbpA abrogates the formation of large aggregates of the substrate simultaneously inhibiting ClpB–DnaK-dependent refolding because of its tight association with polypeptides in aggregates that prevents their processing by the ClpB and DnaK/DnaJ/GrpE chaperones. By the way, IbpB interacts with aggregates via IbpA and alleviates the IbpA-mediated inhibitory effect.32
In vitro, the purified IbpB has been shown to form 2–3 MDa oligomers,1,33 while IbpA forms fibril-like structures.34 In the mixture of these two proteins, IbpB blocks fibril formation by the IbpA.24 The removal of 11 C-terminal amino acids in IbpA abrogated the fibrils formation and the protein was unable to stabilize substrates. In contrast, the N-terminal truncation led to thinner fibrils formation and to the inability to interact with substrates and prevent their aggregation,24 apparently suggesting that fibril formation could be an important structural feature of sHSPs involved in its chaperone-like activity.
While in some (often symbiotic) bacteria there are up to 12 different sHSPs proteins,2 to the date only few bacterial sHSPs are described. Besides IbpA and IbpB from E. coli, four proteins are identified in Agrobacterium tumefaciens (HspC, HspL, HspAT1 and HspAT2), and one sHSP in phytopathogen Xanthomonas.19,24,35,36 Among archaea, genomes of Deinococcus radiodurans and Sulfolobus solfataricus carry two genes encoding sHsp proteins, while only one small heat shock protein is found in Methanococcus jannaschii and Sulfolobus tokodaii.27,37,38 In contrast, there are several (mostly pathogenic) bacterial genomes that do not encode any sHSPs at all.2
While the inactivation of either IbpA or IbpB led to rather moderate effects on the growth and viability of E. coli at high temperatures,39 the presence of sHSPs in mycoplasmas, microorganisms with significantly reduced genomes, indicates their fundamental importance in the stress resistance of bacterial cells. The phytopathogenic mycoplasma Acholeplasma laidlawii contains only one single gene encoding sHSP.40,41 Under heat shock conditions, the amount of AlIbpA protein in A. laidlawii cells increases up to 7% of the total cellular proteins and could appear a key factor that determines the temperature adaptation capacity of this bacterium.40,41 In vitro AlIbpA was shown to exhibit the chaperone-like activity by preventing the DTT- or heat-induced aggregation of insulin40 as well as of various cellular proteins from E. coli and A. laidlawii itself.42,43
In the following we unravel the roles of both N- and C-terminal regions of AlIbpA for self-oligomerization, substrate binding and the chaperone function regulation. Our data indicate that N-terminus behaves as an auto-inhibitor and activity regulator of the C-terminus which in turn is required for the chaperone function and protein oligomerization in fibrillar form.
Materials and methods
Strains and plasmids
A. laidlawii PG8 was obtained from the collection of the Institute of Cytology, RAS, and cultivated in Mycoplasma broth with Supplement G (Oxoid) and 1% glucose. E. coli XL-10 Gold cells were used for cloning procedures. E. coli BL21 (dcmompThsdS(rB− mB−)galλ(DE3)) was used for proteins overexpression.Plasmids used in this study were obtained by cloning of the truncated ibpA genes into pET15b vector and are listed in Table S1.† pET15b-IbpA that provides the expression of N terminally His6-tagged AlIbpA was obtained previously.40 The truncated and mutant ibpA genes were amplified from A. laidlawii genomic DNA by using primer pairs as shown in Table S1† to obtain a given plasmid. Sequences of primers used are shown in Table S2.† The pET15b vector was digested with the restriction endonuclease BamHI. The digested plasmid and the PCR product were assembled using an isothermal, single-reaction method for assembling multiple overlapping DNA molecules as described previously.44 Point mutations leading to the substitution of F11F12 by N11N12 on the N-terminus were introduced by using the QuikChange Site Directed Mutagenesis Kit (Stratagene).
Purification of His6-tagged proteins
E. coli BL21(DE3) cells were transformed with plasmids present in Table S1† carrying various ibpA genes. The cells were grown at 37 °C in 500 ml of LB medium supplemented with 100 μg ml−1 ampicillin. After growth of the culture to an optical density (OD600) of 0.8, proteins expression was induced by the addition of IPTG (until final concentration 1 mM). Four hours after induction, cells were harvested by centrifugation and stored at −20 °C.All proteins with N-terminal His6-tag peptide were purified using affinity chromatography on Ni-NTA sepharose gravity-flow columns (Qiagen) following the protocol described previously.45 E. coli cell free extracts were prepared from the frozen cells (see above) by resuspending the cells in disruption buffer DB [50 mM Tris–HCl pH 7.4, 50 mM KCl, 500 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 2 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride (PMSF)] and broken by sonication. Unbroken cells and debris were removed after 10 min centrifugation at 8.000 rpm. The cell-free extracts were centrifuged again for 20 min at 15.000 rpm. The supernatant was passed through a 1 ml His-select cartridge column that was pre-equilibrated with washing buffer (50 mM Tris–HCl pH 8.0, 500 mM NaCl, 10 mM imidazole) and unbound proteins were washed out with 15 ml washing buffer. Finally, the affinity-bound proteins were eluted with 8 ml elution buffer (50 mM Tris–HCl pH 8.0, 500 mM NaCl, 500 mM imidazole). All proteins were purified to apparent electrophoretic homogeneity and dialyzed against PBS pH 7.4 containing 100 mM NaCl and used immediately for experiments or stored in ice no more than 4 days. Protein concentration was measured with the Roti-Quant protein assay reagent (Carl Roth) and bovine serum albumin as a standard.
Chromatography
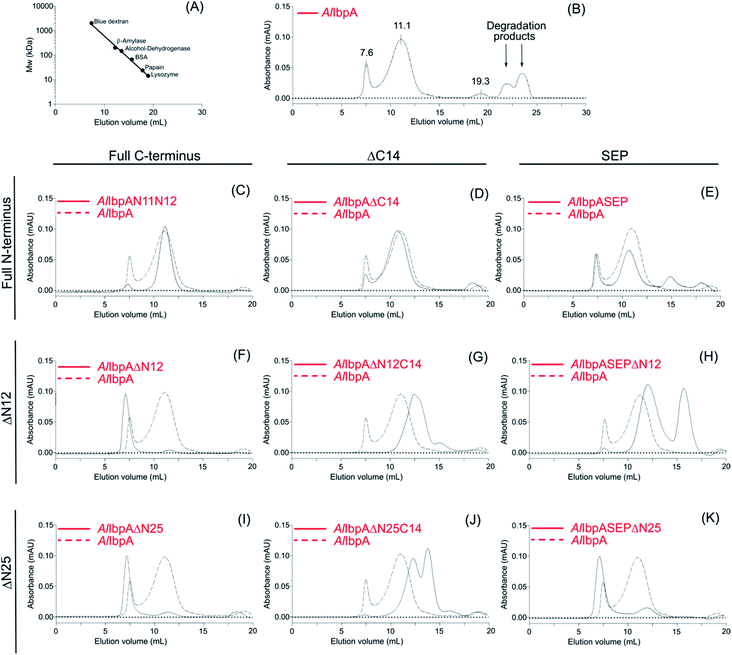
The analytical size-exclusion chromatography was performed on Superdex 200 10/300 column (Sigma) at flow rate of 0.5 ml min−1 on Waters Breeze 2 equipment with the absorbance detection at 280 nm. The protein sample (1 mg ml−1) was centrifuged for 5 min at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm to remove any sediment and 100 μl were injected on the column. The running buffer consisted of 50 mM NaCl, 50 mM KCl, 50 mM K2HPO4 pH 7.4. The apparent molecular weights of proteins were estimated after calibration of the column with following proteins: blue dextran (2000 kDa), β-amylase (200 kDa), alcohol-dehydrogenase (147 kDa), BSA (66 kDa), papain (23,4 kDa), lysozyme (14,3 kDa).
000 rpm to remove any sediment and 100 μl were injected on the column. The running buffer consisted of 50 mM NaCl, 50 mM KCl, 50 mM K2HPO4 pH 7.4. The apparent molecular weights of proteins were estimated after calibration of the column with following proteins: blue dextran (2000 kDa), β-amylase (200 kDa), alcohol-dehydrogenase (147 kDa), BSA (66 kDa), papain (23,4 kDa), lysozyme (14,3 kDa).
In vitro cross-linking of proteins
The chemical cross-linking by glutaraldehyde (GA) was performed as described in46 with modifications. Proteins were dialyzed overnight in a PBS pH 7.4. The reaction mixture consisted of 1.5 μg μl−1 of dialyzed AlIbpA protein and 0.05% (w/w) freshly prepared solution of glutaraldehyde. The reaction was performed at 25 °C for 20 minutes and then stopped by the addition of 100 mM Tris pH 7.4. The samples were further analyzed by immunoblotting with AlIbpA-specific antibodies obtained previously40 and developed by using anti-rabbit antibodies conjugated with peroxidase (SigmaAldrich cat. no A0545).Pull down assay
For the protein–protein interaction assays, the His6-tagged AlIbpA proteins (0.4 mg) and bovine insulin (Sigma) (0.4 mg) were diluted in 300 μl of buffer A (50 mM Tris–HCl pH 8.0, 200 mM NaCl) and incubated at 37 °C for 30 min.45 Afterwards, the protein mix was loaded to Ni-NTA sepharose (Qiagen) equilibrated by 10 column volumes (10 × 0.2 ml) of buffer A following by washing 4 times by 5 volumes of the same buffer. Proteins were eluted by buffer B (buffer A supplemented with 500 mM imidazole). The samples were collected, precipitated with TCA and separated with 17.5% SDS-PAGE.Surface plasmon resonance assay
SPR experiments were performed by using a BIAcore T200 biosensor system (GE Healthcare Life Sciences). The running buffer contained 10 mM Na2HPO4, 150 mM NaCl, pH 7.4. Purified His6-IbpA proteins were immobilized on the Ni2+-loaded NTA (nitrilotriacetate) sensor chip to FC2 (flow cell 2) in a volume of 10 μl with a flow rate 10 μl min−1 to receive a binding signal of approximately 2000 RU, which corresponds to a surface concentration change of 2 ng mm−2. The transcription factor TnrA from B. subtilis (His6-TnrA20) with deleted protein-binding domain45 was injected onto FC1 and served as a control for unspecific interactions. Then the alcohol dehydrogenase (SigmaAldrich, cat. no 55689) at the concentration of 1.0 mg ml−1 was loaded on the AlIbpA-loaded chip surface pre-washed with 3 mM EDTA. Injection of the protein sample was performed with a flow rate of 10 μl min−1 in the running buffer and the response difference (FC2 − FC1) was recorded. To load fresh AlIbpA-His6 protein on the NTA sensor chip, bound proteins were first removed by injecting 10 μl of 0.5 M imidazol pH 7.0. The data were fitted using the BiaEvaluation and GraphPad Prism software.Chaperone-like activity assay
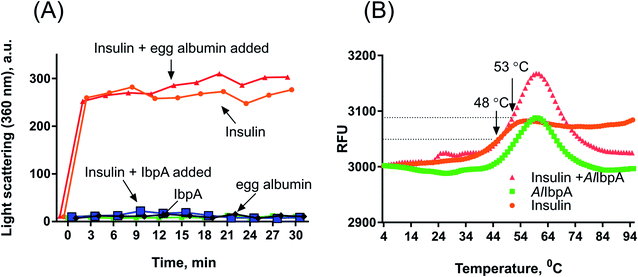
The chaperone-like activity was investigated by measuring the capacity of AlIbpA to suppress the heat-shock-induced aggregation of bovine insulin.Bovine insulin as a model substrate and egg albumine (Sigma, USA) as a control were used in final concentrations 0.5 mg ml−1. Final concentrations of mycoplasmal sHSP were 0.5 mg ml−1 (AlIbpA itself, AlIbpA in mixture with insulin). Experiments were carried out at 55 °C for 30 min in total volume of 100 μl (PBS 1×, pH 7.4). Samples (1 μl) were taken every 3 min and light scattering measurements were done with NanoDrop 2000 (Thermo Scientific, USA) at the wave length of 360 nm.
The protein aggregation was also assessed by measuring the fluorescence of the SYPRO Orange, which associates with denatured proteins. The mixtures of various AlIbpA proteins (0.4 mg) with insulin (0.4 mg) in total volume of 30 μl were heated in PBS from 10 °C to 95 °C with a temperature increment by 1 °C per 1 min in the presence of 10 μM SYPRO Orange, the fluorescence was detected by using FAM-filter set detection on Bio-Rad CFX96 thermocycler. The data analysis was performed as described in ref. 47 and 48 by determining the temperature leading to fluorescence increase by 50% of maximal signal (Tm 50).
Transmission electron microscopy
The AlIbpA samples were negatively stained on grids coated with collodion films that were discharged in UV (1 min) prior to the application of 5 μl of the protein solution (1–10 μg ml−1 in PBS, pH 7.4). The protein solution was blotted off after 2 min, immediately followed by adding and blotting off after 2 min of 3 μl 2% aqueous uranyl acetate. Negatively stained grids were visualized in a Libra 120 electron microscope (Zeiss, Germany) at 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–40
000–40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 magnification.
000 magnification.
Bioinformatics
The multiple alignment of proteins amino acid sequences was performed by using Clustal Omega web server49 and manually refined. The protein sequences were derived from NCBI database: the nonphotosynthetic alga Polytomella parva (Polyt), green photosynthetic alga Chlamydomonas reinhardtii (Cr; XP_001703658.1), land plants Physcomitrella patens (Physco; BAF36548.1), Arabidopsis thaliana (At; NP_192099.1), Oryza sativa japonica (Os; Os05g0133100), and Solanum lycopersicum (Sl; AAR14689.1), red algae Porphyra purpurea (Pp; NP_053864.1), Porphyra umbilicalis (Pu; AFC39923.1), and Pyropia yezoensis (Py; AGH27579.1), cyanobacteria Synechococcus elongatus PCC 7942 (Sy; P0A3F4.1), Synechocystis sp. PCC 6803 (Sc; CAA66127.1), and Escherichia coli (Ec; CAQ32926.1).The models of tertiary structures of sHSPs were obtained by using Phyre2 (ref. 50) or I-TASSER51,52 servers and also analyzed with in-house developed algorithm.53 The superposition of sHSP molecules was obtained with SuperPose online software.54
Statistical analysis
All experiments were performed in biological triplicates with three repeats in each run. The data were analyzed and graphically visualized using GraphPad Prism version 6.00 for Windows (GraphPad Software, USA, https://www.graphpad.com). The comparison with the control has been performed using the non-parametric Kruskal–Wallis one-way analysis of variance test. Significant differences were reported at p < 0.05. Densitometry was performed by using the Bio-Rad software for XRS + gel documentation system. The analysis of elution profiles was performed using Waters Breeze 2 software. The analysis of TEM images was based on the shape-based algorithm of globular vs. fibrillar structures selection55 with subsequent sub-populations quantification using an in-house developed algorithm and software.56Results
Analysis of amino acid sequence of AlIbpA
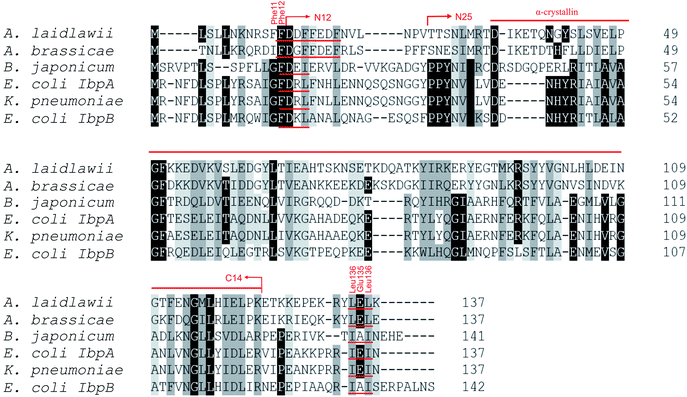
Most bacterial sHSPs exhibit common conserved structure and consist of α-crystalline domain flanked by variable N-terminal and C-terminal regions.14,17,18 AlIbpA has maximum identity with Hsp20 proteins from various Clostridia and Faecalibacterium species (45%) and exhibits only 18% and 20% identity with IbpA and IbpB from E. coli (EcIbpA and EcIbpB), respectively (Fig. 1, Table S3†). The central α-crystalline domain in AlIbpA is highly conserved and it's in silico-reconstructed structure demonstrates high similarity with both EcIbpA and EcIbpB despite of the low identity of their amino acid sequences (Fig. S1 and Table S3†). It comprises seven anti-parallel β-strands forming a hydrophobic β-sandwich which could tend to spontaneous oligomerization similarly to other sHSPs.16Both N- and C-terminal regions of AlIbpA contain all sHSP-specific motifs, while differ significantly from the well-characterized IbpA and IbpB from E. coli (Fig. 1 and S1, Table S3†). In particular, the N-terminus of AlIbpA is four amino acid residues shorter compared to that one of EcIbpA and EcIbpB, according to the most probable models obtained with either Phyre2 (ref. 50) or I-TASSER51,52 servers. Moreover, it does not exhibit the α-helical structure, albeit containing two tandem (W/F)(D/F)PF-like motifs (Fig. 1). Notably, similar double N-terminal (W/F)(D/F)PF-like motifs with high abundance of Phe-residues are highly conserved among Hsp20 proteins from various Acholeplasma spp known to the date (Fig. S2†) that likely appears a specific feature of sHSPs from these bacteria. Since the N-terminal (W/F)(D/F)PF-like motifs are required for the oligomerization and for the chaperone-like activity of sHSPs,16 their existence in the N-terminus of AlIbpA suggests that the protein might implement chaperone-like activity in the same manner like the other sHSPs do.16 Next, the C-terminus of AlIbpA contains LEL amino acid residues apparently constituting the conserved V/IXI/V motif, which was shown to be necessary for the interaction with the α-crystalline domain of EcIbpA during oligomerization, as its truncation completely abolished the chaperone-like activity of the protein.24 This fact allows assuming that AlIbpA might exhibit chaperone-like activity while being in the oligomeric form.
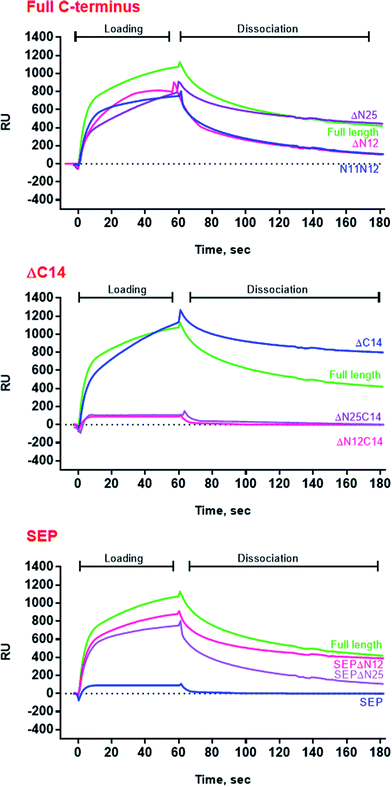
For further assessment of the roles of both N- and C-termini of AlIbpA in the oligomerization and thereby also in the chaperone-like activity, various truncated recombinant proteins have been constructed. The removed parts from either N- or C-termini are shown in Fig. 1. In particular, AlIbpAΔN12 lacks the first 12 amino acid residues and carries the truncated first (W/F)(D/F)PF-like motif, which is probably required for the oligomerization and thus also for the chaperone-like activity. In AlIbpAN11N12 both F11F12 are substituted by N11N12 leading to the damage to the motif while keeping the complete N-terminus length. AlIbpAΔN25 lacks both (W/F)(D/F)PF-like motifs. In order to understand the role of the C-terminal LEL motif, 14 amino acids flanking the α-crystalline domain from the C-terminus were removed by obtaining AlIbpAΔC14.
It has been shown previously24 that the IEI-motif is necessary for the interaction with the α-crystalline domain of EcIbpA during oligomerization via binding with glutamic acid at position 62 in the α-crystalline domain. In AlIbpA this position corresponds to lysin according the alignment (Fig. 1), and both electrostatic and hydrophobic interactions are possible. Therefore the AlIbpASEP with L134 and L136 replaced by polar amino acids S134 and P136 (substitution of the LEL motif by SEP) have been constructed to keep the length of the C-terminus (Fig. 1). Thus, the first leucine residue was replaced by polar serine, while the second leucine residue was replaced by proline to obtain the turn of the chain and provide a steric barrier for the interaction of C-terminus with the α-crystallin.
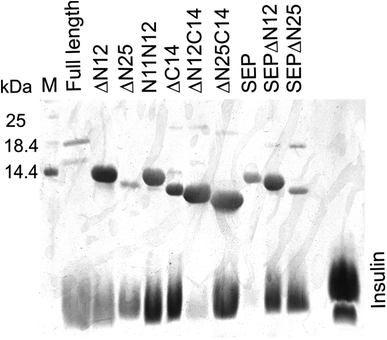
Additionally, AlIbpAΔN12C14, AlIbpAΔN25C14, AlIbpASEPΔN12 and AlIbpASEPΔN25 with truncated both N- and C-termini have been constructed. The recombinant wild-type and the truncated AlIbpA were expressed in E. coli and purified to the apparent electrophoretic homogeneity (Fig. S3†).
Evaluation of oligomers formation
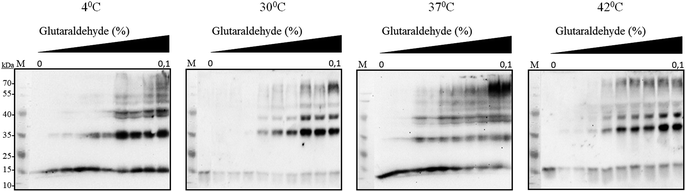
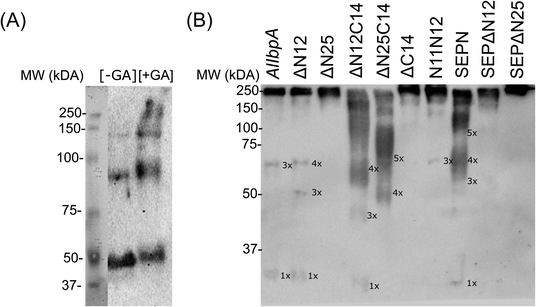
It has been reported earlier that sHSPs exhibit their chaperone-like activity while being in huge oligomers consisting typically between 9 and 48 subunits.1,5,6 Therefore the oligomerization of AlIbpA has been analyzed in vivo at various temperatures by chemical cross-link.A. laidlawii were grown in liquid medium until the late exponential growth phase. Then the aliquots of culture were incubated for 1 h at various temperatures as indicated, fixed with glutaraldehyde and analyzed with immunoblotting. These data clearly indicated that AlIbpA is present in the cells as aggregates/multimers regardless of the temperature (Fig. 2). Next, both in vivo and in vitro cross-link of recombinant full-length AlIbpA confirmed that the protein is present as oligomers at 37 °C (Fig. 3).
Surprisingly, the removal of neither N- nor C-termini of AlIbpA did not abolish the oligomerization, while the double removal of both N- and C-termini led to the decrease of the oligomerization degree (Fig. 3B). Similarly, the removal of 12 N-terminal amino acid residues from the protein with mutated C-terminal LEL-motif abrogated the large multimers formation, while di-, tri- and tetramers were still observed. These data suggest that the protein forms oligomers by either N- or C-terminus, in contrast to EcIbpA, which was shown to be in low-oligomeric state when either N- or C-terminus had been removed.24
The role of N- and C-termini in substrates binding and chaperone-like activity of AlIbpA
To evaluate the importance of N- and C-termini of AlIbpA for the substrate binding, the interaction of various proteins with AlIbpA has been assessed in vitro with the Surface Plasmon Resonance (SPR). Only alcohol dehydrogenase (ADH) and in a lesser extent the bovine insulin (SigmaAldrich cat no. I6634) were able to interact with AlIbpA in SPR assay (see Fig. S4†).To further characterize the functional role of N- and C-termini of AlIbpA, its chaperone-like activity has been also evaluated in vitro. It has been shown previously that AlIbpA demonstrates the chaperone-like activity in vitro by preventing the temperature-induced denaturation of the bovine insulin.40 Taking into account the fact that the denaturation temperature (Tm 50) of ADH and other proteins exceeded that of AlIbpA, the bovine insulin has been chosen as a model substrate for the chaperone-like activity assays (see Table S4† for values). For that, the Tm 50 of insulin in the presence of either full-length or truncated AlIbpA proteins was determined by using the fluorescent SYPRO-orange dye as described in Materials and methods (see Table S4† for values). Additionally, the interaction of AlIbpA proteins with insulin was analyzed by using pull-down assay. For that, AlIbpA proteins and insulin were mixed in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 weights ratio and after 30 min of incubation at 37 °C were purified on Ni-NTA agarose followed by SDS-PAGE analysis (Fig. 4, Table 1).
1 weights ratio and after 30 min of incubation at 37 °C were purified on Ni-NTA agarose followed by SDS-PAGE analysis (Fig. 4, Table 1).
| AlIbpA variant | Protein structure | bΔTm 50, °C | cInsulin binding | dADH binding KD, μM | eDistribution of various oligomeric fractions, % | |||
|---|---|---|---|---|---|---|---|---|
| eIbpA | fIbpA + insulin | I peak | II peak | 1×-4×-mers | ||||
a * – denotes statistically significant difference with full-length AlIbpA.b Determined by measuring the sypro-orange fluorescence.c Determined as ratio of AlIbpA and insulin bands density on in vitro pull-down assay SDS-PAGE (see ESI Fig. S5): “−” – no interaction, “+” – 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, “++” 1 1 ratio, “++” 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and higher.d Calculated from the ADH-to-AlIbpA surface binding curve slopes obtained with SPR (NA – not available because of no interaction).e Calculated as fractions of areas under the peaks on the elution profile.f The Tm 50 value of full-length AlIbpA is accepted as zero.g The Tm 50 value of insulin in absence of full-length AlIbpA is accepted as zero. 2 and higher.d Calculated from the ADH-to-AlIbpA surface binding curve slopes obtained with SPR (NA – not available because of no interaction).e Calculated as fractions of areas under the peaks on the elution profile.f The Tm 50 value of full-length AlIbpA is accepted as zero.g The Tm 50 value of insulin in absence of full-length AlIbpA is accepted as zero. |
||||||||
| Full length |  |
0g | +5 ± 0.8 | ++ | 1.6 ± 0.3 | 23 ± 10.3 | 74 ± 17.8 | 3 ± 0.2 |
| ΔN12 |  |
+3 ± 0.3* | +10 ± 2.3* | + | 3.5 ± 1.1 | 88 ± 17.1 | 10 ± 1.1 | 3 ± 0.1 |
| ΔN25 |  |
+3 ± 0.4* | +10 ± 2.1* | ++ | 2.2 ± 0.7 | 77 ± 21.3 | 12 ± 2.4 | 11 ± 2.1 |
| N11N12 |  |
+5 ± 0.8* | +11 ± 2.4* | ++ | 0.5 ± 0.1 | 8 ± 1.7 | 88 ± 12.2 | 4 ± 0.7 |
| ΔC14 |  |
−7 ± 0.6* | −3±1.0* | ++ | 0.7 ± 0.1 | 7 ± 4.1 | 86 ± 23.4 | 7 ± 2.3 |
| ΔN12C14 |  |
−5 ± 0.7* | −6±1.2* | — | NA | 1 ± 0.1 | 86 ± 22.8 | 14 ± 10.3 |
| ΔN25C14 |  |
−13 ± 2.2* | −9±1.1* | + | NA | 1 ± 0.2 | 50 ± 13.5 | 49 ± 10.3 |
| SEP |  |
−2 ± 0.1 | +2 ± 0.2* | — | NA | 18 ± 4.4 | 56 ± 7.8 | 26 ± 5.9 |
| SEPΔN12 |  |
−1 ± 0.3 | +3 ± 0.5 | + | 7.2 ± 1.4 | 2 ± 0.8 | 59 ± 10.1 | 39 ± 9.9 |
| SEPΔN25 |  |
+1 ± 0.2 | +5 ± 0.9 | ++ | 3.2 ± 0.5 | 69 ± 15.5 | 23 ± 3.2 | 8 ± 2.0 |
Despite of weak interaction observed by the SPR, the insulin was co-eluted with the full-length AlIbpA (Fig. 4, lane 1) suggesting the interaction of proteins under conditions used. In agreement with earlier data, AlIbpA was able to prevent the temperature-induced denaturation of the bovine insulin when measuring either the light scattering of proteins mixture during 30 min at 55 °C (Fig. 5A) or the fluorescence of the SYPRO Orange during proteins mixture heating from 10 °C to 95 °C (Fig. 5B). The Tm 50 of insulin was increased by 5 °C (from 48 °C to 53 °C) in presence of AlIbpA (Fig. 5B) confirming the presence of chaperone-like activity.
Next the chaperone-like activity and substrate binding capacity of various truncated AlIbpA proteins has been evaluated (Table 1). The removal of either 12 or 25 amino acids from the N-terminus as well as the substitution of F11F12 by N11N12 led to the increase of Tm 50 of the protein mixture by 5 °C compared to the full-length protein, while interaction with both insulin and ADH was not affected (Fig. 4 and 6, Table 1). These data suggest that (W/F)(D/F)PF-like motifs are apparently not required for the interaction with substrate proteins as well as for the protein stabilization. Moreover, N-terminus seems to suppress the chaperone-like activity of the full-length protein.
The deletion of the C-terminus significantly decreased the temperature stability of AlIbpA itself and its mixture with insulin (Table 1). Nevertheless, both insulin and ADH were still able to bind with AlIbpAΔC14, apparently, interacting with its N-terminus, while no protection of the substrate protein from the heat-induced denaturation could be observed (Fig. 4 and 6, Table 1). The removal of both 12 N-terminal and 14 C-terminal amino acids completely abrogated the interaction of AlIbpA with both insulin and ADH and also no denaturation prevention could be observed. AlIbpA lacking 25 N-terminal and 14 C-terminal amino acids was characterized with the lowest stability and chaperone-like activity. Interestingly, some binding with insulin could be detected, probably by its direct interaction with the α-crystalline domain (Fig. 4).
Next, the substitution of the LEL motif by SEP did not affect significantly the stability of the protein, while abolished both the chaperone-like activity of AlIbpASEP and its interaction with both insulin and ADH (Fig. 4 and 6, Table 1). Interestingly, the stability, substrate binding and chaperone-like activity of both AlIbpASEPΔN12 and AlIbpASEPΔN25 increased in comparison with AlIbpASEP and were comparable with those of the full-length protein while remaining significantly lower than in AlIbpAΔN12 and AlIbpAΔN25.
Taken together these data clearly demonstrate that both N- and C-termini determine the interaction of AlIbpA with substrate proteins. In particular, the C-terminus seems to be responsible for the chaperone-like activity of the protein, while the N-terminus could play a regulatory role.
N-terminal domain provides formation of the globular structure and represses fibril formation
Results of the chaperone-like activity (Table 1) revealed that the interaction with substrates differs between the full-length and the truncated AlIbpAs regardless of their oligomerization leading to the assumption of inter-molecular alternations in the oligomer structure. To corroborate this hypothesis, the oligomerization level of both full-length and all truncated modifications of AlIbpA proteins have been assessed with size-exclusion chromatography in a thermostatic chamber at 30 °C (Fig. 7).The full-length AlIbpA was eluted in two main peaks at 7.6 ml and 11.1 ml corresponding to the oligomers of ∼1700 kDa and ∼400 kDa, respectively (Fig. 7B). First two peaks corresponding to the high molecular weight oligomers of the protein were harvested and immediately subjected to the in vitro cross-linking with glutaraldehyde, negatively stained on grids and analyzed with transmission electron microscopy (Fig. 8). As indicated by the microscopy data, the full-length AlIbpA is represented by the heterogeneous mixture of both globular and fibrous structures. After gel-filtration, in the first peak the mixture of fibrils and protein agglomerates was eluted, while the second peak corresponds to globules consisting of apparently 24 monomers (estimated molecular weight 451 kDa).
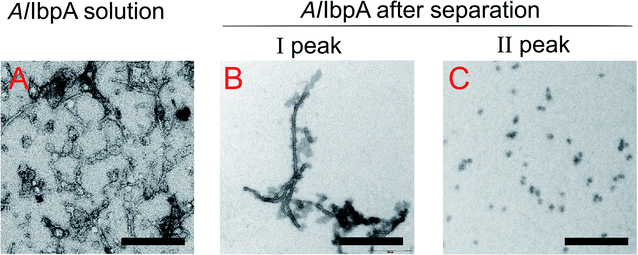 | ||
| Fig. 8 Transmission electron microscopy of cross-linked full-length AlIbpA before (A) and after separation on size-exclusion column (B and C). I and II – first and second peaks, respectively, on AlIbpA elution profile shown on Fig. 7. The bar corresponds to 200 nm. | ||
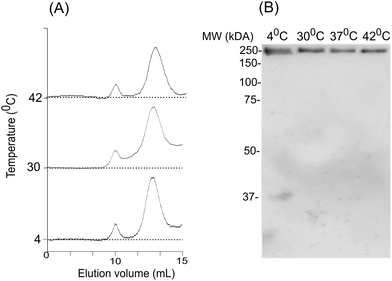
To assess the temperature-dependent inter-molecular rearrangements of sHSP oligomers, the full-length AlIbpA was cross-linked after 1 h incubation at different temperatures (4 °C, 30 °C and 42 °C) before the chromatography, to avoid the inter-molecular reorganizations during the gel-filtration. The elution profiles of the protein did not differ significantly from the untreated one suggesting relative stability of AlIbpA oligomeric forms (Fig. 9A). The SDS-PAGE analysis of cross-linked proteins also confirmed that recombinant protein existed as huge oligomers independently of the temperature (Fig. 9B). These data suggest that the full-length AlIbpA in solution is represented by a mixture of 24-mer globules of ∼400 kDa and high-molecular weight fibrils/agglomerates with about 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio regardless of the temperature (Table 1).
1 ratio regardless of the temperature (Table 1).
Next, the truncated proteins were analyzed also with transmission electron microscopy in order to discover their oligomeric structure (Fig. 10). Either removal of 14 amino acids from the C-terminus or truncation of the LEL motif barely affected the ratio of globules and huge multimers in the protein solution (Fig. 7D and E), while no fibrils could be observed with both AlIbpASEP and AlIbpAΔC14 being present as agglomerates in the first peak fraction (Fig. 10B and C). In marked contrast, while the truncation of the first two F-residues did not affect the globules formation (Fig. 7C and 10A), the removal of either 12 or 25 amino acid residues from the N-terminus of protein completely abolished the globules formation and the protein was present predominantly as long fibrils (Fig. 7F, I and 10D, G). The removal of both N- and C-termini of the protein led to a significant decrease of the oligomers molecular weight to ∼200 kDa (Fig. 7G, J and 10E, H). Similarly, in agreement with in vitro cross-link data (Fig. 3B), the removal of 12 N-terminal amino acid residues and the truncation of the LEL-motif also suppressed the oligomerization of the AlIbpAΔN12C14 (Fig. 7G and 10E). Remarkably, after removal of 25 N-terminal amino acid residues and mutation in the LEL-motif AlIbpASEPΔN25 could be observed in form of fibers (Fig. 7K and 10I). Apparently, in this case either the α-crystalline domain tends to form oligomers by unspecific hydrophobic interactions, or other C-terminal motif also provides the fibrils formation in absence of functionally active LEL-motif.
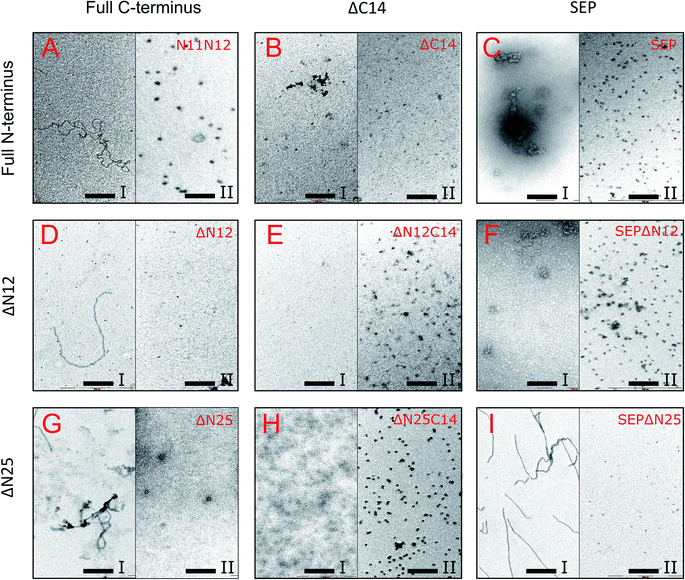 | ||
| Fig. 10 Transmission electron microscopy of mutated and truncated AlIbpA proteins after separation on size-exclusion column. I and II – first and second peaks, respectively, shown on Fig. 7 were collected and immediately subjected to cross-linking with 0.05% glutaraldehyde. 5 μl of solution were transferred to the grids and negatively stained. The bar corresponds to 200 nm. | ||
These data clearly demonstrate that both N- and C-termini in AlIbpA participate in the protein oligomerization. The C-terminus seems to be necessary for the fibrous form formation, while the N-terminus seems to be responsible for the globules formation and in turn apparently acts as a suppressor of the transition into fibrous form.
Discussion
Small heat shock proteins (sHSPs) represent the first line of cellular defense against stress-induced protein aggregation, and their tertiary structures are nearly identical in various organisms from bacteria to mammals. Despite significant variations between amino acid sequences, these proteins consist of the central α-crystalline domain flanked with N- and C-terminal regions.5,14,17–19 While the main role for substrates binding and prevention of their aggregation likely belongs to the central α-crystalline domain, the removal of either N- or C-terminal domains generally leads to the loss of the chaperone-like activity, suggesting their necessity for the sHSPs functionality.18,24,25,57 Nevertheless, exact mechanisms of how either N- or C-terminal domains regulate the activity of the protein remains questionable. In this work we attempted to unravel the functional role of both N- and C-terminal domains in sHSP AlIbpA from Acholeplasma laidlawii, a ubiquitous phytopathogen of the genus Mollicutes.The Mollicutes represent simplest bacteria characterized by a complete lack of cell walls and drastically reduced genome size. Among them, presumably only those species that are able to survive in the environment carry the gene encoding for the sHSP-like protein with classical α-crystalline domain in the genomes (Fig. S2†) suggesting that these proteins are essential for the survival of mycoplasmas under environmental conditions.40 AlIbpA was the first well-described mycoplasmal sHSP isolated from the free-leaving mycoplasma Acholeplasma laidlawii.40,41 Under the heat-shock conditions, AlIbpA becomes the one of the most abundant proteins in A. laidlawii confirming its importance for the stress resistance of this bacterium, although the deletion of sHSP genes in E. coli did not drastically affect the viability of the cells at elevated temperatures.39
The structural organization of AlIbpA is similar to other sHSPs and includes the central α-crystalline domain flanked with N- and C-termini (Fig. 1). The N-terminus of the protein contains double (W/F)(D/F)PF-like motif (Fig. 1), which was previously shown to be involved and even required for their oligomerization and chaperone-like activity of other sHSPs.12,16,22,24 The truncation of the N-terminus generally abrogates both the interaction with substrates proteins and the oligomerization of sHSPs, leading to only dimers formation, particularly for the IbpA from E. coli24 and for the HSP26 from S. cerevisiae.22,23 In marked contrast, the deletion of either 12 or 25 amino acid residues from the N-terminus of AlIbpA or partial truncation of the first (W/F)(D/F)PF-like motif affected neither oligomerization (Fig. 3B) nor substrates binding (Fig. 4 and 6) of AlIbpAΔN12, AlIbpAΔN25 and AlIbpAN11N12. Moreover, the protein stability and capability to prevent the temperature-induced aggregation of insulin even increased in comparison with the full-length protein (Table 1). Further analysis of the oligomerization revealed that both AlIbpAΔN12 and AlIbpAΔN25 are present predominantly in fibrous form (Fig. 7F, I and 10D, G), while the full-length protein represents a mixture of 24-mer globules, fibrils and protein agglomerates with the prevalence of globules (Fig. 7B and 8). Similarly, AlIbpAN11N12 is present predominantly as globules consisting of apparently 24 monomers with low fraction of long fibrils as indicated by both gel-filtration and TEM data (Fig. 7C and 10A). Of note, one could assume that the presence of globules on TEM image of the first elution peak of full-length AlIbpA (Fig. 7I peak) might be a consequence of the fast reorganization of the protein quaternary structure in contrast to the truncated versions of AlIbpA which are predominantly in either fibrillar or globular state.39
At the C-terminus of AlIbpA the conserved LEL-motif participating in oligomerization and chaperone-like activity of sHSPs has been identified (Fig. 1).14,18,25 The removal of the C-terminus of AlIbpA completely abrogated the chaperon-like activity of AlIbpAC14, while, in contrast to EcIbpA, its interaction with insulin and ADH could nevertheless be observed (Fig. 4 and 6, Table 1). Only double removal of both N- and C-termini completely abrogated both the chaperon-like activity and the substrate proteins binding, suggesting that the N-terminus also participates in the substrate binding, while the chaperon-like activity is apparently provided by the C-terminus only. From the structural point of view, AlIbpAΔC14 was observed predominantly as globules (peak II) or their aggregates (peak I), while simultaneous truncations of both N- and C-termini led to the low-level oligomerization of AlIbpAΔN12C14 and AlIbpAΔN25C14 with the formation of di- and tetramers (Fig. 3B and 7G, J) likely due to the α-crystalline self-oligomerization.
It has been shown previously that the purified EcIbpA and EcIbpB from E. coli can be also present as fibrils and huge agglomerates, respectively,1,33,34 while in their mixture EcIbpB blocks the fibril formation by EcIbpA.24 Therefore, it could be speculated that the N-terminal extension of AlIbpA acts as autoinhibitory motif suppressing the fibril formation, similarly as EcIbpB abrogates the fibrillation of EcIbpA (Fig. 7 and 10) while also negatively regulating the chaperone-like activity of the protein (see ΔTm 50 data on Table 1). Consequently, the removal of the N-terminus could open the α-crystalline domain for the interaction with substrates.
The speculative explanation of fibrils formation by AlIbpAΔN12 and AlIbpAΔN25 could be the following. The globular form of sHSPs is considered as the inactive form of sHSPs in the cell (used to store the protein) in which a spatial access to the α-crystalline for the interaction with both substrates and the C-terminus is limited.58 Under stress conditions, the rearrangement of subunits occurs, thereby opening the α-crystalline for the interaction with the C-terminal LEL motif of the neighboring subunit of AlIbpA. Apparently, in the presence of two (W/F)(D/F)PF-like motifs in AlIbpA preferential globular structure formation governed by the hydrophobic (W/F)(D/F)PF-like motifs in full-length protein blocking the fibrils formation and substrates binding, could be hypothesized. This assumption fits well with more pronounced chaperone-like activity of AlIbpAΔN12 and AlIbpAΔN25 (see Table 1). By the way, AlIbpAN11N12 appeared also as globules (Fig. 7C and 10A), while more pronounced chaperone-like activity of protein could be observed in comparison with the full-length protein (Table 1). Apparently, the truncation of only one (W/F)(D/F)PF-like motif appears insufficient for the opening of the α-crystalline domain for the interaction with the C-terminus and fibrils formation, while the substrate binding is less limited (Fig. 4 and 6).
Next, to confirm the role of the C-terminal LEL-motif for the AlIbpA functions, LEL was substituted by SEP. The resulting AlIbpASEP was neither able to bind insulin and ADH nor able to prevent the aggregation of insulin (Fig. 4 and 6, Table 1). Furthermore, AlIbpASEP appeared in aggregates on the first peak (Fig. 10C), that is in agreement with earlier reported C-terminal IXI motif in sHSPs interaction with a hydrophobic groove on the α-crystalline domain of a neighboring dimer thereby facilitating the formation of higher-order oligomers.14,25 Altogether these data confirm the necessity of the LEL-motif for the fibrils formation and thereby also for the chaperone-like activity of AlIbpA similarly to other sHSPs.14,18,25 In the second peak the globules of AlIbpASEP were observed (Fig. 10C) supporting the hypothesis that the N-terminus is responsible for the globular structure formation.
Nevertheless, any amino acids besides the LEL motif on the C-terminus seem to be required for the protein stabilization, since the substitution of LEL by SEP did not reduce the protein stability (Table 1). Moreover, the removal of the N-terminal amino acids from the AlIbpASEP suppresses the mutation of the LEL-motif and restores the insulin binding and the chaperone-like activities of the resulting proteins that appears even more pronounced for AlIbpASEPΔN25 than for AlIbpASEPΔN12 (Fig. 4, Table 1). In marked contrast, AlIbpASEPΔN12 was not able to form high-order oligomers, while AlIbpASEPΔN25 was present as fibrils (Fig. 3, 7H, K and 10I). These data suggest that both N-terminal (W/F)(D/F)PF-like motifs are involved in the globular structure formation, allowing fine-tuned activation of the protein depending on the stress force. From the other side, not only the C-terminal LEL motif but also other amino acid residues are involved in fibrils formation by AlIbpA, in contrast to other sHSPs, confirming that the AlIbpA activity regulation is governing by complex intramolecular mechanisms thereby providing the viability of A. laidlawii under environmental conditions.
Conclusions
Taken together, our data demonstrate non-trivial features of the N-terminal domain of sHSP AlIbpA from A. laidlawii. We suggest that the N-terminus provides the formation of approximately 24-mer globules of AlIbpA while also behaves as an autoinhibitor and activity regulator of the C-terminus. In turn, the C-terminus is required for the chaperone-like activity and protein oligomerization into the fibrous form. Furthermore, our data (with the exception of AlIbpAN11N12) show clear association of the fibrils formation by AlIbpA and its ability to prevent the temperature-induced aggregation of insulin (see Table 1, Fig. 7 and 10). Thus, while the full-length protein is present as a mixture of agglomerates, fibrils and globules demonstrating moderate chaperone-like activity on insulin, the stability and activity of truncated AlIbpA versions appearing as fibrils appears significantly higher. Thus, in contrast to E. coli sHSPs IbpA and IbpB which work in strong cooperation and thus apparently regulate their activity, the AlIbpA represents a first sHSP20 protein where the competition between N- and C- termini governs the shift of the protein quaternary structure to either fibrous or globular form thereby representing the molecular mechanism of the AlIbpA function regulation.Author contributions
Conceptualization, A. K. and I. V.; methodology, A. K., V. Ch. and I. V.; software, M. B.; validation, L. Ch., A. K. and I. V.; formal analysis, I. V.; investigation, L. Ch.; resources, A. K. and I. V.; data curation, M. B.; writing—original draft preparation, A. K. and I. V.; writing—review and editing, A. K., I. V. and M. B.; visualization, L. Ch., A. K. and I. V.; supervision, A. K. and I. V.; project administration, A. K. and I. V.; funding acquisition, I. V.Funding
This research was funded by the Russian Science Foundation (project no. 17-74-20065 to Innokentii Vishnyakov) while being performed in the framework of the Russian Government Program of the Competitive Development of Kazan Federal University (Liliya Chernova and Airat Kayumov). The APC was funded by the Russian Science Foundation (project no. 17-74-20065 to Innokentii Vishnyakov).Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors gratefully acknowledge Dr Dmitry Bogolyubov and Dr Evgeniya Pechnikova for their help with transmission electron microscopy, Prof. Vadim Ivanov for providing anti-IbpA antibodies and Andrey Runov for the assistance in NanoDrop measurements. We are also grateful to Prof. Sergei Borchsenius for illuminating discussions during the interpretation of the experimental results and Prof. Dr Kai Thormann from the Institut für Mikro-und Molekularbiologie, Justus-Liebig Universität Gieβen for technical support.References
- W. W. Jiao, M. D. Qian, P. L. Li, L. Zhao and Z. Y. Chang, J. Mol. Biol., 2005, 347, 871–884 CrossRef CAS PubMed.
- M. Haslbeck and E. Vierling, J. Mol. Biol., 2015, 427, 1537–1548 CrossRef CAS PubMed.
- M. Haslbeck, T. Franzmann, D. Weinfurtner and J. Buchner, Nat. Struct. Mol. Biol., 2005, 12, 842–846 CrossRef CAS PubMed.
- C. Slingsby and A. R. Clark, Structure, 2013, 21, 193–194 CrossRef CAS PubMed.
- R. Van Montfort, C. Slingsby and E. Vierling, Protein Folding in the Cell, 2002, vol. 59, pp. 105–156 Search PubMed.
- S. Mishra, S. A. Chandler, D. Williams, D. P. Claxton, H. A. Koteiche, P. L. Stewart, J. L. P. Benesch and H. S. McHaourab, Structure, 2018, 26, 1116–1126 CrossRef CAS PubMed.
- S. Carra, S. Alberti, P. A. Arrigo, J. L. Benesch, I. J. Benjamin, W. Boelens, B. Bartelt-Kirbach, B. Brundel, J. Buchner, B. Bukau, J. A. Carver, H. Ecroyd, C. Emanuelsson, S. Finet, N. Golenhofen, P. Goloubinoff, N. Gusev, M. Haslbeck, L. E. Hightower, H. H. Kampinga, R. E. Klevit, K. Liberek, H. S. McHaourab, K. A. McMenimen, A. Poletti, R. Quinlan, S. V. Strelkov, M. E. Toth, E. Vierling and R. M. Tanguay, Cell Stress Chaperones, 2017, 22, 601–611 CrossRef CAS PubMed.
- M. Haslbeck, S. Weinkauf and J. Buchner, J. Biol. Chem., 2019, 294, 2121–2132 CrossRef CAS PubMed.
- T. Fleckenstein, A. Kastenmuller, M. L. Stein, C. Peters, M. Daake, M. Krause, D. Weinfurtner, M. Haslbeck, S. Weinkauf, M. Groll and J. Buchner, Mol. Cell, 2015, 58, 1067–1078 CrossRef CAS PubMed.
- M. Matuszewska, D. Kuczynska-Wisnik, E. Laskowska and K. Liberek, J. Biol. Chem., 2005, 280, 12292–12298 CrossRef CAS PubMed.
- J. A. Aquilina, J. L. P. Benesch, L. L. Ding, O. Yaron, J. Horwitz and C. V. Robinson, J. Biol. Chem., 2004, 279, 28675–28680 CrossRef CAS PubMed.
- I. Santhanagopalan, M. T. Degiacomi, D. A. Shepherd, G. K. A. Hochberg, J. L. P. Benesch and E. Vierling, J. Biol. Chem., 2018, 293, 19511–19521 CrossRef CAS PubMed.
- T. Grousl, S. Ungelenk, S. Miller, C. T. Ho, M. Khokhrina, M. P. Mayer, B. Bukau and A. Mogk, J. Cell Biol., 2018, 217, 1269–1285 CrossRef CAS PubMed.
- S. P. Delbecq and R. E. Klevit, FEBS Lett., 2013, 587, 1073–1080 CrossRef CAS PubMed.
- S. P. Delbecq, J. C. Rosenbaum and R. E. Klevit, Biochemistry, 2015, 54, 4276–4284 CrossRef CAS PubMed.
- E. Basha, H. O'Neill and E. Vierling, Trends Biochem. Sci., 2012, 37, 106–117 CrossRef CAS PubMed.
- K. C. Giese, E. Basha, B. Y. Catague and E. Vierling, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 18896–18901 CrossRef CAS PubMed.
- R. Bakthisaran, R. Tangirala and C. M. Rao, Biochim. Biophys. Acta, Proteins Proteomics, 2015, 1854, 291–319 CrossRef CAS PubMed.
- M. V. Sudnitsyna, E. V. Mymrikov, A. S. Seit-Nebi and N. B. Gusev, Curr. Protein Pept. Sci., 2012, 13, 76–85 CrossRef CAS PubMed.
- S. E. Jackson, Mol. Chaperones, 2013, 328, 69–98 Search PubMed.
- E. T. McDonald, M. Bortolus, H. A. Koteiche and H. S. McHaourab, Biochemistry, 2012, 51, 1257–1268 CrossRef CAS PubMed.
- M. Haslbeck, A. Ignatiou, H. Saibil, S. Helmich, E. Frenzl, T. Stromer and J. Buchner, J. Mol. Biol., 2004, 343, 445–455 CrossRef CAS PubMed.
- T. Stromer, E. Fischer, K. Richter, M. Haslbeck and J. Buchner, J. Biol. Chem., 2004, 279, 11222–11228 CrossRef CAS PubMed.
- J. Strozecka, E. Chrusciel, E. Gorna, A. Szymanska, S. Zietkiewicz and K. Liberek, J. Biol. Chem., 2012, 287, 2843–2853 CrossRef CAS PubMed.
- S. Y. Pasta, B. Raman, T. Ramakrishna and C. M. Rao, Mol. Vis., 2004, 10, 655–662 CAS.
- S. Studer and F. Narberhaus, J. Biol. Chem., 2000, 275, 37212–37218 CrossRef CAS PubMed.
- A. Bepperling, F. Alte, T. Kriehuber, N. Braun, S. Weinkauf, M. Groll, M. Haslbeck and J. Buchner, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 20407–20412 CrossRef CAS PubMed.
- P. A. Lund, Adv. Microb. Physiol., 2001, 44, 93–140 CrossRef CAS PubMed.
- S. A. Bissonnette, I. Rivera-Rivera, R. T. Sauer and T. A. Baker, Mol. Microbiol., 2010, 75, 1539–1549 CrossRef CAS PubMed.
- L. Veinger, S. Diamant, J. Buchner and P. Goloubinoff, J. Biol. Chem., 1998, 273, 11032–11037 CrossRef CAS PubMed.
- A. Mogk, C. Schlieker, K. L. Friedrich, H. J. Schofeld, E. Vierling and B. Bukau, J. Biol. Chem., 2003, 278, 31033–31042 CrossRef CAS PubMed.
- E. Ratajczak, S. Zietkiewicz and K. Liberek, J. Mol. Biol., 2009, 386, 178–189 CrossRef CAS PubMed.
- J. R. Shearstone and F. Baneyx, J. Biol. Chem., 1999, 274, 9937–9945 CrossRef CAS PubMed.
- E. Ratajczak, J. Strozecka, M. Matuszewska, S. Zietkiewicz, D. Kuczynska-Wisnik, E. Laskowska and K. Liberek, FEBS Lett., 2010, 584, 2253–2257 CrossRef CAS PubMed.
- Y. L. Tsai, Y. R. Chiang, C. F. Wu, F. Narberhaus and E. M. Lai, PLoS One, 2012, 7(11), e49685 CrossRef CAS PubMed.
- E. Hilario, F. J. M. Martin, M. C. Bertolini and L. Fan, J. Mol. Biol., 2011, 408, 74–86 CrossRef CAS PubMed.
- L. Liu, J. Y. Chen, B. Yang, F. H. Wang, Y. H. Wang and C. H. Yun, Structure, 2015, 23, 2066–2075 CrossRef CAS PubMed.
- K. Takeda, T. Hayashi, T. Abe, Y. Hirano, Y. Hanazono, M. Yohda and K. Miki, J. Struct. Biol., 2011, 174, 92–99 CrossRef CAS PubMed.
- J. G. Thomas and F. Baneyx, J. Bacteriol., 1998, 180, 5165–5172 CrossRef CAS PubMed.
- I. E. Vishnyakov, S. A. Levitskii, V. A. Manuvera, V. N. Lazarev, J. A. Ayala, V. A. Ivanov, E. S. Snigirevskaya, Y. Y. Komissarchik and S. N. Borchsenius, Cell Stress Chaperones, 2012, 17, 171–180 CrossRef CAS PubMed.
- I. E. Vishnyakov and S. N. Borchsenius, Microbiology, 2013, 82, 653–667 CAS.
- I. E. Vishnyakov, M. I. Bogachev, I. Salafutdinov, S. N. Borchsenius and A. R. Kayumov, Bionanoscience, 2016, 6, 437–442 CrossRef.
- A. R. Kayumov, M. I. Bogachev, V. A. Manuvera, V. N. Lazarev, A. V. Sabantsev, T. O. Artamonova, S. N. Borchsenius and I. E. Vishnyakov, Mol. Biol., 2017, 51, 131–141 CrossRef CAS.
- D. G. Gibson, L. Young, R. Y. Chuang, J. C. Venter, C. A. Hutchison and H. O. Smith, Nat. Methods, 2009, 6, 343–345 CrossRef CAS PubMed.
- A. Kayumov, A. Heinrich, K. Fedorova, O. Ilinskaya and K. Forchhammer, FEBS J., 2011, 278, 1779–1789 CrossRef CAS PubMed.
- K. Hauf, A. Kayumov, F. Gloge and K. Forchhammer, J. Biol. Chem., 2016, 291, 3483–3495 CrossRef CAS PubMed.
- F. H. Niesen, H. Berglund and M. Vedadi, Nat. Protoc., 2007, 2, 2212–2221 CrossRef CAS PubMed.
- K. Huynh and C. L. Partch, Curr. Protoc. Protein Sci., 2015, 79, 28 Search PubMed.
- F. Sievers, A. Wilm, D. Dineen, T. J. Gibson, K. Karplus, W. Z. Li, R. Lopez, H. McWilliam, M. Remmert, J. Soding, J. D. Thompson and D. G. Higgins, Mol. Syst. Biol., 2011, 7(1), 539 CrossRef PubMed.
- L. A. Kelley, S. Mezulis, C. M. Yates, M. N. Wass and M. J. E. Sternberg, Nat. Protoc., 2015, 10, 845–858 CrossRef CAS PubMed.
- J. Y. Yang and Y. Zhang, Nucleic Acids Res., 2015, 43, W174–W181 CrossRef CAS PubMed.
- C. X. Zhang, P. L. Freddolino and Y. Zhang, Nucleic Acids Res., 2017, 45, W291–W299 CrossRef CAS PubMed.
- M. I. Bogachev, A. R. Kayumov, O. A. Markelov and A. Bunde, Sci. Rep., 2016, 6, 22286 CrossRef CAS PubMed.
- R. Maiti, G. H. Van Domselaar, H. Zhang and D. S. Wishart, Nucleic Acids Res., 2004, 32, W590–W594 CrossRef CAS PubMed.
- M. I. Bogachev, V. Y. Volkov, G. Kolaev, L. Chernova, I. Vishnyakov and A. Kayumov, Bionanoscience, 2019, 9, 59–65 CrossRef.
- M. I. Bogachev, V. Y. Volkov, O. A. Markelov, E. Y. Trizna, D. R. Baydamshina, V. Melnikov, R. R. Murtazina, P. V. Zelenikhin, I. S. Sharafutdinov and A. R. Kayumov, PLoS One, 2018, 13(5), e0193267 CrossRef PubMed.
- J. A. Carver, A. B. Grosas, H. Ecroyd and R. A. Quinlan, Cell Stress Chaperones, 2017, 22, 627–638 CrossRef CAS PubMed.
- A. Mogk, C. Ruger-Herreros and B. Bukau, Annu. Rev. Microbiol., 2019, 73, 89–110 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra10172a |
| ‡ The authors contributed equally to this manuscript as senior authors. |
| This journal is © The Royal Society of Chemistry 2020 |