Open Access Article
Open Access ArticleA ligation-triggered and protein-assisted fluorescence anisotropy amplification platform for sensitive and selective detection of small molecules in a biological matrix†
Meizi Chen‡
*a,
Bing Wan‡b,
Wei Duc,
Hongbo Hua,
Long Zenga,
Xintong Duana,
Jia Liua,
Zixiang Weia,
Li Tange and
Yongbo Peng *ad
*ad
aDepartment of General Internal Medicine, The First People's Hospital of Chenzhou, Chenzhou 423000, P. R. China. E-mail: chenmeizi11@163.com; pengyongbo2000@126.com
bDepartment of Respiratory and Critical Care Medicine, The Affiliated Jiangning Hospital of Nanjing Medical University, Nanjing 211100, P. R. China
cDepartment of Cardiology, The First People's Hospital of Chenzhou, Chenzhou 423000, Hunan, P. R. China
dInstitute of Chinese Medicine, Hunan Academy of Chinese Medicine, Hunan University of Chinese Medicine, Changsha 410208, P. R. China
eRadiation Oncology Center, Chongqing University Cancer Hospital/Chongqing Cancer Institute, Chongqing 400030, P. R. China
First published on 8th June 2020
Abstract
Effective detection of biomolecules is important for biological research and medical diagnosis. We here propose a ligation-triggered and protein-assisted fluorescence anisotropy amplification platform for sensitive and selective detection of small biomolecules in a complex biological matrix. In the proposed method, in the presence of target small molecules, FAM-labeled DNA 1 and biotin-labeled DNA2 were ligated to produce an integrated DNA. As a result, taking advantage of the extraordinary strong interaction between biotin and streptavidin, we employed a novel mass amplification strategy for sensitive detection of small molecules through fluorescence anisotropy. The method could detect ATP from 0.05 to 1 μM, with a detection limit of 41 nM, and detect NAD+ from 0.01 to 1 μM, with a detection limit of 6.7 nM. Furthermore, ligase-specific dependence of different cofactors provides good selectivity for the detection platform. As a result, the new platform has a broad spectrum of applications both in bioanalysis and biomedical fields.
1. Introduction
Simple, fast and direct monitoring of small biomolecules (for example, adenosine triphosphate (ATP), nicotinamide adenine dinucleotide (NAD+), etc.) is important for biological study, clinical diagnosis and forensic investigations.1–4 Currently, fluorescence-based sensing systems have been widely used to detect biomolecules with high sensitivity and selectivity.5–8 Although these approaches are sensitive and accurate, they still face challenges in practical applications. For example, fluorescence-based sensing systems require precise conformational change of the probe molecule upon target binding to correctly report the interaction. Second, methods like FRET or fluorescence quenching need two or more dyes for labeling to report target molecules. Moreover, the aforementioned approaches require sophisticated/expensive instruments, qualified experimenters and time-consuming manipulations.1 More importantly, traditional fluorescence-based detection methods are challenged by background fluorescence signals from complex biological matrixes, which would mask report signals and influence analytical results.9As an alternative signal output method, fluorescence anisotropy (FA) is a reliable choice to circumvent the above problems. FA is related to the phenomenon that upon excitation with polarized light, it is sensitive to the changes of molecular size or molecular weight. Since FA only requires single dye labelling, and does not need complex probe design, it has been widely used for study of interactions between different molecules. Meanwhile, as a ratiometric approach, the fluorescence intensity of a fluorophore is measured by two axes: horizontal (IH) and vertical (IV). As the polarization value (P) is defined by (IV − IH)/(IV + IH), it measures the orientation of the fluorescence emission (i.e., horizontal, vertical) rather than the fluorophore concentration. As a result, FP is minimally affected by solution opacity or color, which influences the intensity rather than the orientation of the fluorescence.10,11 Although FA is not sensitive to small molecules for their poor molecular mass, FA has also been successfully developed for detection of a series of important small molecules such as ATP, adenosine, cocaine, ochratoxin A, and L-tyrosinamide. Such as, Yang's group developed an anisotropy aptamer probes-based molecular mass amplifying for detection of small molecules. In their design, probes were activated to bind to anisotropy amplifiers upon binding with the target. Consequently, the formation of the probe/target complex considerably increased the molecular mass and FA value.12 Based on this strategy, a series of aptamer structure switch FA methods using high-mass or macromolecules (e.g. nanomaterials, proteins)-assisted design and dye-labeled functional nucleic acid probes have been proposed for FA assay of small molecules.12–21
While these reports make possible to construct fluorescence anisotropy sensor for sensing small molecules in biological matrix, unfortunately, limited by the aptamer's inherent affinity, some critical limits still need be solved. First, while the dissociation constant (Kd) of aptamer to small molecules were micromolar range, most of them exhibit poor sensitivity;22 second, part of the aptamers have weak selectivity to distinguish analogues of the target small molecules, take the anti-ATP aptamer as an example, it can both binding with ATP and adenosine, as a result, the reported ATP aptasensors cannot distinguish ATP from its analogues, such as adenosine, adenosine monophosphate (AMP), and adenosine diphosphate (ADP);23 third, although it was claimed that all the targets can get their matched aptamer, there still some important biological small molecules such as nicotinamide adenine dinucleotide (NAD+) need non-aptamer based detection method.
In this work, based on ligation-triggered and protein-assisted mass amplification, we constructed a fluorescence anisotropy sensing platform for sensitive and selective small biomolecules detection in complex biological matrix. Take ATP and NAD+ as two model molecules, we demonstrated the sensing performance by the proposed platform. The results indicated that our FA platform can achieve high sensitive detection of ATP (LOD: 41 nM) and NAD+ (LOD: 6.7 nM). Meanwhile, the ligase-triggered scheme can effectively distinguish the target from its analogues, exhibiting excellent selectivity. More importantly, because FA is less affected by complex matrix, ATP and NAD+ were directly detected in cell media. All the results indicated that the new FA platform has a wide applications for sensitive, rapid, and selective detection of small biomolecules in complex biological matrix.
2. Experimental section
2.1 Materials
Streptavidin, ATP, ADP, AMP, adenosine, CTP, GTP, UTP, NAD+, NADH, NADP, and NADPH were purchased from Sigma-Aldrich (Shanghai, China). Other reagents were purchased from Shanghai Chemical Reagent Corporation (Shanghai, China). DNA oligonucleotides were synthesized from Takara Biotechnology Co. Ltd. (Dalian, China) and the sequences were listed in Table 1. For DNA2, 5′ 6-FAM (fluorescein) was labeled on it by the DNA synthesizer directly.| Name | Sequence (5′-3′) |
|---|---|
| DNA1 | Biotin-GTG ACA TGC CGA |
| DNA2 | PO4−-ACC GAC GTC ACA-FAM |
| C-DNA | GGA TGC GGT TGT GAC GTC GGT TCG GCA TGT CAC GCG CGT |
| Invasive DNA | ACG CGC GTG ACA TGC CGA ACC GAC GTC ACA ACC GCA TCC |
2.2 Fluorescence measurements
Fluorescence measurements were carried out on a FluoroMax-4 Spectrofluorometer using the L-format configuration (Horiba Jobin Yvon, Paris, France). Excitation was set as 490 nm and emission was set as 520 nm. The fluorescence anisotropy value was tested from the Spectrofluorometer.2.3 Experimental procedure
20 μL ligase reaction buffer containing 150 nM DNA1, 150 nM DNA2 and 100 nM C-DNA was first incubated at 95 °C for 5 min and then cooled to room temperature. Subsequently, 2 U DNA ligase (T4) and different concentration of ATP were added to the buffer to initiate the ligation reaction at 25 °C for 0.5 h. Then the buffer was heated at 85 °C for 15 min to stop the reaction. Subsequently, 300 nM of invasive DNA were added to replace the ligated DNA 1 and DNA 2, and the sample was diluted to 100 μL and added 10 μg mL−1 SA and incubated 10 min for the following anisotropy assays. The NAD+ assay was similar to ATP assay by using 3 U E. coli DNA ligase.3. Results and discussion
3.1 Detection mechanism
Based on the literatures,24 fluorescein is the most popular fluorophores in FA assays for several reasons, including excellent spectral properties (such as high quantum yield of 92% and fluorescent lifetime of 4.05 ns), chemical stability and adaptability, relative inexpensiveness and commercial availability. As a result, 5′-6-FAM fluorescein was used in our method. The working principle is shown in Fig. 1. Take the ATP as the model target, in the T4 DNA ligase reaction system, after addition of ATP (A), FAM-labeled DNA 1 and biotin-labeled DNA2 were ligated to produce an integrated DNA by T4 DNA ligase, and the ligated DNA was then integrated with streptavidin by the biotin-SA binding. As a result, a FAM-biotin/SA complex will increase the mass of the complex, thus get an increased FA signal. As a control, in the absence of ATP (B), DNA 1 and DNA2 cannot be ligated to produce an integrated DNA, with the introduction of invasive, two fragments can be liberated from template C-DNA and the fluorophore-labelled DNA1 was unconnected with the biotin-labelled DNA2, thus get a weak FA signal.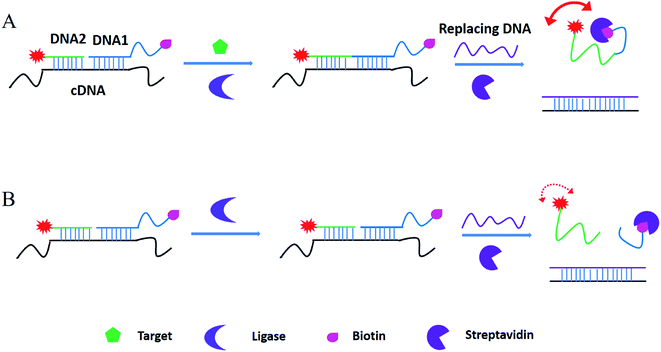 | ||
| Fig. 1 Working principle of ligation-triggered and protein-assisted fluorescence anisotropy sensing platform. | ||
3.2 Feasibility analysis
According to the modified Perrin eqn (1), the FA value of a rotating molecule is proportional to the viscosity of solvent (η), size (![[small nu, Greek, macron]](https://www.rsc.org/images/entities/i_char_e0ce.gif) ) and molecular mass (Mr) of the molecule.
) and molecular mass (Mr) of the molecule.
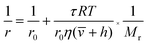 | (1) |
![[small nu, Greek, macron]](https://www.rsc.org/images/entities/i_char_e0ce.gif) is the specific volume of the molecule, and h is the hydration radius, T is the temperature in K, R is the molar gas constant (8.31 J mol−1 K−1), and η is the viscosity in poise (P). When the changes of R, T, η are negligible, the FA value is proportional to molecular mass (Mr) or volume, and is inversely proportional to fluorescence lifetime (τ).25 To demonstrate the concept, we first compare the performance of the sensing system with and without the amplifier: SA. The molecular mass of DNA2 (5′-P-ACC GAC GTC ACA-FAM-3′) is 4249, after ligated to DNA1, the molecular mass of integrated DNA (5′-Biotin-GTG ACA TGC CGA ACC GAC GTC ACA-FAM-3′) is 8383, causing a detectable si‡gnal output for 1.97-fold mass increase. In order to further improve the sensitivity, SA with a high molecular mass (53
is the specific volume of the molecule, and h is the hydration radius, T is the temperature in K, R is the molar gas constant (8.31 J mol−1 K−1), and η is the viscosity in poise (P). When the changes of R, T, η are negligible, the FA value is proportional to molecular mass (Mr) or volume, and is inversely proportional to fluorescence lifetime (τ).25 To demonstrate the concept, we first compare the performance of the sensing system with and without the amplifier: SA. The molecular mass of DNA2 (5′-P-ACC GAC GTC ACA-FAM-3′) is 4249, after ligated to DNA1, the molecular mass of integrated DNA (5′-Biotin-GTG ACA TGC CGA ACC GAC GTC ACA-FAM-3′) is 8383, causing a detectable si‡gnal output for 1.97-fold mass increase. In order to further improve the sensitivity, SA with a high molecular mass (53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) was chosen as the amplifier protein, as a consequence, after the SA binding with biotin-labelled integrated DNA, the mass of the probe can increase from 4249 to 57
000) was chosen as the amplifier protein, as a consequence, after the SA binding with biotin-labelled integrated DNA, the mass of the probe can increase from 4249 to 57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 249, generating 13-fold mass change
249, generating 13-fold mass change 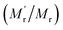 , which would cause a significantly increase of the FA value. We then studied the influence of SA on fluorescence lifetime and fluorescence intensity change. As shown in Fig. S1A of the ESI,† the lifetime increased from 4.1 ns to 6.9 ns and the lifetime ratio (τ′/τ) was 1.7. For fluorescence intensity, it has a weak increase (91 to 99). Taking the molecular mass ratio
, which would cause a significantly increase of the FA value. We then studied the influence of SA on fluorescence lifetime and fluorescence intensity change. As shown in Fig. S1A of the ESI,† the lifetime increased from 4.1 ns to 6.9 ns and the lifetime ratio (τ′/τ) was 1.7. For fluorescence intensity, it has a weak increase (91 to 99). Taking the molecular mass ratio 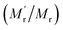 as 13, and the lifetime ratio (τ′/τ) as 1.7, while the fluorescence intensity change and other factors were negligible, the method could obtain a 7.6-fold FA signal increase
as 13, and the lifetime ratio (τ′/τ) as 1.7, while the fluorescence intensity change and other factors were negligible, the method could obtain a 7.6-fold FA signal increase 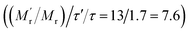 .
.
To validate whether the experimental result was consistent with the aforementioned theoretical calculation, we performed a titration experiment. As shown in Fig. 2, in the presence of SA, the FA value increased dramatically with the concentration of ATP from 0, 0.1 to 10 μM. As a comparison, in the absence of SA, the FA value was too weak to detect ATP with the concentration of 0.1 μM, and even the concentrations of ATP increased from 1 to 10 μM, there was still slight signal increase, confirming that the protein-assisted signal amplification can improve the FA-based detection sensitivity for ATP assay.
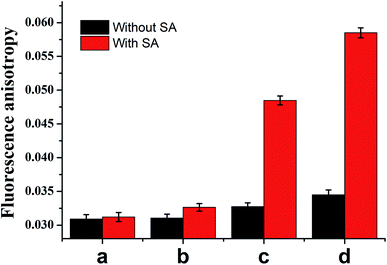 | ||
| Fig. 2 Comparison of FA value with (red column) and without (black column) streptavidin after addition different concentrations of ATP, the ATP is 0 (a), 0.1 (b), 1 (c) and 10 μM respectively. | ||
On this basis, we also optimized the experiment parameters, containing the ratio of DNA 1/DNA 2 to C-DNA, the amount of T4 DNA ligase, SA and reaction time. As shown in Fig. S2 of the ESI,† 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 of DNA 1/DNA 2 to C-DNA (A), 2 U T4 DNA ligase (B), 10 μg mL−1 SA (C), and 30 min reaction time (D) were chosen respectively.
1 of DNA 1/DNA 2 to C-DNA (A), 2 U T4 DNA ligase (B), 10 μg mL−1 SA (C), and 30 min reaction time (D) were chosen respectively.
3.3 Detection sensitivity and selectivity
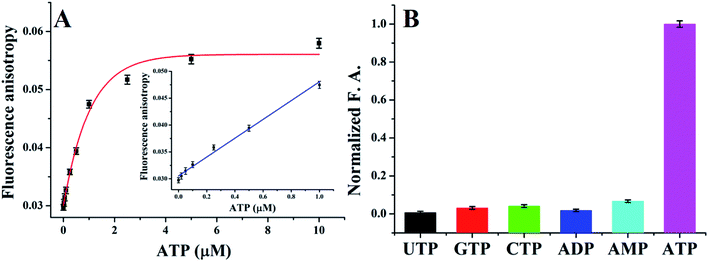
Fig. 3A plots the FA value with various concentrations of ATP, as the ATP concentration increased, the FA value increased accordingly. A linear relationship between the FA value and the ATP concentration was observed in the range of 0.05–1 μM (y = 0.0305 + 0.0175x, R2 = 0.9848, insert of Fig. 3A). A 41 nM detection limit was calculated, which is 2–3 orders of magnitude more sensitive than many fluorescence or FA-based aptasensors (see Table 3).Currently, aptasensor has been successfully developed for FA sensing of ATP,15,26–29 however, the anti-ATP aptamer has no selectivity to distinguish analogues such as ADP, AMP, and adenosine while they have the same aptamer.23 Fortunately, as a source of energy, ATP is easily distinguished from AMP, ADP, and adenosine when used in ligase reaction. In order to test the selectivity of T4 DNA ligase, a variety of ATP analogs, including ADP, AMP, CTP, GTP and UTP, were used as controls, as shown in Fig. 3B, no distinct FA value changes were observed, demonstrating an excellent selectivity of the presented method.
3.4 Detection in complex biological samples
The previous data indicated that the new platform has excellent performance in laboratory buffer system. However, when used in biological samples, the detection platform needs to tolerate matrix interferences. To test the performance of the presented FA platform, in this work, take cell media as a model biological samples, we test the performance of the new platform. As listed in Table 2, different concentrations of ATP were spiked in the cell media buffer containing the optimal probes, the data reveals that the recoveries are distributed between 92.5% and 105.3%, suggesting the presented platform has excellent assay performance for ATP detection in biological matrix.| Sample | Added (nM) | Founded (nM) | Recovery (%) |
|---|---|---|---|
| 1 | 50 | 52.4 | 104.8 |
| 2 | 100 | 105.3 | 105.3 |
| 3 | 200 | 185.0 | 92.5 |
| 4 | 500 | 483.0 | 96.6 |
| 5 | 1000 | 991.3 | 99.1 |
3.5 Generality
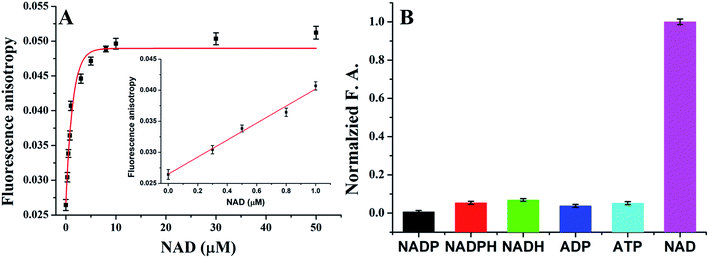
Thus far, we have demonstrated that the presented platform has excellent detection performance for ATP detection in biological matrix. Because different ligase used different cofactors, replace T4 DNA ligase with other ligases, the platform should be also used for detection other biomolecules. In order to test the generality of the platform, we test the performance for NAD+ detection. After getting optimized reaction parameters, we investigated the sensitivity and selectivity for detection of NAD+. As shown in Fig. 4A, as increase the concentrations of NAD+, the fluorescence anisotropy increased from 0.02644 to 0.05122 in the range of 0 to 50 μM. A linear relationship between the FA value and the NAD+ concentration within the range of 0.01 to 1 μM (y = 0.0265 + 0.0137x, R2 = 0.9786), and a limit of detection of 6.7 nM was calculated.As described in the previous, the NAD+ detection was also found to be highly selective. A series of NAD+ analogs including NADP, NADPH, NADH, ADP, and ATP were employed as controls to test the detection selectivity. As shown in Fig. 4B, there were no distinct FA value changes observed, indicating an excellent selectivity of the method. Therefore, the sensing universality of this strategy is well proved.
4. Conclusions
In conclusions, based on the small molecules-dependent ligation reaction and biotin-SA amplifier, we have proposed a ligation-triggered and protein-assisted fluorescence anisotropy amplification platform for sensitive and selective detection of biomolecules in biological matrix. The proposed method can detect ATP and NAD+ with concentration as low as 41 and 6.7 nM respectively. Meanwhile, for the DNA ligation reaction has high specific biomolecule-dependence, the work exhibits high selectivity toward the target cofactor from its analogues. As listed in Table 3, compared with aptamer-based methods, the new method exhibited both the high sensitivity and excellent selectivity. Meanwhile, because fluorescence anisotropy cannot be affected by matrix interferences, the biomolecules can be detected directly in biological matrix. Based on the DNA ligation reaction sensing systems, the mass amplified fluorescence anisotropy platform may find a broad spectrum of applications in both bioanalysis and biomedical fields.| Materials used/signal output | Target/detection limit | Selectivity differentiation of target and it's analogues | Ref. |
|---|---|---|---|
| Ligase and PCR-assisted signal amplification/fluorescence | ATP/100 pM | Yes | 8 |
| NAD+/10 pM | |||
| Ligase and DNAzyme-assisted signal/amplification fluorescence | ATP/100 pM | Yes | 7 |
| NAD+/50 pM | |||
| Aptamer and HCR-assisted signal amplification/FA | ATP/0.5 μM | No | 16 |
| Aptamer and protein-assisted signal amplification/FA | ATP/0.5 μM | No | 12 |
| Aptamer and protein-assisted signal amplification/FA | ATP/0.5 μM | No | 22 |
| Aptamer, nuclease and protein-assisted signal amplification/FA | Adenosine/0.5 μM | No | 30 |
| Ligase and protein-assisted signal amplification/FA | ATP/36 nM | Yes | This work |
| NAD+/6.7 nM |
Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
This work was supported by Hunan Natural Science Foundation Youth Fund Project (2019JJ50021); Key Research and Development Project of Hunan Province (2019 NK2101); Natural science Foundation of Jiangsu Province (BK20191351), the Natural Science Foundation Project of CQ CSTC (cstc2017jxjl130015, cstc2018jcyjAX0460).References
- P. B. Dennis, A. Jaeschke, M. Saitoh, B. Fowler, S. C. Kozma and G. Thomas, Science, 2001, 294, 1102–1105 CrossRef CAS PubMed.
- A. Wilkinson, J. Day and R. Bowater, Mol. Microbiol., 2001, 40, 1241–1248 CrossRef CAS PubMed.
- Q. Zhang, D. W. Piston and R. H. Goodman, Science, 2002, 295, 1895–1897 CAS.
- H. Wang, Z. Feng, Y. Qin, J. Wang and B. Xu, Angew. Chem., 2018, 57, 4931–4935 CrossRef CAS PubMed.
- M. H. Lee, J. S. Kim and J. L. Sessler, Chem. Soc. Rev., 2015, 44, 4185–4191 RSC.
- W. Li, C. X. Yang and X. P. Yan, Chem. Commun., 2017, 53, 11469–11471 RSC.
- L. M. Lu, X. B. Zhang, R. M. Kong, B. Yang and W. Tan, J. Am. Chem. Soc., 2011, 133, 11686–11691 CrossRef CAS PubMed.
- X. Zhang, Y. Wang and X. Zhou, Anal. Chem., 2019, 91, 1665–1670 CrossRef CAS PubMed.
- J. Liu, Z. Cao and Y. Lu, Chem. Rev., 2009, 109, 1948–1998 CrossRef CAS PubMed.
- D. S. Smith and S. A. Eremin, Anal. Bioanal. Chem., 2008, 391, 1499–1507 CrossRef CAS PubMed.
- D. M. Jameson and J. A. Ross, Chem. Rev., 2010, 110, 2685–2708 CrossRef CAS PubMed.
- L. Cui, Y. Zou, N. Lin, Z. Zhu, G. Jenkins and C. J. Yang, Anal. Chem., 2012, 84, 5535–5541 CrossRef CAS PubMed.
- J. Ruta, S. Perrier, C. Ravelet, J. Fize and E. Peyrin, Anal. Chem., 2009, 81, 7468–7473 CrossRef CAS PubMed.
- Y. Li, L. Sun and Q. Zhao, Talanta, 2017, 174, 7–13 CrossRef CAS PubMed.
- S. Perrier, V. Guieu, B. Chovelon, C. Ravelet and E. Peyrin, Anal. Chem., 2018, 90, 4236–4248 CrossRef CAS PubMed.
- B. Yang, X. B. Zhang, L. P. Kang, G. L. Shen, R. Q. Yu and W. Tan, Anal. Chem., 2013, 85, 11518–11523 CrossRef CAS PubMed.
- H. Ye, N. Duan, H. Gu, H. Wang and Z. Wang, Mikrochim. Acta, 2019, 186, 173 CrossRef PubMed.
- Y. X. Jiang, J. N. Tian, K. Hu, Y. C. Zhao and S. L. Zhao, Microchim. Acta, 2014, 181, 1423–1430 CrossRef CAS.
- J. A. Cruz-Aguado and G. Penner, Anal. Chem., 2008, 80, 8853–8855 CrossRef CAS PubMed.
- Z. Zhu, T. Schmidt, M. Mahrous, V. Guieu, S. Perrier, C. Ravelet and E. Peyrin, Anal. Chim. Acta, 2011, 707, 191–196 CrossRef CAS PubMed.
- D. Q. Qiao, J. G. Xu, P. Z. Qin, L. Yao, J. F. Lu, S. Eremin and W. Chen, Anal. Chem., 2018, 90, 7171–7175 CrossRef CAS PubMed.
- Y. Li and Q. Zhao, Anal. Chem., 2019, 91, 7379–7384 CrossRef CAS PubMed.
- D. E. Huizenga and J. W. Szostak, Biochemistry, 1995, 34, 656–665 CrossRef CAS PubMed.
- H. Y. Zhang, S. P. Yang, K. De Ruyck, N. V. Beloglazova, S. A. Eremin, S. De Saeger, S. X. Zhang, J. Z. Shen and Z. H. Wang, Trac. Trends Anal. Chem., 2019, 114, 293–313 CrossRef CAS.
- Y. Li, Y. Sun, J. Y. Ye, F. P. Pan, B. Peng, H. Q. Li, M. M. Zhang and Y. Xu, Anal. Methods, 2020, 12, 687–692 RSC.
- S. Perrier, C. Ravelet, V. Guieu, J. Fize, B. Roy, C. Perigaud and E. Peyrin, Biosens. Bioelectron., 2010, 25, 1652–1657 CrossRef CAS PubMed.
- Z. Zhu, C. Ravelet, S. Perrier, V. Guieu, E. Fiore and E. Peyrin, Anal. Chem., 2012, 84, 7203–7211 CrossRef CAS PubMed.
- J. Liu, C. Wang, Y. Jiang, Y. Hu, J. Li, S. Yang, Y. Li, R. Yang, W. Tan and C. Z. Huang, Anal. Chem., 2013, 85, 1424–1430 CrossRef CAS PubMed.
- J. N. Tian, Y. Wang, S. Chen, Y. X. Jiang, Y. C. Zhao and S. L. Zhao, Microchim. Acta, 2013, 180, 203–209 CrossRef CAS.
- L. P. Kang, B. Yang, X. B. Zhang, L. Cui, H. M. Meng, L. Mei, C. C. Wu, S. L. Ren and W. H. Tan, Anal. Chim. Acta, 2015, 879, 91–96 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra09621c |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |