Open Access Article
Open Access ArticleComparative transcriptomic analysis reveals the regulatory effects of inorganic nitrogen on the biosynthesis of Monascus pigments and citrinin†
Jia-Li Hong‡
 ac,
Li Wu‡b,
Jin-Qiang Luac,
Wen-Bin Zhouabc,
Ying-Jia Caoabc,
Wen-Long Lvd,
Bin Liu
ac,
Li Wu‡b,
Jin-Qiang Luac,
Wen-Bin Zhouabc,
Ying-Jia Caoabc,
Wen-Long Lvd,
Bin Liu *ac,
Ping-Fan Raob,
Li Nib and
Xu-Cong Lv
*ac,
Ping-Fan Raob,
Li Nib and
Xu-Cong Lv *abc
*abc
aCollege of Food Science, Fujian Agriculture and Forestry University, No. 15 Shangxiadian Rd, Cangshan District, Fuzhou, Fujian 350002, China. E-mail: xucong1154@163.com; liubin618@hotmail.com
bInstitute of Food Science and Technology, College of Biological Science and Technology, Fuzhou University, Fuzhou, Fujian 350108, P. R. China
cNational Engineering Research Center of JUNCAO Technology, Fujian Agriculture and Forestry University, Fuzhou, Fujian 350002, P. R. China
dDepartment of Radiation Oncology, The First Affiliated Hospital of Fujian Medical University, Fuzhou, Fujian 350005, P. R. China
First published on 3rd February 2020
Abstract
Monascus spp. and its secondary metabolites have been widely applied in foods and medicines for thousands of years in eastern Asia. Nitrogen sources are essential nutrients for the growth and metabolism of Monascus spp. Our previous study found that inorganic nitrogen sources (especially NH4Cl and NH4NO3) promoted the biosynthesis of Monascus pigments (MPs) and inhibited the production of citrinin. The objective of the present study was to investigate the regulatory mechanism of inorganic nitrogen on the biosynthesis of MPs and citrinin by the comparative transcriptional approach (RNA sequencing combined with RT-qPCR). Results indicated that the submerged fermentation of M. purpureus M3103 with NH4Cl or NH4NO3 as the sole nitrogen source can significantly increase the yields of MPs (especially for Monascus orange and red pigments) and decrease citrinin production, compared with the organic nitrogen source (peptone group). Comparative transcriptomic profiling by RNA sequencing found that the numbers of differentially expressed genes (DEGs) between different experimental groups—M group (peptone group) vs. ML group (NH4Cl group), and M group (peptone group) vs. MX group (NH4NO3 group), were 722 and 1287, respectively. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed that genes involved in carbon and nitrogen metabolism, biosynthesis of amino acids were up-regulated by NH4Cl and NH4NO3, which would produce more biosynthetic precursors for MPs. Whereas, the inorganic nitrogen source (both of NH4Cl and NH4NO3) down-regulated the expression levels of genes involved in tyrosine metabolism. In addition, NR analysis indicated that the essential genes and transcription factors involved in the biosynthesis pathway of citrinin were down-regulated by NH4Cl and NH4NO3. These results indicated that NH4Cl or NH4NO3 as a nitrogen source for M. purpureus M3103 can significantly promote the precursor synthesis of Monascus pigments, but reduce the transcription of polyketide synthase for citrinin, and therefore significantly increase Monascus pigments production and decrease citrinin formation. These findings will facilitate a comprehensive understanding of the regulatory mechanisms of inorganic nitrogen in the biosynthesis of secondary metabolites in M. purpureus, and would benefit the application of M. purpureus in the production of MPs.
1. Introduction
Red yeast rice is a kind of rice koji made from rice as the main raw material, and fermented by the filamentous fungus Monascus. It has been widely applied in traditional Chinese medicine, food additives, wine fermentation and other fields with a long history in eastern Asia.1,2 Monascus spp. can synthesize abundant useful secondary metabolites, such as pigments, dimerumic acid, γ-GABA, monacolin K, citrinin, and so on.3 Monascus pigments (MPs), a mixture of polyketones, are widely utilized as food colorants in various foods in China.4 Over the past few years, MPs have been reported to exhibit antimicrobial,5 hypolipidaemic,6 anti-atherosclerotic7 and photosensitizer activities.8 However, most strains of Monascus spp. also produce mycotoxin citrinin with strong nephrotoxic, hepatotoxic9 and teratogenic10 effects on animals and humans, which greatly limits the application of Monascus fermented products. Therefore, the safety of Monascus fermented products has attracted much attention in many countries, especially Japan and European countries.11 Thus, how to promote the biosynthesis of functional MPs but reduce the content of citrinin has become an urgent problem in the industrial production and applications of Monascus fermented products.The regulation of the fermentation process is important and the most commonly used approach to improve the quality and safety of Monascus related products. In recent years, many researchers have tried to control the fermentation process to increase the production of MPs with low-yield or no citrinin, mainly through physical and chemical methods. The physical methods used to regulate the production of MPs and citrinin mainly focus on controlling oxygen supply,12 temperature,13 pH14 and light15 during the fermentation process. Apparently, regulation of fermentation process through physical methods is difficult to block citrinin biosynthesis completely in Monascus spp. Regulation of fermentation process by chemical methods are mainly through the further supplementation of carbon and nitrogen sources, and other exogenous chemicals to the fermentation substrate.16–18 Over the past several decades, a considerable amount of studies have investigated the effects of different types of carbon and nitrogen sources on the production and secretion of secondary metabolites by Monascus spp. During the fermentation process, carbon source can form the carbon scaffolds of the secondary metabolites. It was found that mannitol had the best effect in enhancing the MPs production.19 By contrast, nitrogen source is essential for the biosynthesis of amino acids through the ammonia pathway and other nitrogen-containing organic compounds, such as Monascus red pigments (rubropunctamine [C21H23NO4] and monascorubramine [C23H27NO4]).
There are significant differences in the ability of utilizing inorganic and organic nitrogen sources for secondary metabolism by Monascus spp. Previous studies found that the addition of inorganic nitrogen source significantly increased the yields of Monascus red and yellow pigments, compared with the same amount of organic nitrogen source during the solid-state fermentation. Ammonium chloride (NH4Cl) has been proved to be the most favorable for the biosynthesis of MPs, followed by ammonium nitrate (NH4NO3).20,21 Our previous experimental results also found that inorganic nitrogen (especially NH4Cl and NH4NO3) not only significantly promoted the biosynthesis of MPs, but also significantly inhibited the production of citrinin. In recent years, the biosynthetic pathways involved in MPs and citrinin have been gradually revealed and understood.22,23 However, the regulatory mechanisms of inorganic nitrogen on the biosynthesis of pigments and citrinin in Monascus spp. have not been systematically investigated.
With the rapid development of high-throughput sequencing technology, comparative transcriptomics analysis based on RNA sequencing (RNA-seq) has been widely applied to investigate gene expression profiles under different types of conditions.24,25 In this study, RNA-seq was used to study the regulatory mechanisms of inorganic nitrogen on the biosynthesis of pigments and citrinin in high pigment producing Monascus purpureus M3103. Besides, RT-qPCR was also used to detect the relative expression of MPs and citrinin related genes. The transcriptional levels of the three experimental groups were compared by transcriptome analysis. Results would enhance our knowledge of the molecular mechanisms behind the biosynthesis of secondary metabolites in Monascus spp.
2. Materials and methods
2.1 Strain and culture conditions
The high pigment producing strain Monascus purpureus M3103 (isolated from Hongqu provided by Lianjiang Hongqu Factory (Lianjiang county, Fujian Province, China)) could be obtained from the collection of Food Biotechnology Laboratory, Fujian Agriculture and Forestry University (Fuzhou, China). M. purpureus M3103 used in this study was obtained by strain screening from red yeast rice of Fujian Province in China. The strain was inoculated on potato dextrose agar (PDA) slant and incubated at 28 °C for 7 days, and then kept at 4 °C.2.2 Inoculum preparation and fermentation mediums
To prepare the inoculum, a 5 day-old mycelium of M. purpureus M3103 on a PDA slant was washed by 10 mL dH2O and then transferred to the fermentation medium by adjusting the initial pH to 5.00 and the concentration of suspension spores at about 1 × 105 spores per mL. The inoculum was incubated in a 500 mL Erlenmeyer flask containing 100 mL of medium for 7 days in a rotary shaker at 180 rpm under 30 °C. The fermentation mediums using peptone, NH4Cl, and NH4NO3 as nitrogen sources were named as M, ML, and MX, respectively. After fermentation for 7 days, the harvested Monascus mycelia were immediately frozen in liquid nitrogen and stored at −80 °C until the RNA extraction. Fermentation mediums: M (peptone group): 5% soluble starch, 0.4% fishmeal peptone (Sinopharm Chemical Reagent Co., Ltd, China), 0.1% 7H2O·MgSO4, 0.25% KH2PO4, 100 mL distilled water; ML (NH4Cl group): 5% soluble starch, 0.4% NH4Cl (moles of nitrogen was 7.5 mmol), 0.1% 7H2O·MgSO4, 0.25% KH2PO4, 100 mL distilled water; MX (NH4NO3 group): 5% soluble starch, 0.4% NH4NO3 (moles of nitrogen was 10.0 mmol), 0.1% 7H2O·MgSO4, 0.25% KH2PO4, 100 mL distilled water. These mediums were sterilized at 121 °C for 20 min.2.3 Determination of Monascus pigment and citrinin
The dry mycelium of Monascus (0.100–0.105 g) and the fermentation broth (0.1 mL) were transferred into a tube and extracted with 1 mL 70% ethanol in 200 W ultrasonic bath for 30 min at 50 °C, and then shaken on a rotary shaker at 200 rpm for 1.5 h. The supernatant was analyzed by ultraviolet spectrophotometer. The pigment concentration was measured by the corresponding absorbance (390, 465 and 510 nm for yellow, orange and red pigments, respectively). Subsequently, the supernatant liquid was filtered with a 0.22 μm pore size filter into a 1 mL vial to be used for citrinin determination by HPLC-FLD.26,27 The excitation and emission wavelengths were 331 nm and 500 nm, respectively. ZORBAX SB-C18 column (150 mm × 4.6 mm, 5 μm; Agilent, USA) was used at 30 °C, and isocratic elution was performed for 20 min using acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1
water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v; adjusted pH to 2.5 by acetic acid) as the mobile phase at 1 mL min−1.27
1, v/v; adjusted pH to 2.5 by acetic acid) as the mobile phase at 1 mL min−1.27
2.4 UPLC-Orbitrap MS analysis of the composition of Monascus pigments
The main components of MPs were identified using a water UPLC system (Waters, MA, USA) and a Thermo Scientific TM Orbitrap mass spectrometer (Thermo, MA, USA). The mobile phase A: water (0.1% formic acid), and the mobile phase B: methanol (0.1% formic acid). A T3 column (2.1 mm × 100 mm, 3 μm) (Waters, USA) was kept at 30 °C. The gradient elution: 98% A–90% A at 0–0.5 min, 99% A–37.5% A at 0–0.5 min, 37.5% A–16.8% A at 0.5–3.5 min, 16.8% A–7% A at 3.5–10.5 min, 7% A–0.1% A at 10.5–13 min, and 0.1% A at 13–14 min; finally, equilibrated with 99% A for 3 min. The flow rate: 0.40 mL min−1, and the injection volume: 5 μL. The mass spectrometry detection: positive ion electrospray mode. The scan range: 100 to 1000 m/z, and the source temperature: 120 °C. The capillary voltage: 3.2 kV at ESI+. The collision energy was 15 to 40 eV for the energy scan.2.5 RNA extraction and of high-throughput sequencing
Total RNA was extracted by using Spin Column Fungal Total RNA Purification Kit (Sangon Biotech, code no. B518659). Measurement of the concentration and integrity of the final RNA samples were referenced in ref. 28. The total RNA of samples were used for cDNA library construction and high-throughput sequencing at Biomarker Technologies Corporation (Beijing, China) on the Illumina HiSeq™ 2500 sequencing platform (Illumina™, San Diego, CA, USA) with 150 bp paired-end reads.2.6 Sequence reads assembly, mapping and annotation
Raw data (raw reads) were provided in FASTQ format. The clean reads were obtained by removing reads containing adapter, reads containing ploy-N and low quality reads from raw data. The clean reads were mapped to M. purpureus YY-1 genomic sequence (NCBI accession number: samn08978766) using TopHat (http://ccb.jhu.edu/software/tophat/). Q20, Q30, GC-content and sequence duplication level of the clean data were also calculated. The transcript or gene expression levels were estimated by fragments per kilobase of transcript per million fragments mapped.29 The sequencing data were deposited to the Sequence Read Archive (SRA) database at the National Center for Biotechnology Information (NCBI) with accession number SRR8380849 (ML group), SRR8380850 (MX group) and SRR8380851 (M group).2.7 Quantification and differential expression analysis of transcripts
Differential expression analysis of two groups was performed using the DESeq R package (1.10.1). DESeq provided statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting P values were adjusted using the Benjamini and Hochberg's approach for controlling the false discovery rate. Genes with an adjusted P-value < 0.01 found by DESeq were assigned as differentially expressed.2.8 GO and KEGG enrichment analysis of differentially expressed genes (DEGs)
Gene Ontology (GO) enrichment analysis of the DEGs was implemented by the GOseq R packages based Wallenius non-central hyper-geometric distribution,30 which can adjust for gene length bias in DEGs. KEGG31 is a database resource for understanding high-level functions and utilities of the biological system, such as the cell, the organism and the ecosystem, from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies (http://www.genome.jp/kegg/). KOBAS software was used to test the statistical enrichment of differential expression genes in KEGG pathways.322.9 Expression level of pigments and citrinin biosynthesis-related genes by RT-qPCR
Quantitative real-time reverse transcription polymerase chain reaction (RT-qPCR) analysis of pigments biosynthetic gene and citrinin biosynthetic gene. cDNA synthesis was performed using the PrimeScript™ RT reagent kit with gDNA Eraser (Takara, Dalian, China). Real-time qPCR was conducted to monitor gene expression levels using the SYBR® Premix Ex Taq™ II (Takara, Dalian, China). All primers used in this study were listed in Table S1.† RT-qPCR was performed in duplicate using AB7300 Real-Time PCR System (Applied Biosystems, USA). The relative levels of target mRNAs were determined using the 2−ΔΔCt method and were normalized to the β-actin mRNA signals in each sample.2.10 Statistical analysis
All results are expressed as the means ± standard deviation (SD) (n = 6). GraphPad Prism 7 was used for all analyses. The statistical significance was determined by one-way analysis of variance (ANOVA) with Duncan's multiple test.3. Results
3.1 Effects of inorganic nitrogen on the growth and the production of MPs and citrinin
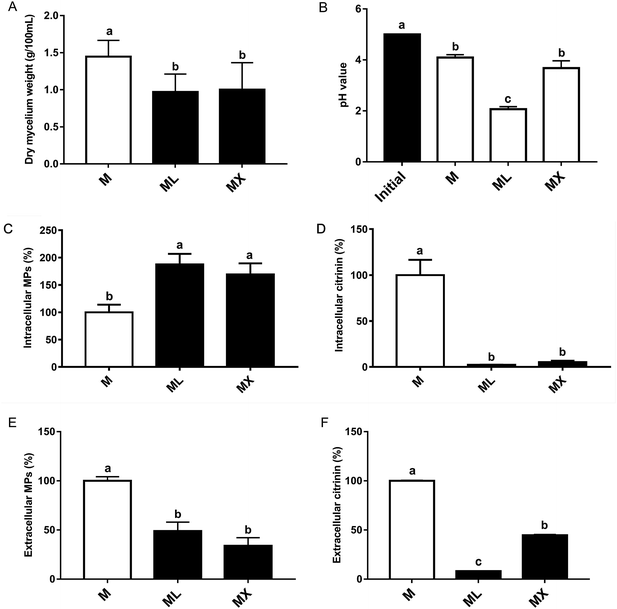
After 7 days of cultivation, there are significant difference in Monascus biomass among the 3 groups. The dried cell weight concentration was 1.49%, 1.20% and 1.11% in M, ML and MX group, respectively (Fig. 1A). The pH values of the final products of M, ML and MX groups were 4.09, 2.07 and 3.68, respectively (Fig. 1B). The effects of inorganic nitrogen (NH4Cl or NH4NO3) on the yields of MPs and citrinin in fermentation of Monascus were evaluated. The relative yields of MPs and citrinin were shown in Fig. 1. The intracellular MPs production by NH4Cl and NH4NO3 were nearly 87.6% and 69.2% higher than the M group (P < 0.05), respectively (Fig. 1C). While the extracellular pigments by NH4Cl and NH4NO3 were nearly 51.1% and 66.2% less than the M group (P < 0.01), respectively (Fig. 1E). Surprisingly, the yields of intracellular citrinin by NH4Cl and NH4NO3 were significantly lower than the peptone group (P < 0.01) (Fig. 1D). Which was consistent with the result of extracellular citrinin production (Fig. 1F). Thus, the above findings indicated that inorganic nitrogen source (NH4Cl and NH4NO3) can significantly promote the biosynthesis of MPs and inhibited the production of citrinin in the submerged fermentation of Monascus, which would greatly reduce the risk of Monascus-related products and would provide a foundation for the study on the regulatory mechanisms of the biosynthesis of pigments and citrinin in M. purpureus.3.2 Effects of inorganic nitrogen on the chemical composition of MPs
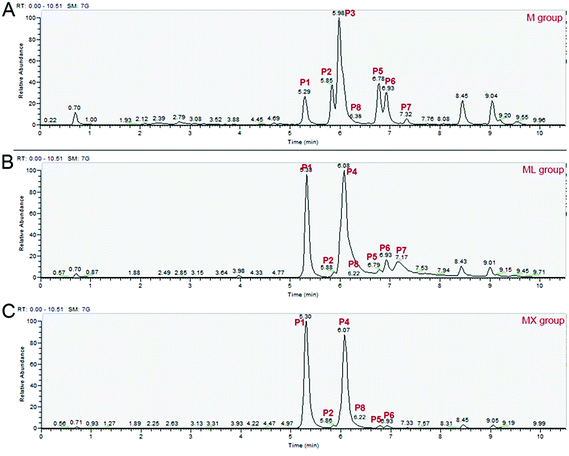
In order to better understand the effects of inorganic nitrogens (NH4Cl and NH4NO3) on the composition of MPs, UPLC-MS/MS was used to detect the differences of the composition of MPs among the M, ML and MX groups. As shown in Fig. 2, S1 and Table S2,† a total of 8 different MPs components were detected, including four yellow pigments (monasfluore A, monascin, monasfluore B and ankaflavin), two orange pigments (rubropunctatin and monascorubrine) and two red pigments (rubropunctamine and monascorubramine). Compared with other pigment components, the peak of rubropunctatin (P8) is not obvious in the total ion chromatogram due to its low relative abundance in total pigment (Fig. 2). Therefore, the specific ion of rubropunctatin (m/z 355.1572) was selectively extracted from the total ion chromatogram to eliminate the interference of other pigments with high abundance.The pigment composition in the fermentation products of ML group and MX group was similar (Fig. 2B and C). It is worth noting that the production of rubropunctamine and monascorubramine (two red pigments) were significantly increased by inorganic nitrogens (NH4Cl and NH4NO3). These findings indicated that inorganic nitrogens (NH4Cl and NH4NO3) could significantly increase the yields of Monascus red pigments (mainly rubropunctamine and monascorubramine) and change the composition of MPs compared with organic nitrogen (peptone).
3.3 High-throughput sequencing and de novo assembly
To better explore the regulatory mechanisms of inorganic nitrogens (NH4Cl and NH4NO3) on the biosynthetic pathways of pigments and citrinin in M. purpureus, high-throughput sequencing was used to investigate the transcription. A total of 222.98 million clean readings were obtained from the Illumina high-throughput sequencing platform after quality control and data filtering. The GC% of the sequencing data from 9 samples was 42.56%, and the average quality score more than 30 reading percentages was not less than 92.31%, indicating that the accuracy and quality of the sequencing data were sufficient for further analysis. General sequencing statistics were shown in ESI Table S3.† From the comparison results, the alignment efficiency of reads and reference genomes of 9 samples ranged from 86.27% to 91.17%, as shown in Table 1. The Pearson correlation coefficient was used as an indicator of biological relevance.33 The strong correlation (R2 > 0.95) between biological replicates indicated that the samples from three different groups had good repeatability (ESI Fig. S2†).| Samples | Total reads | Mapped reads | Uniq. mapped reads | Multiple mapped reads | Reads map to ‘+’ | Reads map to ‘−’ |
|---|---|---|---|---|---|---|
| ML1 | 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 452 452![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 392 392 |
42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 087 087![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 728 (88.69%) 728 (88.69%) |
40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 402 402![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 206 (85.14%) 206 (85.14%) |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 685 685![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 522 (3.55%) 522 (3.55%) |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 944 944![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 939 (44.14%) 939 (44.14%) |
21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 029 029![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 533 (44.32%) 533 (44.32%) |
| ML2 | 53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 871 871![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 824 824 |
47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 670 670![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 074 (88.49%) 074 (88.49%) |
45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 781 781![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 220 (84.98%) 220 (84.98%) |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 888 888![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 854 (3.51%) 854 (3.51%) |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 726 726![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 820 (44.04%) 820 (44.04%) |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 799 799![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 139 (44.18%) 139 (44.18%) |
| ML3 | 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 230 230![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 106 106 |
37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 294 294![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 124 (86.27%) 124 (86.27%) |
35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 845 845![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 197 (82.92%) 197 (82.92%) |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 448 448![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 927 (3.35%) 927 (3.35%) |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 529 529![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 453 (42.86%) 453 (42.86%) |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 649 649![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 921 (43.14%) 921 (43.14%) |
| MX1 | 51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 126 126![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 592 592 |
46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 609 609![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 669 (91.17%) 669 (91.17%) |
44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 905 905![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 008 (87.83%) 008 (87.83%) |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 704 704![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 661 (3.33%) 661 (3.33%) |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 202 202![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 380 (45.38%) 380 (45.38%) |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 297 297![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 756 (45.57%) 756 (45.57%) |
| MX2 | 49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 137 137![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 078 078 |
44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 565 565![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 319 (90.70%) 319 (90.70%) |
42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 961 961![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 449 (87.43%) 449 (87.43%) |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 603 603![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 870 (3.26%) 870 (3.26%) |
22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 191 191![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 415 (45.16%) 415 (45.16%) |
22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 267 267![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 643 (45.32%) 643 (45.32%) |
| MX3 | 52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 234 234![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 716 716 |
47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 158 158![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 784 (90.28%) 784 (90.28%) |
45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 456 456![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 437 (87.02%) 437 (87.02%) |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 702 702![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 347 (3.26%) 347 (3.26%) |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 451 451![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 892 (44.90%) 892 (44.90%) |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 579 579![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 692 (45.14%) 692 (45.14%) |
| M1 | 52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 707 707![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 596 596 |
46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 529 529![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 510 (88.28%) 510 (88.28%) |
44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 936 936![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 750 (85.26%) 750 (85.26%) |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 592 592![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 760 (3.02%) 760 (3.02%) |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 142 142![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 723 (43.91%) 723 (43.91%) |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 257 257![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 067 (44.12%) 067 (44.12%) |
| M2 | 44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 711 711![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 892 892 |
39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 734 734![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 351 (88.87%) 351 (88.87%) |
38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 332 332![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 129 (85.73%) 129 (85.73%) |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 402 402![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 222 (3.14%) 222 (3.14%) |
19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 770 770![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 883 (44.22%) 883 (44.22%) |
19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 832 832![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 956 (44.36%) 956 (44.36%) |
| M3 | 56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 123 123![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 470 470 |
49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 317 317![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 018 (87.87%) 018 (87.87%) |
47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 531 531![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 324 (84.69%) 324 (84.69%) |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 785 785![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 694 (3.18%) 694 (3.18%) |
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 544 544![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 156 (43.73%) 156 (43.73%) |
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 635 635![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 313 (43.89%) 313 (43.89%) |
3.4 Analysis of differentially expressed genes (DEGs)
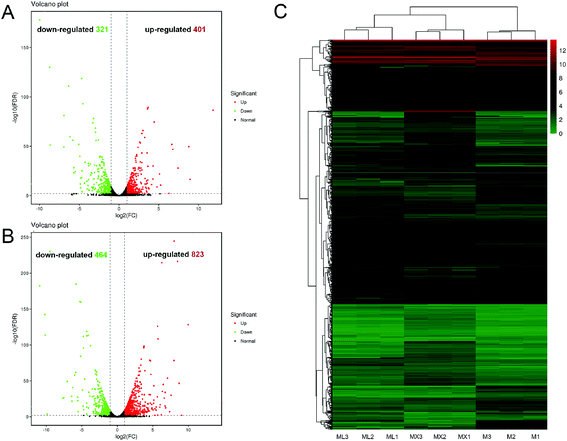
In order to reveals the regulatory mechanisms inorganic nitrogens (NH4Cl and NH4NO3) on the secondary metabolism of M. purpureus, comparative transcriptomic analysis was carried out. The fragments per kilobase of exon model per million mapped reads (FPKM) were calculated as the normalized expression values of each annotated gene. Venn diagram illustrated that 722, 1287 and 523 DEGs was found between the two sample groups: “M vs. ML”, “M vs. MX” and “ML vs. MX”, respectively (ESI Fig. S3†). The number of unique DEGs belonging to “M vs. MX” group (506) was much greater than the number belonging to “M vs. ML” group (145) and “ML vs. MX”group (92). The differences in gene expression levels and statistical significance between the two sample groups (“M vs. ML” and “M vs. MX”) were shown in the volcano plots, respectively (Fig. 3A and B). The number of up-regulated and down-regulated DEGs in “M vs. ML” groups were 401 and 321, respectively (Fig. 3A), while the DEGs in “M vs. MX” groups were 823 up-regulated and 464 down-regulated (Fig. 3B). Hierarchical clustering analysis of the screened differentially expressed genes was also presented in Fig. 3C.3.5 Gene annotation and GO functional classification
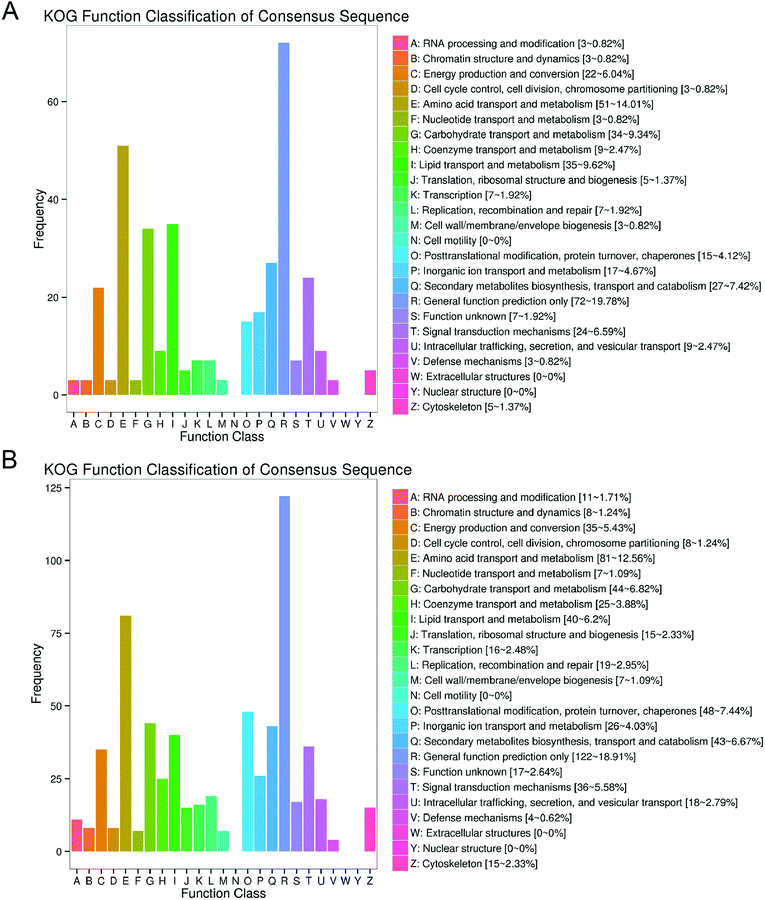
Annotated numbers of the DEGs by different functional databases were shown in Table 2. A total of 1892 DEGs were annotated by eight frequently used functional databases, including Clusters of Orthologous Groups (COG), Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), Eukaryotic Orthologous Groups (KOG), nonredundant (Nr), Pfam, Swiss-Prot and evolutionary genealogy of genes: Nonsupervised Orthologous Groups (eggNOG). In order to further evaluate the RNA-seq data, KOG was used to classify DEGs. The DEGs annotated by KOG were divided into 25 functional categories (Fig. 4A and B). 19.78% (72 genes) and 18.91% (122 genes) DEGs in “M vs. ML” and “M vs. MX” groups were annotated as “General Functional Prediction Only”, which was the most frequently annotated functional category. “Amino acid transport and metabolism” was the second frequently annotated, accounting for 14.01% (51 genes) DEGs in “M vs. ML” and 12.56% (81 genes) DEGs in “M vs. MX”. “Lipid transport and metabolism” was the third and sixth frequently annotated functional category in “M vs. ML” (9.62%, 35 genes) and M vs. MX” (6.20%, 40 genes), respectively. “Carbohydrate transport and metabolism” was the fourth frequently annotated functional category with 9.34% (34 genes) DEGs in “M vs. ML” and 6.82% (44 genes) DEGs in “M vs. MX”. It is interesting to note that 7.42% (27 genes) DEGs in “M vs. ML” and 6.67% (genes) DEGs in “M vs. MX” were annotated as the class Q (secondary metabolites biosynthesis, transport and catabolism), but the specific secondary metabolic pathways involved in the biosynthesis of pigments and citrinin need to be further analyzed through the KEGG metabolic pathway.| DEG set | Total | COG | GO | KEGG | KOG | NR | Pfam | Swiss-Prot | eggNOG |
|---|---|---|---|---|---|---|---|---|---|
| M vs. ML transformants | 682 | 306 | 403 | 187 | 322 | 681 | 539 | 402 | 671 |
| M vs. MX transformants | 1210 | 527 | 697 | 356 | 577 | 1208 | 918 | 707 | 1188 |
| Total | 1892 | 833 | 1100 | 543 | 899 | 1889 | 1457 | 1109 | 1859 |
Furthermore, according to the data from the NR database (ESI Data S1†), we found that gene5930, gene8071 and gene283 were annotated as “citrinin polyketide synthase”, “citrinin biosynthetic transcription CtnC” and “citrinin biosynthesis transcriptional activator CtnR”, respectively. Compared with the M group, the transcription levels of gene5930, gene8071 and gene283 were down-regulated by 1.60/2.98, 3.20/5.79, and 1.18/1.50 log2FC in the ML/MX group, respectively. Conversely, genes related to pigments biosynthesis (such as “non-reducing polyketide synthase” and “Zn cluster transcription factor”) were up-regulated by 1.33 and 2.22 log2FC in the ML and MX groups. The above findings indicated that inorganic nitrogen source (NH4Cl and NH4NO3) can significantly down-regulate the genes involved in citrinin biosynthetic pathway to reduce citrinin production, and up-regulate the genes involved in pigments biosynthetic pathway to enhance pigment production.
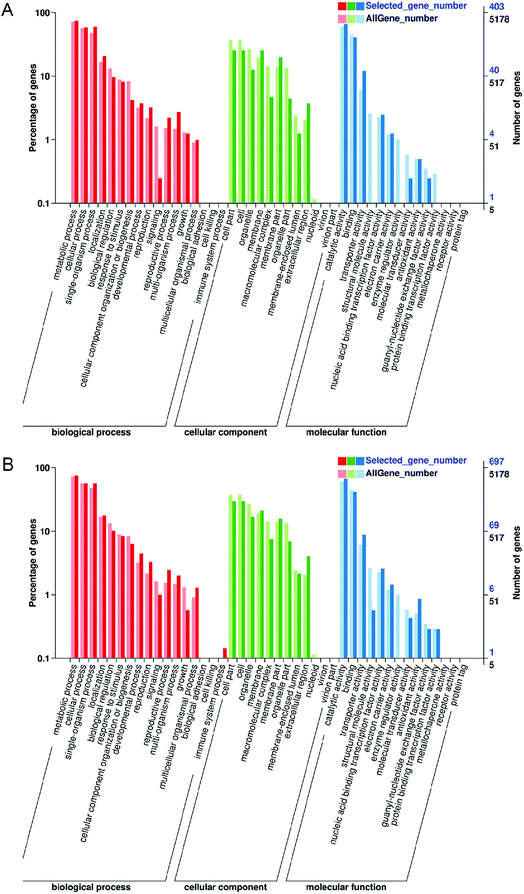
GO is an international standardized classification system for gene functions, which can describe the properties of genes and gene products in organisms. GO enrichment analysis of the three groups revealed that DEGs were classified into 43 functional groups, including biological processes (17 groups), cellular components (12 groups) and molecular functional classification (14 groups) (Fig. 5A and B; ESI Data S2†). “Catalytic activity” was the largest term of “molecular function”, and “metabolic process” was the largest term of “biological process” in “M vs. ML” and “M vs. MX”. “cell” and “cell part” were the largest terms of “cellular component” in “M vs. MX” and “M vs. ML”.
3.6 KEGG pathway enrichment of differentially expressed genes (DEGs)
Transcriptome data were further analyzed by KEGG metabolic pathway, with 75 and 99 metabolic pathways in M vs. ML and M vs. MX, respectively (ESI Data S3†). As shown in Fig. 6, the 20 most significant KEGG pathways for enrichment were revealed. “Biosynthesis of amino acids” was the pathway term with the largest number of DEGs enriched in M vs. ML and M vs. MX (Fig. 6A and B). According to the corrected P-value (qvalue), KEGG pathways in which significantly up-regulated and down-regulated enrichment in M vs. ML and M vs. MX were selected, respectively (Fig. 6A and B). “Biosynthesis of amino acids” was significantly enriched in ML and MX groups. Genes involved in aromatic amino acid metabolism pathway (which associated with “phenylalanine, tyrosine and tryptophan biosynthesis, gene7778”), and branched chain amino acid metabolism (such as “valine, leucine and isoleucine biosynthesis, gene4635”) were up-regulated in ML and MX groups, whereas genes involved in “tyrosine metabolism” were down-regulated in ML and MX groups, which can produce the biosynthetic precursors of pigments (ESI Data S4†). “Carbon metabolism” and “glycolysis/gluconeogenesis” were significantly up-regulated enrichment, and “phenylalanine metabolism” was significantly down-regulated enrichment in “M vs. ML”. “Phenylalanine, tyrosine and tryptophan biosynthesis”, “nitrogen metabolism” and “glycine, serine and threonine metabolism” were significantly up-regulated enrichment in “M vs. MX” (ESI Data S4†).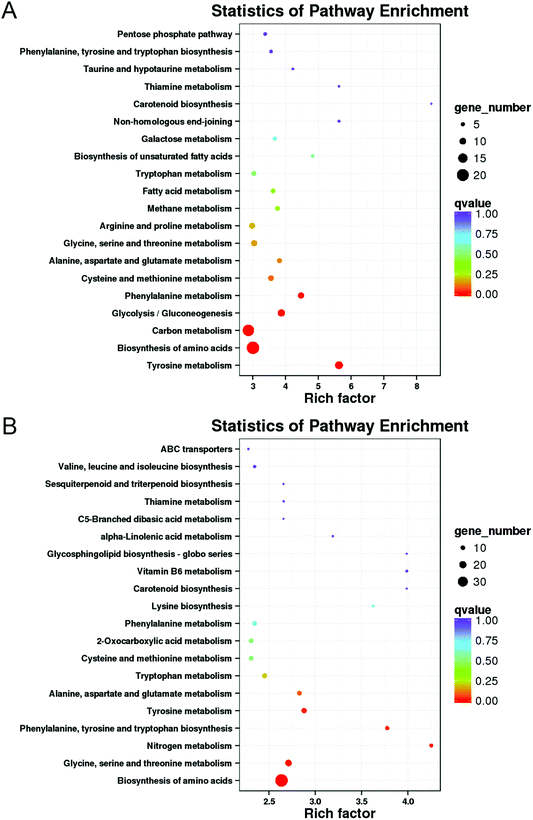 | ||
| Fig. 6 KEGG pathway enrichment of “M vs. ML” (A) and “M and MX” (B). The enrichment factor indicates the number of DEGs relative to the percentage of all annotated genes involved in the pathway. | ||
Expression profile of 61 DEGs in differential transcription levels involved in biosynthesis of amino acids and carbon metabolism pathway in ML and MX group compared to those in the M group (P < 0.01) (ESI Data S5†). Based on gene annotation, enzyme commission number (EC.number) and KEGG Orthology (KO), the transcription levels of gene1276, gene5012, gene6768, gene601, gene5806, gene1572, gene4323 involved in glycolysis/gluconeogenesis pathway in ML/MX group increased by 1.21/1.08, 1.41/0.89, 1.10/1.16, 3.46/2.97, 1.04/1.01, 1.23/0.77 and 1.09/0.78 log2FC, respectively, which encoded glucose-6-phosphate isomerase (EC.5.3.1.9), 6-phosphofructokinase (EC.2.7.1.1/2.7.1.11), glyceraldehyde 3-phosphate dehydrogenase (EC.1.2.1.12), phosphoglycerate kinase (EC.2.7.2.2), 2,3-bisphosphoglycerate-independent phosphoglycerate mutase (EC.5.4.2.12), enolase (EC.4.2.1.11), pyruvate kinase (EC.2.7.1.40), respectively. Depending on these genes enriched in glycolysis/gluconeogenesis pathway, ML/MX group converts starch and sucrose to pyruvate, which can produce more biosynthetic precursors of pigments. The transcription levels of three genes (gene1276, gene3064, gene5012) involved in pentose phosphate pathway increased in ML/MX group by 1.21/1.08, 1.22/1.70, 1.41/0.89 log2FC, respectively, which encoded glucose-6-phosphate isomerase (EC.5.3.1.9), gluconokinase (EC.2.7.1.12), 6-phosphofructokinase (EC.2.7.1.1/2.7.1.11), respectively. Depending on these genes enriched in pentose phosphate pathway, ML/MX group converts starch and sucrose to aromatic amino acid and NADPH, which can provide precursors of pigments and a reducing agent for pigment synthesis. The transcription levels of five genes (gene4426, gene517, gene1078, gene5124, gene5672) involved in the biosynthesis of amino acids pathway (such as glycine, serine and threonine metabolism, cysteine and methionine metabolism and phenylalanine, tyrosine and tryptophan biosynthesis) in ML/MX group increased by 2.69/3.83, 1.57/2.76, 0.95/2.66, 3.25/4.73, and 2.54/2.59 log2FC, respectively, which encoded O-acetylhomoserine/O-acetylserine sulfhydrylase (EC.2.5.1.49/2.5.1.47), aspartate aminotransferase (EC.2.6.1.1), phosphoserine aminotransferase (EC.2.6.1.52), tryptophan synthase (EC.4.2.1.20), L-serine dehydratase/L-threonine ammonia-lyase (EC.4.3.1.17/4.3.1.19), respectively. Depending on these genes enriched in amino acids pathway, ML/MX group converts serine and L-cysteine to tryptophan and pyruvate, which can produce more biosynthetic precursors of pigments.
Transcription factors play a critical role in the regulation of gene expression patterns that control the overall metabolism, including secondary metabolic pathways.34,35 Therefore, the expression levels of transcription factor genes in M. purpureus during pigment and citrinin production were analyzed. Based on Pfam annotation, 216 genes were identified as transcription factor-encoding candidates, among which 35 and 48 showed significantly different expression levels in the ML and MX group compared to those in M group (ESI Data S6†). Gene283 annotated as “citrinin biosynthesis transcriptional activator CtnR” in the ML/MX group were down-regulated with the log2FC value 1.18/1.50, respectively. While those related to pigment biosynthesis (such as gene4184 annotated as “Zn cluster transcription factor”, gene6903 annotated as “C6 transcription factor” or “Fungal Zn(2)–Cys(6) binuclear” in the ML/MX group, gene6670 annotated as “Fungal Zn(2)–Cys(6) binuclear” in the MX group) was up-regulated by 2.22/2.66, 1.17/2.20, 1.80 log2FC, respectively. This observation was inconsistent with the report that CtnA, which contains a typical Zn(II)2Cys6 DNA binding motif, functions as an activator of pksCT and orf5 and that its disruption leads to the reduction of citrinin production to barely detectable levels.36 Gene5724 encoding a C2H2-type zinc finger in the ML/MX group, which is a key activator of sexual development in Aspergillus nidulans, was up-regulated by 2.07/2.95 log2FC, respectively. Gene2534 (encoding a transcription factor AreA), a key transcription factor of nitrogen metabolism in filamentous fungi, was up-regulated by 0.81/1.36 log2FC in the ML/MX group, respectively. Gene1163 (encoding nitrogen assimilation transcription factor nit-4), a member of the GAL4 family of fungal transcription factors for nitrogen metabolism, was up-regulated by 1.33/1.10 log2FC in the ML/MX group, respectively.
3.7 RT-qPCR of specific genes related to the biosynthesis of MPs and citrinin
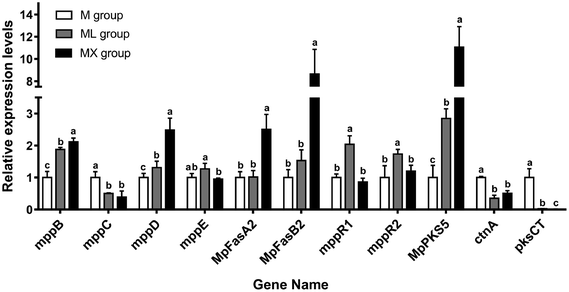
In order to verify the effect of the medium supplemented with NH4Cl or NH4NO3 on the yields of secondary metabolites of Monascus from the molecular level and the results of RNA-seq, the relative expression levels of the genes related to the biosynthesis of MPs and citrinin were also analyzed by RT-qPCR. The genes (mppB, mppC, mppD, mppE, MpFasA2, MpFasB2, mppR1, mppR2 and MpPKS5) related to the biosynthesis of MPs, and the genes (ctnA and pksCT) involved in the biosynthesis of citrinin were detected (Fig. 7). It was found that NH4NO3 treatment resulted in significant elevations in the mRNA levels of mppB, mppD, MpFasA2, MpFasB2 and MpPKS5, compared with the M group (P < 0.05). Moreover, NH4Cl treatment made significant elevations in the mRNA levels of mppB, mppD, mppR1, mppR2 and MpPKS5, compared with the M group (P < 0.05). Both NH4NO3 and NH4Cl treatment resulted in reductions in the mRNA levels of ctnA and pksCT, compared with the M group (P < 0.05). It was noted that the mRNA levels of mppC (the gene responsible for the conversion of orange pigment to yellow pigment) in ML and MX were reduced (P < 0.05). The mRNA levels of mppD (the gene responsible for the conversion of yellow pigment to orange pigment) in ML and MX were increased simultaneously.4. Discussion
Great efforts have been made to improve the fermentation conditions and medium ingredients to enhance the production of MPs in submerged fermentation.13,14 Although a large number of studies have been reported on the effects of different carbon and nitrogen sources on the production of MPs,19,20,37 the dual effects of inorganic nitrogen sources on the production of MPs and citrinin and their regulatory mechanisms are rarely reported. Previous studies have reported the use of RNA-seq technology to study the effects of blue light on Monascus and its related light-regulated gene regulatory networks.15 Some scholars have also explored the biosynthesis pathway of lovastatin in M. purpureus by RNA-seq technique.38 However, there are few studies on the effect of nitrogen sources on the production of MPs and citrinin with submerged fermentation by Monascus through RNA-seq analysis.Nitrogen source is a general term for nitrogen-containing compounds such as proteins and nucleic acids, which can not only affect the growth of mould, but also induce or inhibit the production of secondary metabolites in fungi. Previous studies stated that the best nitrogen sources used for good pigment production are NH4Cl, NH4NO3 and peptone.39,40 Therefore, in this study, we use NH4Cl, NH4NO3 and peptone as nitrogen sources to study their effects on the production of MPs and citrinin by liquid fermentation of Monascus. Compared with peptone, NH4Cl and NH4NO3 can significantly increase the yields of MPs and inhibit the production of citrinin. Previous studies have shown that organic nitrogen sources are more likely to promote the growth of Monascus than inorganic nitrogen source but unfavourable for pigment production, probably because when Monascus grows too fast, it affects the transmission of nutrients and affects the synthesis of secondary metabolites.41,42 When Monascus is grown with different nitrogen sources, the Monascus pigment components have different characteristics, which is consistent with previous studies when nitrogen source was ammonium nitrate, which significantly increased the production of red pigments, and when peptone was used as nitrogen source, the intracellular pigments was mainly yellow pigments, and a small amount of red pigments.14,41 Nitrogen sources have the same ammonium ion but different negative ions (such as NH4Cl and NH4NO3). Different nitrogen sources have different effects on growth and secondary metabolism.43 These results indicated that different ions may affect the biosynthesis of MPs or citrinin, and also reveal the complexity and specificity of the nitrogen source for the regulation of the biosynthesis of secondary metabolites from Monascus.
According to the previous research reported by Chen et al.,2,44 different Monascus pigments are produced by the same pigment intermediate through three synthetic pathways. Two red pigments (rubropunctamine and monascorubramine) are produced in a non-catalyzed affinity ammonia reaction by orange pigments (rubropunctatin and monascorubrin). The yellow pigments (monasfluore A, monasfluore B) and (monascin, ankaflavin) are competitively synthesized by two enzyme-catalyzed reaction pathways.2,14,44 In this study, the red pigment synthetic pathways were preferred in the ML and MX groups, which is consistent with the production of red pigments in Penicillium sp. AZ.45 Interestingly, the accumulation of yellow and orange pigments in Penicillium sp. AZ can be regulated by the restriction of inorganic ammonium salt in fermentation medium.45
To better understand the effects of NH4Cl and NH4NO3 on the production of MPs and citrinin by the liquid fermentation of Monascus, we used transcriptome sequencing to explore the differential gene expression of Monascus purpureus M3103. In filamentous fungi, the transcription factors AreA, AreB, MeaB, TamA and NmrA regulate the nitrogen source catabolism in combination.46,47 The nit-4 family transcription factor (gene1163) and the GATA family transcription factor AreA (gene2543) were detected in our transcription data. Nit-4, a member of the GAL4 family of fungal transcription factors, encodes a protein with an amino-terminal Cys6/Zn2 domain that provides sequence-specific DNA binding.46,48 AreA is considered as the major regulator of a large number of genes involved in uptake and metabolism of non-favored N sources in fungi,49 and acts as a positive regulator promoting the use of secondary nitrogen sources and participating in certain secondary metabolites of many clusters of many culture under nitrogen-limiting conditions.46,49 Compared with M group, the production of secondary metabolites of pigments and citrinin in ML and MX were significantly changed. And the utilization of secondary nitrogen sources were promote by upregulating AreA after the large consumption of ammonium nitrogen in the MX group. These results were consistent with other literature reports.49,50
The sequencing data of Monascus mycelium in NH4Cl group, NH4NO3 group and peptone group were compared. According to the KEGG pathway, nitrogen sources played an important role in the primary metabolism and secondary metabolism of Monascus. When NH4Cl was used as nitrogen source, amino acid metabolism (including glycine, serine and threonine metabolism), carbon metabolism (glycolysis/glyconeogenesis, pentose phosphate pathway), and lipid metabolism were increased, and phenylalanine metabolism and tyrosine metabolism were attenuated. However, when NH4NO3 was used as nitrogen source, nitrogen metabolism and amino acid metabolism (such as phenylalanine, tyrosine and tryptophan biosynthesis, glycine, serine and threonine metabolism, valine, leucine and isoleucine metabolism) were enhanced, and tyrosine metabolism was attenuated. Glycine plays an important role in cell division.51 Recent research has shown that serine is an important glycolysis fuel and a product of oxidative decarboxylation of glycine, providing a reducing power (NADH) and a carbon precursor for anabolism,52 and affects intracellular redox metabolic activity. Serine, glycine and threonine metabolism provided energy for the TCA cycle by pyruvate and acetyl-CoA.53 This indicated that NH4Cl as nitrogen source can enhance partial amino acid metabolism to provide energy for the growth of Monascus. It is worth noting that phenylalanine, tyrosine tryptophan are aromatic amino acids, which can be converted into several biological compounds (including 5-hydroxytryptamine, heteroauxin, dopamine, melanin, etc.) through a series of oxidation reactions and participate in the biological physiology and growth process,2 and which can produce the biosynthetic precursors of pigments. Valine, leucine and isoleucine are branched chain amino acids can be degrade to produce acetyl-CoA and methyl-CoA,54 which can be further catalyzed by polyketide synthase to form type II polyketides, the precursors of synthetic MPs.55 This indicated that both NH4NO3 and NH4Cl attenuated the growth ability of Monascus, but promoted the secondary metabolic process of Monascus.
Further, according to the data of the NR database, it was found that when NH4Cl was used as nitrogen source, the number of genes related to the polyketide synthase pathway and citrinin biosynthesis transcription factor were down-regulated. Similarly, such a phenomenon was found in Monascus with NH4NO3 as nitrogen source. This further confirmed that when using NH4NO3 or NH4Cl as nitrogen source, the yields of MPs were increased and the yield of citrinin was decreased. It is noteworthy that the relative expression levels of MpPKS5, mppR1, mppR2, mppB and mppD, were significantly up-regulated in the ML group compared with the M group. MpPKS5 and mppD were the key genes in the polyketide synthase (PKS) pathway of Monascus pigment synthesis. And knocking out MpPKS5 resulted in very low production of MPs.56 Amine oxidoreductase encoded by mppD was responsible for the conversion of yellow pigment to orange pigment. MppR1 was a specific transcription factor of Monascus with fungal Zn(2)–Cys(6) binuclear cluster domain, and ankyrin repeat protein encoded by mppR2 was an important regulator of MPs biosynthesis.57,58 MppB encoding fatty acyltransferase in the polyketide pathway producted pigments condensed by esterification: a short-chain fatty acid (C6 or C8) and a hexaketide synthesized.59 The up-regulation of these gene expression levels was consistent with the up-regulation of the yield of MPs. The expression level of pigment-related genes (mppC and mppD) were regulated by inorganic nitrogen sources to produce red pigment oxidized from yellow pigment and orange pigment. Moreover, there was no significant difference in the relative expression levels of genes MpFasA2 and MpFasB2 between ML group and M group, which indicated that the addition of NH4Cl did not change the expression of these genes in liquid fermentation. MpFasA2 and MpFasB2 encoded the synthesis of short-chain fatty acyl coenzyme A, which was the key gene in the fatty acid pathway of MPs.2 This study demonstrated that the addition of ammonium chloride in liquid fermentation stimulated the expression of genes related to the polyketide synthase (PKS) pathway in the synthesis of MPs, but did not significantly affect the expression of genes related to the fatty acid pathway. The relative expression levels of pksCT and ctnA were also significantly down-regulated. Both ctnA and pksCT were very important genes related to citrinin biosynthesis.60 The inhibitory effects of ctnA and pksCT on transcriptional expression of MX group were better than ML group, resulting in a dramatic decrease in citrinin production. These conclusions can be verified in previous NR database data and KEGG metabolic pathways.
Several novel genes involved in pigment and citrinin synthesis were found in our study (ESI Data S7†). Such as (newGene369, newGene370), (newGene371, newGene372, newGene_112, newGene281, newGene1307) and newGene_1228, annotated as non-reducing polyketide synthase, conidial yellow pigment biosynthesis polyketide synthase and polyketide synthase dehydratase, respectively, all of them may be constituted a gene cluster of polyketides. NewGene_227, newGene_979, newGene_1137, newGene_802, newGene_218 and newGene_219, annotated as pyruvate decarboxylase, pyruvate dehydrogenase, putative cytochrome p450 protein, cytochrome c oxidase subunit 7A, and FAD dependent oxidoreductase, may be involved in the biosynthesis, transport and catabolism of secondary metabolites. NewGene_185, newGene 339, newGene 510 and newGene1227, annotated as fatty acid synthase alpha subunit, 3-oxoacyl-[acyl-carrier-protein] synthase, 3-ketoacyl-CoA ketothiolase and zinc-binding dehydrogenase, may involved in lipid transport and metabolism. NewGene_623 and newGene1144 annotated as citrinin polyketide synthase and Zn(2)–Cys(6) binuclear cluster domain, were closely related to the citrinin biosynthesis.
5. Conclusion
In this study, the effects of inorganic nitrogen on the secondary metabolism and gene transcription of Monascus spp. in liquid fermentation were studied, which provides a better understanding of the links between primary and secondary metabolism in Monascus spp., and is expected to lay a foundation for future studies on the molecular mechanisms underlying the effects of different substances on secondary metabolism in Monascus spp. Transcriptome analysis highlighted differences not only in gene expression of pigments and citrinin biosynthesis related genes, but also shed light on regulatory effects on genes involved in other energetic metabolisms and biological processes, such as carbon and nitrogen metabolism, biosynthesis of amino acids and so on.Conflicts of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.Acknowledgements
This work was financially supported by National Natural Science Foundation of China (No. 31601466), International cooperation and exchange project of Fujian Agriculture and Forestry University (No. KXG15001A), Project of China Postdoctoral Science Foundation (2016T90591), Natural Science Foundation of Fujian Province in China (2016J01095), fund for outstanding young scientific talents of Fujian Agriculture and Forestry University of China (XJQ201607).References
- X. Lv, R. Jia, J. Chen and W. Zhou, et al., Evaluation of different PCR primers for denaturing gradient gel electrophoresis (DGGE) analysis of fungal community structure in traditional fermentation starters used for Hong Qu glutinous rice wine brewing, Int. J. Food Microbiol., 2017, 255, 58–65 CrossRef CAS PubMed.
- W. Chen, R. Chen, Q. Liu and Y. He, Orange, red, yellow: biosynthesis of azaphilone pigments in Monascus fungi, Chem. Sci., 2017, 8(7), 4917–4925 RSC.
- W. Chen, Y. He, Y. Zhou, Y. Shao, Y. Feng, M. Li and F. Chen, Edible filamentous fungi from the species Monascus: early traditional fermentations, modern molecular biology, and future genomics, Compr. Rev. Food Sci. Food Saf., 2015, 14(5), 555–567 CrossRef CAS.
- L. Dufossé, P. Galaup, A. Yaron, S. M. Arad, P. Blanc, K. N. C. Murthy and G. A. Ravishankar, Microorganisms and microalgae as sources of pigments for food use: a scientific oddity or an industrial reality?, Trends Food Sci. Technol., 2005, 16(9), 389–406 CrossRef.
- L. C. Hsu, Y. H. Liang, Y. W. Hsu, Y. H. Kuo and T. M. Pan, Anti-inflammatory properties of yellow and orange pigments from Monascus purpureus NTU 568, J. Agric. Food Chem., 2013, 61(11), 2796–2802 CrossRef CAS PubMed.
- W. Zhou, R. Guo, W. L. Guo and J. Hong, et al., Monascus yellow, red and orange pigments from red yeast rice ameliorate lipid metabolic disorders and gut microbiota dysbiosis in Wistar rats fed on a high-fat diet, Food Funct., 2019, 10(2), 1073–1084 RSC.
- C. L. Lee, Y. P. Hung, Y. W. Hsu and T. M. Pan, Monascin and ankaflavin have more anti-atherosclerosis effect and less side effect involving increasing creatinine phosphokinase activity than monacolin K under the same dosages, J. Agric. Food Chem., 2013, 61(1), 143–150 CrossRef CAS PubMed.
- W. Sang-Aroon, S. Saekow and V. Amornkitbamrung, Density functional theory study on the electronic structure of Monascus dyes as photosensitizer for dye-sensitized solar cells, J. Photochem. Photobiol., A, 2012, 236, 35–40 CrossRef CAS.
- L. Rumora, A. M. Domijan, T. Žanić Grubišić and M. Šegvić Klarić, Differential activation of MAPKs by individual and combined ochratoxin A and citrinin treatments in porcine kidney PK15 cells, Toxicon, 2014, 90, 174–183 CrossRef CAS PubMed.
- A. Ciegler, R. F. Vesonder and L. K. Jackson, Production and biological activity of patulin and citrinin from Penicillium expansum, Appl. Environ. Microbiol., 1977, 33(4), 1004–1006 CrossRef CAS PubMed.
- B. H. Lee and T. M. Pan, Benefit of Monascus-fermented products for hypertension prevention: a review, Appl. Microbiol. Biotechnol., 2012, 94(5), 1151–1161 CrossRef CAS PubMed.
- D. G. Pereira, A. Tonso and B. V. Kilikian, Effect of dissolved oxygen concentration on red pigment and citrinin production by Monascus purpureus ATCC 36928, Braz. J. Chem. Eng., 2008, 25(2), 247–253 CrossRef CAS.
- P. Velmurugan, H. Hur, V. Balachandar and S. Kamala-Kannan, Monascus pigment production by solid-state fermentation with corn cob substrate, J. Biosci. Bioeng., 2011, 112(6), 590–594 CrossRef CAS PubMed.
- K. Shi, D. Song, G. Chen, M. Pistolozzi, Z. Wu and L. Quan, Controlling composition and color characteristics of Monascus pigments by pH and nitrogen sources in submerged fermentation, J. Biosci. Bioeng., 2015, 120(2), 145–154 CrossRef CAS.
- D. Chen, M. Chen, S. Wu, Z. Li, H. Yang and C. Wang, The molecular mechanisms of Monascus purpureus M9 responses to blue light based on the transcriptome analysis, Sci. Rep., 2017, 7(1), 1–10 CrossRef PubMed.
- M. Patrovsky, K. Sinovska and B. Branska, Effect of initial pH, different nitrogen sources, and cultivation time on the production of yellow or orange Monascus purpureus pigments and the mycotoxin citrinin, Food Sci. Nutr., 2019, 7(11), 3494–3500 CrossRef CAS PubMed.
- E. I. Pisareva and A. V. Kujumdzieva, Influence of carbon and nitrogen sources on growth and pigment production by Monascus pilosus C1 strain, Biotechnol. Biotechnol. Equip., 2010, 24(suppl. 1), 501–506 CrossRef.
- H. C. Wong, Y. C. Lin and P. E. Koehler, Regulation of growth and pigmentation of Monascus purpureus by carbon and nitrogen concentrations, Mycologia, 1981, 73(4), 649 CrossRef CAS.
- C. Yang, X. Yang, Z. He, D. Lu, Z. Chen and L. He, Effects of carbon and nitrogen sources on Monascus purpureus S in its secreting pigment, Fujian J. Agric. Sci., 2011, 26(5), 832–836 Search PubMed.
- R. Vidyalakshmi, R. Paranthaman, S. Murugesh and K. Singaravadivel, Stimulation of Monascus pigments by intervention of different nitrogen sources, Global J. Biotechnol. Biochem., 2009, 4(1), 25–28 CAS.
- V. K. Joshi, D. Attri, A. Bala and S. Bhushan, Microbial Pigments, Indian J. Biotechnol., 2003, 2(3), 362–369 CAS.
- C. Agboyibor, W. B. Kong, D. Chen, A. M. Zhang and S. Q. Niu, Monascus pigments production, composition, bioactivity and its application: a review, Biocatal. Agric. Biotechnol., 2018, 16, 433–447 CrossRef.
- Y. He and R. J. Cox, The molecular steps of citrinin biosynthesis in fungi, Chem. Sci., 2016, 7(3), 2119–2127 RSC.
- D. Chen, C. Xue, M. Chen, S. Wu, Z. Li and C. Wang, Effects of blue light on pigment biosynthesis of Monascus, J. Microbiol., 2016, 54(4), 305–310 CrossRef CAS PubMed.
- B. Liang, X. J. Du, P. Li, C. C. Sun and S. Wang, Investigation of citrinin and pigment biosynthesis mechanisms in Monascus purpureus by transcriptomic analysis, Front. Microbiol., 2018, 9, 1374 CrossRef.
- W. Wang, Q. Chen, X. Zhang, H. Zhang, Q. Huang, D. Li and J. Yao, Comparison of extraction methods for analysis of citrinin in red fermented rice, Food Chem., 2014, 157, 408–412 CrossRef CAS PubMed.
- Y. Zheng, Y. Xin and Y. Guo, Study on the fingerprint profile of Monascus products with HPLC-FD, PAD and MS, Food Chem., 2009, 113(2), 705–711 CrossRef CAS.
- J. Jian, W. Wei, G. Yin, A. Hettinghouse, C. Liu and Y. Shi, RNA-seq analysis of interferon inducible p204-mediated network in anti-tumor immunity, Sci. Rep., 2018, 8(1), 1–10 CrossRef CAS PubMed.
- L. Florea, L. Song and S. L. Salzberg, Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues, F1000Research, 2013, 2(188), 1–24 Search PubMed.
- M. D. Young, M. J. Wakefield, G. K. Smyth and A. Oshlack, Gene ontology analysis for RNA-seq: accounting for selection bias, Genome Biol., 2010, 11(2), 1–12 CrossRef PubMed.
- M. Kanehisa, M. Araki, S. Goto and M. Hattori, KEGG for linking genomes to life and the environment, Nucleic Acids Res., 2008, 36, 480–484 CrossRef PubMed.
- X. Mao, T. Cai, J. G. Olyarchuk and L. Wei, Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary, Bioinformatics, 2005, 21(19), 3787–3793 CrossRef CAS PubMed.
- S. K. Schulze, R. Kanwar, M. Gölzenleuchter, T. M. Therneau and A. S. Beutler, SERE: single-parameter quality control and sample comparison for RNA-seq, BMC Genomics, 2012, 13(1), 7–9 CrossRef PubMed.
- E. Gutierrez, A. García-Villaraco, J. A. Lucas and A. Gradillas, Transcriptomics, targeted metabolomics and gene expression of blackberry leaves and fruits indicate flavonoid metabolic flux from leaf to red fruit, Frontiers in Plant Science, 2017, 8, 1–15 CrossRef PubMed.
- D. C. J. Wong and M. José Tomás, Constructing integrated networks for identifying new secondary metabolic pathway regulators in grapevine: recent applications and future opportunities, Frontiers in Plant Science, 2017, 8, 505 Search PubMed.
- T. Shimizu, H. Kinoshita and T. Nihira, Identification and in vivo functional analysis by gene disruption of ctnA, an activator gene involved in citrinin biosynthesis in Monascus purpureus, Appl. Environ. Microbiol., 2007, 73(16), 5097–5103 CrossRef CAS PubMed.
- Z. R. Huang, The regulation mechanisms of soluble starch and glycerol for production of azaphilone pigments in Monascus purpureus FAFU618 as revealed by comparative proteomic and transcriptional analyses, Food Res. Int., 2018, 106, 626–635 CrossRef CAS PubMed.
- C. Zhang, J. Liang, L. Yang, B. Sun and C. Wang, De novo RNA sequencing and transcriptome analysis of Monascus purpureus and analysis of key genes involved in monacolin K biosynthesis, PLoS One, 2017, 12(1), 1–19 Search PubMed.
- P. Juzlová, L. Martinkova and V. Kren, Secondary metabolites of the fungus Monascus: a review, J. Ind. Microbiol. Biotechnol., 1996, 16, 163–170 CrossRef.
- M. H. Chen and M. R. Johns, Effect of pH and nitrogen source on pigment production by Monascus purpureus, Appl. Microbiol. Biotechnol., 1993, 40, 132–138 CrossRef CAS.
- M. Carels and D. Shepherd, The effect of different nitrogen sources on pigment production and sporulation of Monascus species in submerged, shaken culture, Can. J. Microbiol., 1977, 23(10), 1360–1372 CrossRef CAS PubMed.
- L. Pastrana, P. J. Blanc, A. L. Santerre, M. O. Loret and G. Goma, Production of red pigments by Monascus ruber in synthetic media with a strictly controlled nitrogen source, Process Biochem., 1995, 30(4), 333–341 CrossRef CAS.
- B. Wang, X. Han, Y. Bai, Z. Lin and M. Qiu, Effects of nitrogen metabolism on growth and aflatoxin biosynthesis in Aspergillus flavus, J. Hazard. Mater., 2017, 324, 691–700 CrossRef CAS PubMed.
- W. P. Chen, Y. L. Feng and I. Molnár, et al., Nature and nurture: confluence of pathway determinism with metabolic and chemical serendipity diversifies Monascus azaphilone pigments, Nat. Prod. Rep., 2019, 36(4), 561–572 RSC.
- J. Ogihara and K. Oishi, Effect of ammonium nitrate on the production of pp-v and monascorubrin homologues by Penicillium sp. az, J. Biosci. Bioeng., 2002, 93(1), 54–59 CrossRef CAS PubMed.
- B. Tudzynski, Nitrogen regulation of fungal secondary metabolism in fungi, Front. Microbiol., 2014, 5, 656 Search PubMed.
- Z. Lin, J. Wang, Y. Bao, Q. Guo, C. A. Powell and S. Xu, et al., Deciphering the transcriptomic response of fusarium verticillioides in relation to nitrogen availability and the development of sugarcane pokkah boeng disease, Sci. Rep., 2016, 6(1), 29692 CrossRef CAS PubMed.
- G. F. Yuan and G. A. Marzluf, Transformants of Neurospora crassa with the nit-4 nitrogen regulatory gene: copy number, growth rate and enzyme activity, Curr. Genet., 1992, 22(3), 205–211 CrossRef CAS PubMed.
- P. Andreas, L. Johannes and S. Lena, Comparative transcriptome and proteome analysis reveals a global impact of the nitrogen regulators AreA and AreB on secondary metabolism in Fusarium fujikuroi, PLoS One, 2017, 12(4), e0176194 CrossRef PubMed.
- M. Mihlan, V. Homann and T. W. D. Liu, AreA directly mediates nitrogen regulation of gibberellin biosynthesis in Gibberella fujikuroi, but its activity is not affected by NMR, Mol. Microbiol., 2003, 47(4), 975–991 CrossRef CAS PubMed.
- M. Jain, R. Nilsson, S. Sharma and N. Madhusudhan, Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation, Science, 2012, 336(6084), 1040–1044 CrossRef CAS.
- N. E. Flynn, M. E. Patyrak, J. B. Seely and G. Wu, Glycine oxidation and conversion into amino acids in Saccharomyces cerevisiae and Candida albicans, Amino Acids, 2010, 39(2), 605–608 CrossRef CAS PubMed.
- J. z. Ye, X. m. Lin, Z. x. Cheng, Y. b. Su and W. x. Li, Identification and efficacy of glycine, serine and threonine metabolism in potentiating kanamycin-mediated killing of Edwardsiella piscicida, J. Proteomics, 2018, 183(2017), 34–44 CrossRef CAS PubMed.
- G. Reazin, H. Scales and A. Andreasen, Mechanism of major congener formation in alcoholic grain fermentations, J. Agric. Food Chem., 1970, 18(4), 585–589 CrossRef CAS.
- H. Hajjaj, A. Klaébé, M. O. Loret, G. Goma, P. J. Blanc and J. François, Biosynthetic Pathway of Citrinin in the Filamentous Fungus Monascus Ruber as Revealed by 13C Nuclear Magnetic Resonance, Appl. Environ. Microbiol., 1999, 65(1), 311–314 CrossRef CAS PubMed.
- C. H. Lin, T. H. Lin and T. M. Pan, Alleviation of metabolic syndrome by monascin and ankaflavin: the perspective of Monascus functional foods, Food Funct., 2017, 8(6), 2102–2109 RSC.
- Y. Shao, M. Lei, Z. Mao, Y. Zhou and F. Chen, Insights into Monascus biology at the genetic level, Appl. Microbiol. Biotechnol., 2014, 98(9), 3911–3922 CrossRef CAS PubMed.
- B. Balakrishnan, S. Karki and S. H. Chiu, Genetic localization and in vivo characterization of a Monascus azaphilone pigment biosynthetic gene cluster, Appl. Microbiol. Biotechnol., 2013, 97(14), 6337–6345 CrossRef CAS PubMed.
- H. Hajjaj, P. Blanc, E. Groussac, J. L. Uribelarrea, G. Goma and P. Loubiere, Kinetic analysis of red pigment and citrinin production by Monascus ruber as a function of organic acid accumulation, Enzyme Microb. Technol., 2000, 27(8), 619–625 CrossRef CAS PubMed.
- Y. Wan, J. Zhang, H. Han, L. Li, Y. Liu and M. Gao, Citrinin-producing capacity of Monascus purpureus in response to low – frequency magnetic fields, Process Biochem., 2017, 53, 25–29 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra09760k |
| ‡ Co-first authors: Jia-Li Hong and Li Wu contributed equally to this study. |
| This journal is © The Royal Society of Chemistry 2020 |