Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synthesis of new piperazinyl-pyrrolo[1,2-a]quinoxaline derivatives as inhibitors of Candida albicans multidrug transporters by a Buchwald–Hartwig cross-coupling reaction†
Jean Guillon‡
a,
Shweta Nim‡b,
Stéphane Moreaua,
Luisa Rongaa,
Solène Savrimoutoua,
Elisabeth Thiveta,
Mathieu Marchiviec,
Attilio Di Pietrod,
Rajendra Prasade and
Marc Le Borgne *f
*f
aUniv. Bordeaux, INSERM U1212 – UMR CNRS 5320, ARNA Laboratory, UFR des Sciences Pharmaceutiques, F-33076 Bordeaux Cedex, France
bSchool of Life Sciences, Jawaharlal Nehru University, 110067 New Delhi, India
cCNRS, Univ. Bordeaux, Bordeaux INP, ICMCB, UMR 5026, F-33608 Pessac Cedex, France
dDRMP Group, IBCP, UMR 5086 (MMSB), CNRS/Lyon I University, 69367 Lyon, France
eAmity Institute of Integrative Sciences and Health, Amity University, Education Valley, Gurgaon 122413, India
fUniversité de Lyon, Université Claude Bernard Lyon 1, Faculté de Pharmacie – ISPB, EA 4446 Bioactive Molecules and Medicinal Chemistry, SFR Santé Lyon-Est CNRS UMS3453 – INSERM US7, Lyon, France. E-mail: marc.le-borgne@univ-lyon1.fr
First published on 15th January 2020
Abstract
Two series of piperazinyl-pyrrolo[1,2-a]quinoxaline derivatives were prepared via a Buchwald–Hartwig cross-coupling reaction and then evaluated for their ability to inhibit the drug efflux activity of CaCdr1p and CaMdr1p transporters of Candida albicans overexpressed in a Saccharomyces cerevisiae strain. In the initial screening of twenty-nine piperazinyl-pyrrolo[1,2-a]quinoxaline derivatives, twenty-three compounds behaved as dual inhibitors of CaCdr1p and CaMdr1p. Only four compounds showed exclusive inhibition of CaCdr1p or CaMdr1p. Further biological investigations were developed and for example, their antifungal potential was evaluated by measuring the growth of control yeast cells (AD1-8u−) and efflux pump-overexpressing cells (AD-CDR1 and AD-MDR1) after exposition to variable concentrations of the tested compounds. The MIC80 values of nineteen compounds ranging from 100 to 901 μM for AD-CDR1 demonstrated that relative resistance index (RI) values were between 8 and 274. In comparison, only seven compounds had RI values superior to 4 in cells overexpressing Mdr1p. These results indicated substrate behavior for nineteen compounds for CaCdr1p and seven compounds for CaMdr1p, as these compounds were transported via MDR transporter overexpressing cells and not by the AD1-8u− cells. Finally, in a combination assay with fluconazole, two compounds (1d and 1f) have shown a synergistic effect (fractional inhibitory concentration index (FICI) values ≤ 0.5) at micromolar concentrations in the AD-MDR1 yeast strain overexpressing CaMdr1p-protein, indicating an excellent potency toward chemosensitization.
Introduction
Transplantation surgery, cancer chemotherapy, and HIV infections have led to a worldwide rise of the immunocompromised population, and hence also of bacterial and fungal opportunistic infections.1 The fungal genera most often associated with invasive fungal infections include Candida, Aspergillus, and Cryptococcus,2 with opportunistic strains of Candida albicans accounting for approximately 50–60% causes of candidiasis, particularly in immunocompromised patients. The treatment of these Candida infections relies heavily on azole antifungal agents,3 for which widespread and prolonged use has led to the rapid emergence of multidrug resistant (MDR) isolates of C. albicans as well as of non-albicans species.4 Various mechanisms potentially contributing to the development of MDR have been identified, and the induction of genes encoding drug-efflux pumps, like the primary ATP binding cassette (ABC) transporter genes CaCDR1 and CaCDR2 and the secondary major-facilitator superfamily (MFS) transporter gene CaMDR1, has been shown to play a prominent role in the development of resistance to antifungal drugs.5–7 Overexpression of these pump proteins may lead to an increased efflux of drug substrates in MDR clinical isolates.4,8,9The potent modulators of multidrug transporter CaCdr1p such as the immunosuppressants cyclosporin, FK520 and FK506, the natural polyphenol curcumin, the quorum-sensing molecule farnesol, the antabuse drug disulfiram, the antibiotic milbemycin, some synthetic-D-octapeptides, the anti-inflammatory drug ibuprofen and the antibacterial unnarmicins have been displayed to prevent drug extrusion and restore fungicidal synergism with the azoles and other drugs.10–14 Unlike CaCdr1p, there is only a handful number of chemosensitizers in case of CaMdr1p such as verapamil and enniantin B.15,16 Recently, a further screening from a library of synthetic aromatic compounds sharing a cyclobutene-dione moiety was investigated for the discovery of new inhibitors of MFS and ABC transporters of C. albicans. A few specific inhibitors of MFS transporter CaMdr1p were then identified.17 Therefore, the search for novel inhibitors able to block the drug extrusion mediated by these efflux proteins represents an attractive approach to reverse MDR.
The pyrrolo[1,2-a]quinoxaline heterocyclic framework constitutes the basis of an important class of compounds possessing interesting biological activities. These compounds have been reported as key intermediates for the assembly of several heterocycles including antipsychotic agent,18 anti-HIV agent,19 adenosine A3 receptor modulator,20 antiparasitic agents,21–25 and antitumor agents.26–31 We also previously demonstrated that the pyrrolo[1,2-a]quinoxaline heterocyclic scaffold could lead to the preparation of bacterial multidrug resistance pump inhibitors.32,33
In this context and as part of a programme on the development of new efflux pump inhibitors (EPIs), we decided to broaden the structural diversity and used the pyrrolo[1,2-a]quinoxaline moiety as a template for the design of new derivatives 1 and 2 in which a piperazine is incorporated in position 1, 4 or 9 of the heterocyclic core in analogy with the EPI pyrimidine and quinoline derivatives I–III, quinine and MS-209 used in the various multidrug resistance therapies (Fig. 1).34–38
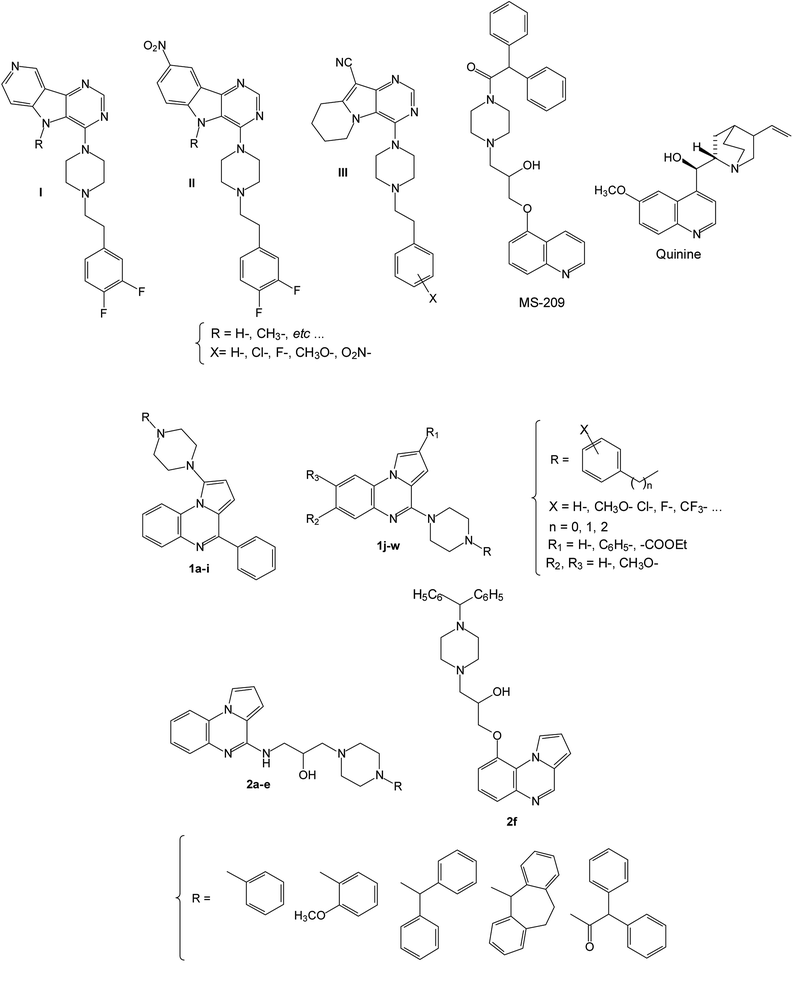 | ||
| Fig. 1 Structures of previously described EPIs I–III, quinine and MS-209, and new synthesized substituted pyrrolo[1,2-a]quinoxaline derivatives 1–2. | ||
Results and discussion
Chemistry
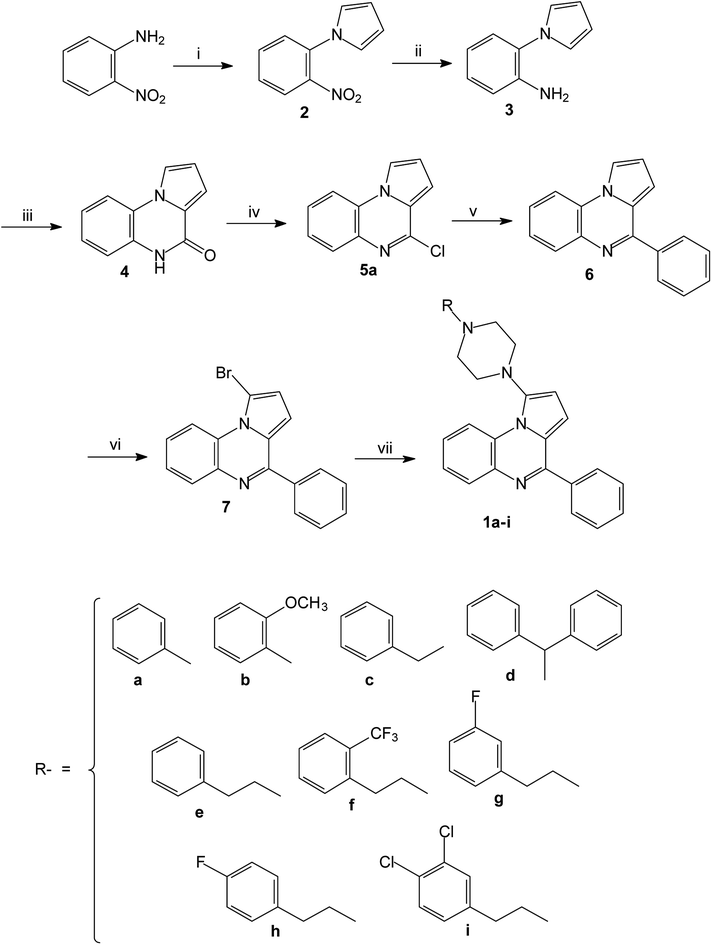
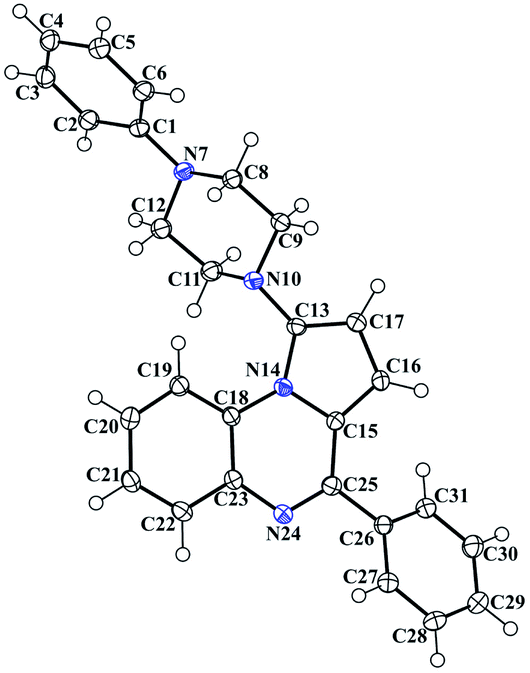
The reported piperazinyl-pyrrolo[1,2-a]quinoxaline derivatives 1a–i were synthesized in five steps from 2-nitroaniline (Scheme 1). Preparation of 1-(2-nitrophenyl)pyrrole 2 was performed according to the Clauson–Kaas reaction run under micro-wave irradiation starting from 2-nitroaniline and 2,5-dimethoxytetrahydrofuran in acetic acid. This pathway partially involved synthetic methodologies already described by our group.21–24,28 The resulting 1-(2-nitrophenyl)pyrrole intermediate 2 was subsequently reduced into the attempted 1-(2-aminophenyl)pyrrole 3 using a sodium borohydride-copper(II) sulfate treatment in ethanol at room temperature. This NaBH4–CuSO4 system was found to be quite powerful in reducing our aromatic nitro group with excellent yield (85%). The reaction of 3 with triphosgene in toluene gave the lactam 4, which was subsequently chlorodehydroxylated with phosphorous oxychloride, leading to the 4-chloropyrrolo[1,2-a]quinoxaline 5a. The 4-phenylpyrrolo[1,2-a]quinoxaline 6 was easily prepared by a direct Suzuki–Miyaura cross-coupling reaction of 4-chloropyrroloquinoxalines 5a with potassium phenyltrifluoroborate performed in the presence of PdCl2(dppf)·CH2Cl2 as a catalyst, cesium carbonate as the base, and THF–H2O as the solvent system.23 Reaction of 6 and one equivalent of N-bromosuccinimide (NBS) afforded the 1-bromo-4-phenylpyrrolo[1,2-a]quinoxaline 7 as the sole reaction product.28 The Buchwald–Hartwig Pd-catalyzed amination of the 1-bromo-4-phenylpyrrolo[1,2-a]quinoxaline 7 using Pd2(dba)3 as catalyst with BINAP as the ligand was then investigated. Under these conditions, various substituted piperazines were successfully coupled with 7 to give the desired piperazinyl-pyrrolo[1,2-a]quinoxaline derivatives 1a–i by using t-BuONa as a base and toluene as the solvent at 100 °C.39–41The 3D structural determinations of 1a and 1h were established by X-ray crystallography (Fig. 2 and 3), and confirmed the structures in the solid state as anticipated on the basis of NMR data.
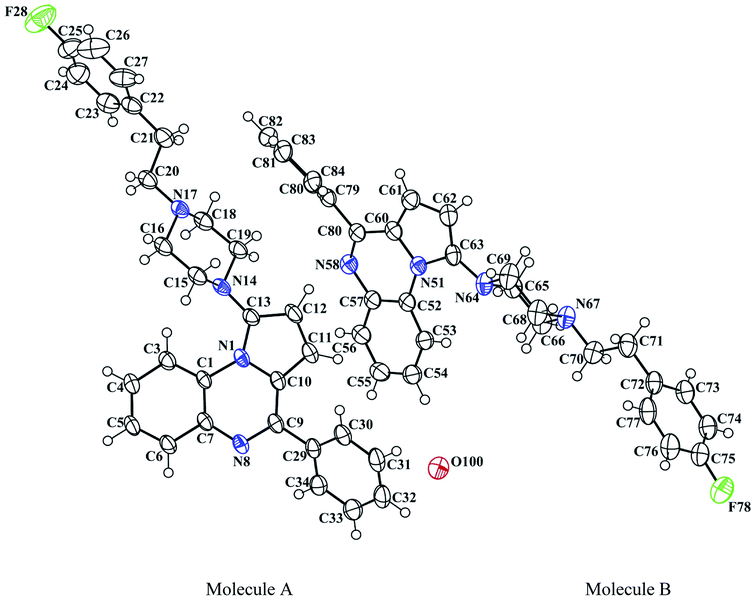 | ||
| Fig. 3 The ORTEP drawing of pyrrolo[1,2-a]quinoxaline 1h (molecules A and B) with thermal ellipsoids at 30% level. | ||
By using the same cross-coupling catalyzed methodology, the Pd-catalyzed coupling of substituted piperazines with the 4-chloropyrrolo[1,2-a]quinoxalines 5a–e30 led to the new 4-(4-substituted-piperazinyl)-4-phenylpyrrolo[1,2-a]quinoxaline 1j–w (Scheme 2). These compounds 1a–w were then converted into their hydrochloride or oxalate salts (Table 1).
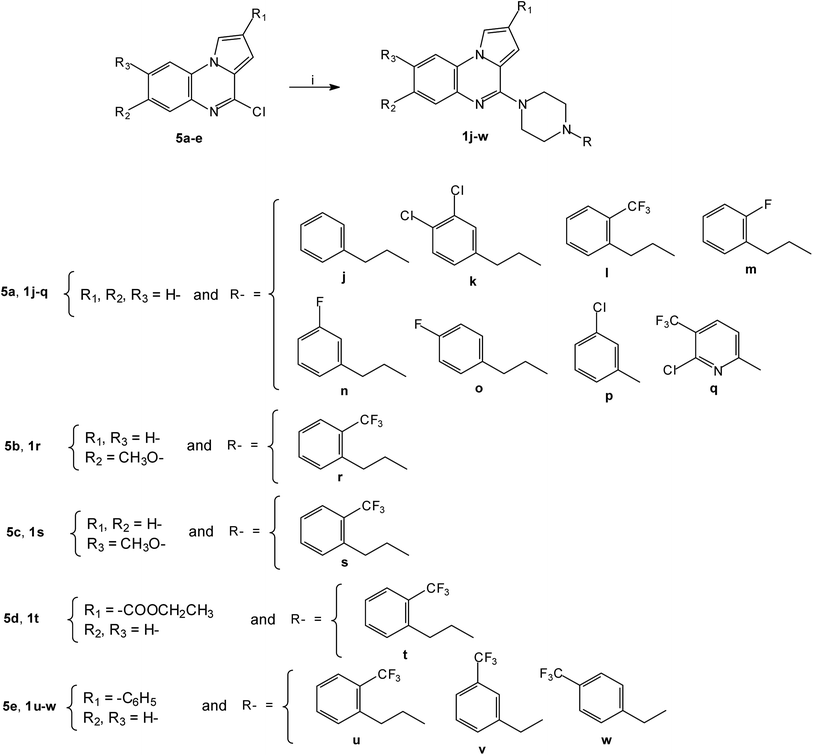 | ||
| Scheme 2 Synthesis of 4-(4-substituted-piperazinyl)-4-phenylpyrrolo[1,2-a]quinoxalines 1j–w; reagents and conditions: (i) R-piperazine, Pd2(dba)3, BINAP, t-BuONa, toluene, 100 °C. | ||
| Compound | a,b,cSalt | dmp (°C) | eYield (%) |
|---|---|---|---|
| a The amines 1a, b were dissolved in 20 mL of anhydrous diethyl ether, and treated with HCl gas. After crystallization, the ammonium chlorides were collected by filtration and were washed with Et2O, then dried under reduced pressure.b The amines 1c–w were dissolved in 30 mL of 2-propanol, heated to boiling, and treated with oxalic acid (4 or 5 equiv., based on the amount of the starting material). The oxalate salts crystallized upon cooling were collected by filtration, and were washed with 2-propanol and Et2O.c The stoichiometry and composition of the salts were determined by elemental analyses (within ± 0.4% of the theoretical values).d Crystallization solvent: 2-PrOH–H2O.e The yields only included the conversions into the ammonium chlorides or oxalates. | |||
| 1a | 3HCl | 114 | 71 |
| 1b | 3HCl | 88 | 68 |
| 1c | (COOH)2, H2O | 172 | 58 |
| 1d | (COOH)2, H2O | 109 | 61 |
| 1e | (COOH)2, H2O | 175 | 52 |
| 1f | (COOH)2, H2O | 156 | 59 |
| 1g | (COOH)2, H2O | 241 | 53 |
| 1h | (COOH)2, H2O | 238 | 64 |
| 1i | (COOH)2, H2O | 93 | 59 |
| 1j | 2(COOH)2 | 203 | 66 |
| 1k | 2(COOH)2 | 144 | 61 |
| 1l | 2(COOH)2 | 189 | 72 |
| 1m | 2(COOH)2 | 236 | 74 |
| 1n | 2(COOH)2 | 228 | 67 |
| 1n | 2(COOH)2 | 215 | 82 |
| 1o | 2(COOH)2 | 194 | 59 |
| 1p | 2(COOH)2 | 190 | 63 |
| 1q | 2(COOH)2 | 189 | 67 |
| 1r | 2(COOH)2 | 212 | 64 |
| 1s | 2(COOH)2 | 225 | 71 |
| 1t | 2(COOH)2 | 190 | 60 |
| 1u | 2(COOH)2 | 180 | 75 |
| 1v | 2(COOH)2 | 251 | 62 |
| 1w | 2(COOH)2 | 253 | 60 |
The series of piperazinylalcohol pyrrolo[1,2-a]quinoxaline derivatives 2a–f was synthesized as previously described by the authors starting from 5a or 8 (Schemes 3 and 4).42
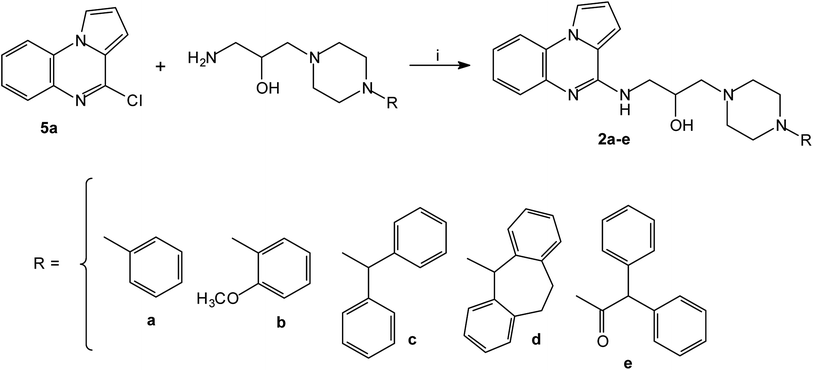 | ||
| Scheme 3 Synthesis of piperazinylalcohol-pyrrolo[1,2-a]quinoxalines 2a–e; reagents and conditions: (i) K2CO3, DMF, 120 °C. | ||
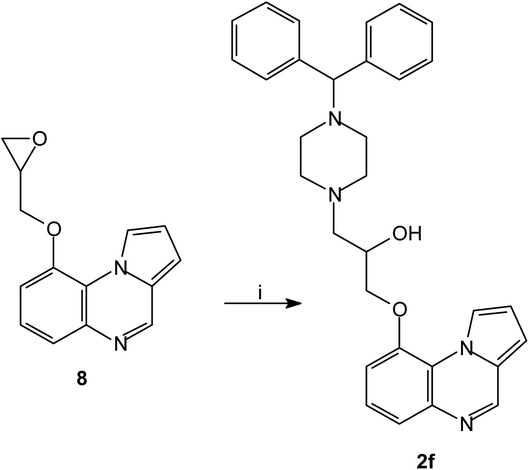 | ||
| Scheme 4 Synthesis of piperazinylalcohol-pyrrolo[1,2-a]quinoxaline 2f; reagents and conditions: (i) (C6H5)2CH-piperazine, isopropanol, Δ. | ||
Biological assays
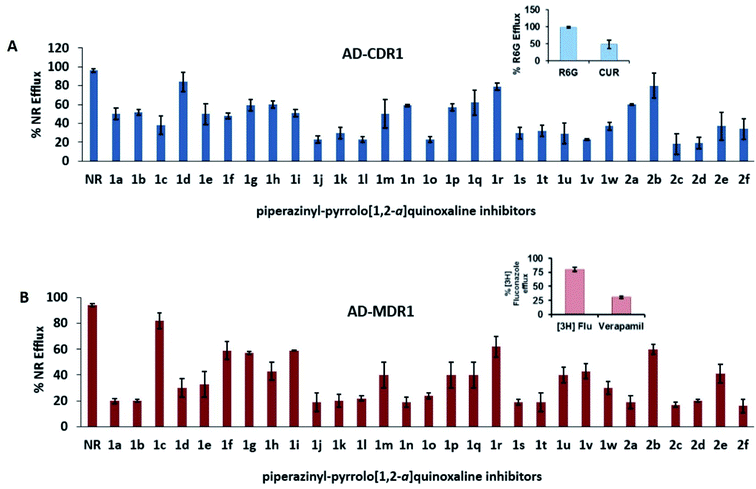
Piperazinyl-pyrrolo[1,2-a]quinoxaline derivatives were evaluated for their ability to inhibit the drug-efflux activity of CaCdr1p and CaMdr1p transporters of C. albicans overexpressed in a Saccharomyces cerevisiae strain. The transport assay was performed by monitoring Nile Red (NR) efflux in cells overexpressing the referred efflux pumps. In this assay, compounds were compared to wild-type control cells, and when the Nile Red efflux was lower than 60%, the compound was indeed considered to have inhibitory activity (Fig. 4). The efflux study performed on twenty-nine compounds revealed the total of twenty-three compounds (1a, 1b, 1e, 1g, 1h, 1i, 1j, 1k, 1l, 1m, 1n, 1o, 1p, 1s, 1t, 1u, 1v, 1w, 2a, 2c, 2d, 2e, 2f) that behaved as dual inhibitors of CaCdr1p and CaMdr1p. Compounds 2c > 2d > 1j = 1l = 1o = 1v revealed the strongest inhibitory activity of Cdr1p efflux pump, ranging from 77 to 82%. Regarding S. cerevisiae cells overexpressing Mdr1p, the most active compounds were 2f > 2c > 1a = 1b = 1j = 1k = 1n = 1s = 1t = 2a = 2d > 1l > 1o ranging from 78 to 84% efflux inhibition. This initial screening identified compounds 1u, 1w and 2e as dual inhibitors with intermediate potency. Compounds 1e, 1g, 1h, 1i, 1m, 1p finally revealed a weak inhibitory activity in both cell lines overexpressing the two types of efflux pumps. Compound 1c and 1f showed exclusive but weak inhibition of CaCdr1p, whereas compounds 1d and 1q demonstrated their exclusive impact on CaMdr1p with efflux inhibition ranged from 29 to 50%. Compounds 1r and 2b are not active on both efflux pumps.The antifungal potential of all piperazinyl-pyrrolo[1,2-a]quinoxaline derivatives was also evaluated by measuring the growth of control yeast cells (AD1-8u−) and efflux pump-overexpressing cells (AD-CDR1 and AD-MDR1) when exposed to variable concentrations of the tested compounds for 48 h. The yeast growth in the absence of inhibitor was considered as 100%. The results were expressed as MIC80, the concentration needed to decrease 80% of cells growth (ESI Table S1†). In case of the control yeast cells (AD1-8u−), compounds did not reveal any significant antifungal activity, as demonstrated by their relative resistance index (RI) values close to 1. However the MIC80 values of nineteen selected compounds (Table 2) ranging from 100 to 901 μM for AD-CDR1 demonstrated that RI values were comprised between 8 and 274. In comparison, only seven compounds (1c, 1e, 1g, 1j, 1k, 1m, 1o) had RI values superior to 4 in cells overexpressing Mdr1p. These results indicate the substrate behavior for both 19 compounds for CaCdr1p and seven compounds for CaMdr1p, as these compounds are transported via MDR transporter overexpressing cells and not by the AD1-8u− cells. Interestingly, all the nineteen compounds were observed to inhibit the Nile Red transport from the AD-CDR1 cells and simultaneously behaved as substrate of CaCdr1p (Table 2). Then it could be suggested that the Nile Red and these compounds seem to share the same drug binding pocket of CDR1, undergoing the kinetics of competitive inhibition. By contrast, in the case of AD-MDR1 cells, the route of efflux transport for Nile Red and these compounds did not overlap as only seven compounds showed substrate behavior. Here the results suggest the presence of an allosteric drug binding pocket for MDR1 and thus following the path of non-competitive kinetics.
| Compounds | Yeast strains | aMIC80 (μM) | bRI |
|---|---|---|---|
| a The MIC80 values of cytotoxicity were determined by measuring the optical density of cultures of each strain in the absence and the presence of a range of concentrations of the different compounds. Yeast growth in the absence of inhibitor was considered as 100%, and the concentration where the growth was decreased to 80% was taken as MIC80. The values are the means ± standard deviations of three independent experiments.b The resistance index (RI) was calculated as the ratio between the MIC80 value determined for the strain overexpressing the transporter relatively to that of the control strain (AD1-8u−). | |||
| 1c | AD1-8u− | 99 ± 10 | 1 |
| CDR1 | 817 ± 87 | 8.25 | |
| MDR1 | 400 ± 55 | 4.04 | |
| 1e | AD1-8u− | 12 ± 1.3 | 1 |
| CDR1 | 810 ± 86 | 67.5 | |
| MDR1 | 789 ± 85 | 65.75 | |
| 1g | AD1-8u− | 50 ± 6 | 1 |
| CDR1 | 803 ± 83 | 16 | |
| MDR1 | 221 ± 31 | 4.2 | |
| 1j | AD1-8u− | 3 ± 0.4 | 1 |
| CDR1 | 100 ± 12 | 33.3 | |
| MDR1 | 25 ± 3 | 8.33 | |
| 1k | AD1-8u− | 3 ± 0.2 | 1 |
| CDR1 | 400 ± 49 | 133.3 | |
| MDR1 | 12 ± 1 | 4 | |
| 1l | AD1-8u− | 6 ± 0.5 | 1 |
| CDR1 | 803 ± 102 | 133.8 | |
| MDR1 | 12 ± 2 | 2 | |
| 1m | AD1-8u− | 12 ± 1.6 | 1 |
| CDR1 | 408 ± 52 | 34 | |
| MDR1 | 53 ± 6.2 | 4.41 | |
| 1n | AD1-8u− | 25 ± 3.1 | 1 |
| CDR1 | 400 ± 53 | 16 | |
| MDR1 | 50 ± 6.2 | 2 | |
| 1o | AD1-8u− | 6 ± 0.71 | 1 |
| CDR1 | 200 ± 31 | 33.3 | |
| MDR1 | 25 ± 3.4 | 4.1 | |
| 1r | AD1-8u− | 6 ± 0.5 | 1 |
| CDR1 | 776 ± 81 | 129.3 | |
| MDR1 | 12 ± 1.7 | 2 | |
| 1s | AD1-8u− | 28 ± 1.7 | 1 |
| CDR1 | 387 ± 47 | 13.8 | |
| MDR1 | 59 ± 4.7 | 2.1 | |
| 1t | AD1-8u− | 94 ± 7.3 | 1 |
| CDR1 | 821 ± 75 | 8.7 | |
| MDR1 | 102 ± 9.3 | 1.08 | |
| 1w | AD1-8u− | 104 ± 8.4 | 1 |
| CDR1 | 901 ± 77 | 8.6 | |
| MDR1 | 187 ± 16 | 1.7 | |
| 2a | AD1-8u− | 47 ± 2.9 | 1 |
| CDR1 | 412 ± 27 | 8 | |
| MDR1 | 91 ± 7.9 | 1.9 | |
| 2b | AD1-8u− | 94 ± 110 | 1 |
| CDR1 | 811 ± 119 | 8.6 | |
| MDR1 | 104 ± 121 | 1.1 | |
| 2c | AD1-8u− | 3 ± 0.1 | 1 |
| CDR1 | 817 ± 77 | 272.3 | |
| MDR1 | 6 ± 0.2 | 2 | |
| 2d | AD1-8u− | 6 ± 0.2 | 1 |
| CDR1 | 831 ± 88 | 138.5 | |
| MDR1 | 5 ± 0.2 | 0.83 | |
| 2e | AD1-8u− | 3 ± 0.1 | 1 |
| CDR1 | 821 ± 86 | 273.6 | |
| MDR1 | 6 ± 0.2 | 2 | |
| 2f | AD1-8u− | 6 ± 0.1 | 1 |
| CDR1 | 804 ± 71 | 134 | |
| MDR1 | 14 ± 1.2 | 2.33 | |
The ability of the compounds to sensitize yeast growth to the antifungal agent fluconazole was evaluated by the checkerboard method.43 In this assay, the control (AD1-8u−) cells and the CaCdr1p- and CaMdr1p-overexpressing cells were grown in the presence of either fluconazole alone or a combination therapy (efflux pump inhibitor plus fluconazole). The results, expressed as the fractional inhibitory concentration index (FICI), are summarized in ESI Table S2.† FICI values ≤ 0.5 indicate synergistic interaction between the inhibitor and the substrate.44 It was observed that the two piperazinyl-pyrrolo[1,2-a]quinoxaline derivatives 1d and 1f with FIC = 0.0076 and 0.25, respectively, displayed strong synergistic effects (FICI = 0.15 and 0.4, respectively) when both are combined with fluconazole (FIC = 0.15) in the AD-MDR1 yeast strain overexpressing the MFS CaMdr1p, decreasing 129-fold the MIC80 of the antifungal agent. High FICI values (≥1) were found for the remaining compounds (ESI Table S2†). Similarly, high FICI values were also found in the AD-CDR1.
The effect of the compounds 1d and 1f was examined by confocal imaging of GFP-tagged Cdr1p and Mdr1p, and revealed the non-effect of these compounds on the intactness of the overexpressing strains AD-CDR1 and AD-MDR1 (Fig. 5).
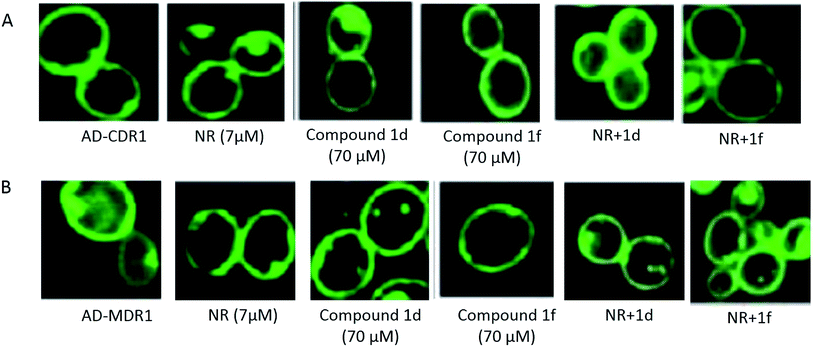 | ||
| Fig. 5 Effect of compounds 1d and 1f on the expression of the GFP-tagged Cdr1p and Mdr1p. Panels (A) AD-CDR1 and (B) AD-MDR1 were evaluated in the presence or in the absence of compounds 1d and 1f. | ||
Finally, compounds 1d and 1f were further evaluated for their ability to chemosensitize the azole-resistant clinical isolate (F5) of C. albicans together with the azole-susceptible strain (F2).45,46 As can be observed in Table 3, when combined with fluconazole, compounds 1d and 1f (FICI = 0.6 and 0.78, respectively) were able to reduce the effective concentration of fluconazole.
| Strain | Compound | aFIC of fluconazole | bFIC of compound | cFICI |
|---|---|---|---|---|
| a Evaluated by the checkerboard method, and expressed as the fractional inhibitory concentration (FIC) values for the fluconazole (=MIC80 of fluconazole in combination/MIC80 of fluconazole alone).b Each compound (=MIC80 of compound in combination/MIC80 of compound alone). The values in brackets are expressed in μM.c FIC index (FICI) value ≤0.5 indicates synergistic interaction between the compound and the fluconazole. | ||||
| AD1-8u− | 1d | 1 (1.5/1.5) | 1 (781/781) | 2 (1 + 1) |
| 1f | 1 (1.5/1.5) | 1 (811/811) | 2 (1 + 1) | |
| AD1-CDR1 | 1d | 0.3 (81/209) | 1 (791/791) | 1.3 (0.3 + 1) |
| 1f | 0.1 (40/209) | 1 (791/791) | 1.1 (0.1 + 1) | |
| AD1-MDR1 | 1d | 0.15 (10/65) | 0.0076 (6.25/812) | 0.15 (0.15 + 0.0076) |
| 1f | 0.15 (10/65) | 0.25 (200/799) | 0.4 (0.15 + 0.25) | |
| F2 | 1d | 1 (13/13) | 1 (618/618) | 2 (1 + 1) |
| 1f | 0.5 (7.5/13) | 1 (400/400) | 1.5 (0.5 + 1) | |
| F5 | 1d | 0.4 (200/418) | 0.2 (150/720) | 0.6 (0.2 + 0.4) |
| 1f | 0.35 (150/418) | 0.43 (350/799) | 0.78 (0.35 + 0.43) | |
About the structure–activity relationships on both series of piperazinyl-pyrrolo[1,2-a]quinoxaline derivatives, the three best dual inhibitors belonging to the series 2 (2c, 2d, 2f) has a common structural feature, namely the presence of a spacer (oxy-propan-2-ol or amino-propan-2-ol) between the tricyclic scaffold and the piperazinyl moiety. On the other hand, exclusive inhibitors of CaCdr1p (1c, 1f) or CaMdr1p (1d, 1q) have the piperazinyl moiety directly linked to the pyrroloquinoxaline. Four of these compounds (2c, 2f, 1d and 2d) have a benzhydryl substituent or related on the piperazine ring. Further pharmacomodulation works will be carried out to extend and deepen our knowledge.
Conclusions
The chemical approach by Buchwald–Hartwig Pd-catalyzed amination of the 1-bromo-4-phenylpyrrolo[1,2-a]quinoxaline 7 or the 4-chloropyrrolo[1,2-a]quinoxalines 5a–e was successfully used to access to new piperazinyl-pyrrolo[1,2-a]quinoxaline derivatives 1a–w. In parallel, six other derivatives containing as well a quinoxaline moiety (compounds 2a–f, previously synthetized42) were added to this study. Then twenty-nine compounds have been selected on their potential to inhibit fungal multidrug resistance pumps as pyrrolo[1,2-a]quinoxaline template was already used and efficient for the inhibition of bacterial efflux pumps.32,33Currently, we broadened our horizon to look into the role of these compounds to inhibit the CaCdr1p and CaMdr1p transporters in pathogenic yeast C. albicans. Our study based on the biological assays corroborated with the piperazinyl-pyrrolo[1,2-a]quinoxaline derivatives to be the putative and promising modulators of efflux pumps in the pathogenic yeast C. albicans. The results of this work have demonstrated that most of the compounds could inhibit the efflux of Nile Red mediated by both the ABC transporter CaCdr1p and the MFS pump CaMdr1p. Some compounds were able to inhibit specifically the efflux of Nile Red without being themselves substrates of the efflux-pump proteins CaMdr1p and CaMdr1p. This assumption was corroborated by the relative resistance index values close to 1 obtained from the cytotoxicity assays, showing that the presence of efflux-pump proteins did not affect the growth and the viability of yeast cells.
In S. cerevisiae cells expressing CaCdr1p and CaMdr1p, the greater inhibitory effect on Nile Red efflux was obtained with compounds 2c–f (5 < MIC80 (μM) < 14) on CaMdr1p. For compounds 2c–e, the best three compounds (MIC80 = 5–6 μM), the main structural feature is the presence of a bulky group R (e.g. compound 2c with a diphenylmethyl moiety).
In the combination assay with fluconazole, the two compounds 1d and 1f have shown a synergistic effect (FICI values ≤ 0.5) at micromolar concentrations in the AD-MDR1 yeast strain overexpressing CaMdr1p-protein, indicating an excellent potency toward chemosensitization. Interestingly, compound 1d showed exclusive and maximum Nile Red efflux inhibition on AD-MDR1 strain and showed excellent chemosensitization in the presence of fluconazole, whereas this was not observed with compound 1f. In this context, it is important to mention that each drug/compound may interact differently with different amino acid residues within the binding pocket of CaMdr1p, which could explain the different behavior of Nile Red and fluconazole with these compounds. As no synergy has been found in the clinical isolate F5 overexpressing CaMdr1p, a significant decreasing of the effective concentration of the antifungal agent was also observed, corroborating the results obtained in the AD-MDR1 strain. It is also interesting to note that compound 1d is the unique active compound bearing a benzhydryl moiety in the sub-series 1.
Finally, this study has shown that piperazinyl-pyrrolo[1,2-a]quinoxaline derivatives are able to reverse antifungal resistance, mediated by efflux pumps belonging to both ABC and MFS superfamilies of transporters of the pathogenic yeast C. albicans. Therefore, at non-inhibitory concentrations, these compounds stand as wise candidates chosen to be potential modulators in MDR reversal. For example, compound 1d could offer a new treatment strategy known as combo-therapy in the use of new azole antifungals recently designed.47,48 Nevertheless further chemical modifications will be carried out to synthetize a second generation of piperazinyl-pyrrolo[1,2-a]quinoxaline derivatives designed specifically as EPIs of the pathogenic yeast C. albicans. Once again, compound 1d will be investigated for assessing the structural importance of its benzhydryl moiety (e.g. nature and position of additional substituents). By similarity, a pharmacomodulation study around compound 2c will be also managed, using rational drug design tools such as 3D structural characteristics of efflux pumps49 and recent chemical features50 to design new piperazinyl-pyrrolo[1,2-a]quinoxaline derivatives as specific EPIs of C. albicans.
Experimental section
Chemistry
1-(4-Phenylpiperazinyl)-4-phenylpyrrolo[1,2-a]quinoxaline (1a). Yellow crystals (48%) mp 203 °C. 1H NMR (CDCl3) δ 9.21 (dd, 1H, J = 8.10 and 2.80 Hz, H-9), 8.02 (dd, 1H, J = 8.10 and 2.80 Hz, H-6), 7.96–7.93 (m, 2H, H-2′ and H-6′), 7.57–7.52 (m, 3H, H-3′, H-4′ and H-5′), 7.47–7.43 (m, 2H, H-7 and H-8), 7.35 (t, 2H, J = 7.70 Hz, H-3′′ and H-5′′), 7.07 (d, 2H, J = 7.70 Hz, H-2′′ and H-6′′), 6.96 (t, 1H, J = 7.70 Hz, H-4′′), 6.92 (d, 1H, J = 4.30 Hz, H-2), 6.59 (d, 1H, J = 4.30 Hz, H-3), 3.78–3.74 (m, 2H, CH2-pip), 3.51–3.47 (m, 2H, CH2-pip), 3.31–3.26 (m, 2H, CH2-pip), 3.18–3.09 (m, 2H, CH2-pip). 13C NMR (CDCl3) δ 156.1 (C-4), 152.6 (C-1′′), 145.1 (C-3a), 139.4 (C-5a), 138.4 (C-1′), 131.2 (C-7), 130.7 (C-8, C-3′′ and C-5′′), 130.2 (C-3′ and C-5′), 130.1 (C-9a), 130.0 (C-2′ and C-6′), 127.7 (C-9), 126.5 (C-6), 123.5 (C-1), 121.8 (C-4′), 117.9 (C-4′′), 117.8 (C-2′′ and C-6′′), 109.9 (C-2), 104.7 (C-3), 53.8 (CH2-pip), 50.6 (CH2-pip). Anal. calcd for C27H24N4: C, 80.17; H, 5.98; N, 13.85. Found: C, 81.04; H, 6.02; N, 13.94.
1-[4-(2-Methoxyphenyl)piperazinyl]-4-phenylpyrrolo[1,2-a]quinoxaline (1b). Yellow crystals (21%) mp 38 °C. 1H NMR (CDCl3) δ 9.23 (dd, 1H, J = 7.85 and 2.40 Hz, H-9), 8.01 (dd, 1H, J = 7.85 and 2.40 Hz, H-6), 7.96–7.94 (m, 2H, H-2′ and H-6′), 7.56–7.53 (m, 3H, H-3′, H-4′ and H-5′), 7.52–7.42 (m, 2H, H-7 and H-8), 7.13–7.06 (m, 2H, H-4′′ and H-5′′), 7.05–7.01 (m, 1H, H-6′′), 6.96 (dd, 1H, J = 8.50 and 2.15 Hz, H-3′′), 6.92 (d, 1H, J = 4.30 Hz, H-2), 6.61 (d, 1H, J = 4.30 Hz, H-3), 3.93 (s, 3H, CH3O), 3.65–3.61 (m, 2H, CH2-pip), 3.49–3.43 (m, 2H, CH2-pip), 3.31–3.12 (m, 4H, CH2-pip). 13C NMR (CDCl3) δ 156.2 (C-4), 153.7 (C-2′′), 145.2 (C-1′′), 142.4 (C-3a), 139.7 (C-5a), 138.7 (C-1′), 131.0 (C-7), 130.8 (C-8), 130.2 (C-9a), 130.1 (C-3′ and C-5′), 129.9 (C-2′ and C-6′), 127.5 (C-9), 126.3 (C-6), 124.8 (C-5′′), 123.5 (C-1), 122.4 (C-4′), 119.7 (C-4′′), 117.9 (C-6′′), 112.7 (C-3′′), 109.5 (C-2), 104.7 (C-3), 56.8 (CH3O), 54.0 (CH2-pip), 52.0 (CH2-pip). Anal. calcd for C28H26N4O: C, 77.39; H, 6.03; N, 12.89. Found: C, 77.41; H, 5.94; N, 13.04.
1-(4-Benzylpiperazinyl)-4-phenylpyrrolo[1,2-a]quinoxaline (1c). Yellow oil (41%). 1H NMR (CDCl3) δ 9.15 (dd, 1H, J = 8.00 and 1.70 Hz, H-9), 8.00 (dd, 1H, J = 8.00 and 1.70 Hz, H-6), 7.96–7.92 (m, 2H, H-2′ and H-6′), 7.54–7.51 (m, 3H, H-3′, H-4′ and H-5′), 7.48–7.45 (m, 2H, H-7 and H-8), 7.43–7.41 (m, 2H, H-2′′ and H-6′′), 7.41–7.28 (m, 3H, H-3′′, H-4′′ and H-5′′), 6.88 (d, 1H, J = 4.30 Hz, H-2), 6.54 (d, 1H, J = 4.30 Hz, H-3), 3.68 (sl, 2H, CH2), 3.33–3.29 (m, 2H, CH2-pip), 3.06–2.97 (m, 4H, CH2-pip), 2.60–2.55 (m, 2H, CH2-pip). 13C NMR (CDCl3) δ 156.2 (C-4), 145.2 (C-3a), 139.8 (C-5a), 139.3 (C-1′′), 138.8 (C-1′), 130.9 (C-7), 130.8 (C-8), 130.6 (C-3′ and C-5′), 130.2 (C-9a), 130.1 (C-2′′ and C-6′′), 129.9 (C-3′′ and C-5′′), 129.8 (C-2′ and C-6′), 128.7 (C-9), 127.5 (C-6), 126.2 (C-4′), 123.4 (C-1), 118.0 (C-4′′), 109.3 (C-2), 104.4 (C-3), 64.6 (NCH2), 54.4 (CH2-pip), 53.7 (CH2-pip). Anal. calcd for C28H26N4: C, 80.35; H, 6.26; N, 13.39. Found: C, 80.09; H, 6.34; N, 13.57.
1-[4-(Diphenylmethyl)piperazinyl]-4-phenylpyrrolo[1,2-a]quinoxaline (1d). Yellow crystals (68%) mp 69 °C. 1H NMR (CDCl3) δ 9.12 (dd, 1H, J = 7.40 and 2.20 Hz, H-9), 8.00–7.92 (m, 3H, H-6, H-2′ and H-6′), 7.55–7.52 (m, 7H, H-3′, H-4′, H-5′, H-2′′ and H-6′′), 7.45–7.41 (m, 2H, H-7 and H-8), 7.37–7.31 (m, 4H, H-3′′ and H-5′′), 7.28–7.21 (m, 2H, H-4′′), 6.90 (d, 1H, J = 4.30 Hz, H-2), 6.55 (d, 1H, J = 4.30 Hz, H-3), 4.41 (s, 1H, CH), 3.09–3.00 (m, 4H, CH2-pip), 2.48–2.41 (m, 2H, CH2-pip). 13C NMR (CDCl3) δ 156.1 (C-4), 145.3 (C-3a), 144.0 (2 C-1′′), 139.8 (C-5a), 138.7 (C-1′), 130.9 (C-7), 130.7 (C-8), 130.2 (C-9a), 130.1 (2 C-3′′, 2 C-5′′, C-3′ and C-5′), 129.9 (C-2′′ and C-6′′), 129.2 (C-2′ and C-6′), 128.7 (C-9), 127.5 (C-6), 126.2 (C-4′), 123.4 (C-1), 118.0 (2 C-4′′), 109.4 (C-2), 104.3 (C-3), 77.9 (NCH), 54.0 (CH2-pip), 53.3 (CH2-pip). Anal. calcd for C34H30N4: C, 82.56; H, 6.11; N, 11.33. Found: C, 83.27; H, 5.82; N, 10.91.
1-(4-Phenethylpiperazinyl)-4-phenylpyrrolo[1,2-a]quinoxaline (1e). Yellow oil (56%). 1H NMR (CDCl3) δ 9.15 (dd, 1H, J = 7.90 and 2.35 Hz, H-9), 7.99 (dd, 1H, J = 7.90 and 2.35 Hz, H-6), 7.96–7.92 (m, 2H, H-2′ and H-6′), 7.56–7.50 (m, 3H, H-3′, H-4′ and H-5′), 7.48–7.45 (m, 2H, H-7 and H-8), 7.37–7.32 (t, 2H, J = 7.20 Hz, H-2′′ and H6′′), 7.33–7.25 (m, 3H, H-3′′, H-4′′ and H-5′′), 6.89 (d, 1H, J = 4.30 Hz, H-2), 6.55 (d, 1H, J = 4.30 Hz, H-3), 3.38–3.34 (m, 2H, CH2-pip), 3.14–3.01 (m, 4H, CH2-pip), 2.95–2.90 (ddd, 2H, J = 12.20, 8.55 and 7.40 Hz, CH2), 2.82–2.76 (ddd, 2H, J = 12.20, 8.55 and 7.40 Hz, CH2), 2.65–2.58 (m, 2H, CH2-pip). 13C NMR (CDCl3) δ 156.1 (C-4), 145.2 (C-3a), 141.3 (C-5a), 139.8 (C-1′′), 138.8 (C-1′), 131.0 (C-7), 130.9 (C-8), 130.2 (C-3′ and C-5′), 130.1 (C-9a), 130.0 (C-2′′ and C-6′′), 129.9 (C-3′′, C-5′′, C-2′ and C-6′), 127.6 (C-9), 127.5 (C-6), 126.2 (C-4′), 123.5 (C-1), 117.9 (C-4′′), 109.4 (C-2), 104.5 (C-3), 61.8 (NCH2), 54.4 (CH2-pip), 53.6 (CH2-pip), 35.0 (CH2). Anal. calcd for C29H28N4: C, 80.52; H, 6.52; N, 12.95. Found: C, 79.96; H, 6.12; N, 12.92.
1-{4-[(2-Trifluoromethyl)phenethyl]piperazinyl}-4-phenylpyrrolo[1,2-a]quinoxaline (1f). Yellow crystals (63%); mp 34 °C. 1H NMR (CDCl3) δ 9.16 (dd, 1H, J = 8.20 and 2.00 Hz, H-9), 8.01 (dd, 1H, J = 8.20 and 2.00 Hz, H-6), 7.97–7.93 (m, 2H, H-2′ and H-6′), 7.68 (d, 1H, J = 7.80 Hz, H-3′′), 7.57–7.48 (m, 5H, H-4′, H-3′, H-5′, H-7 and H-8), 7.46–7.41 (m, 2H, H-4′′ and H-6′′), 7.35 (t, 1H, J = 7.60 Hz, H-5′′), 6.90 (d, 1H, J = 4.25 Hz, H-2), 6.55 (d, 1H, J = 4.25 Hz, H-3), 3.38–3.34 (m, 2H, CH2), 3.13–3.00 (m, 6H, CH2-pip and CH2), 2.81–2.76 (m, 2H, CH2-pip), 2.68–2.61 (m, 2H, CH2-pip). 13C NMR (CDCl3) δ 156.2 (C-4), 145.1 (C-3a), 140.1 (q, J = 1.6 Hz, C-1′′), 139.8 (C-1′), 138.8 (C-5a), 133.2 (C-5′′), 133.1 (C-6′′), 133.0 (C-4′′), 131.0 (C-7), 130.9 (C-8), 130.3 (q, J = 29.5 Hz, C-2′′), 130.2 (C-9a), 130.1 (C-3′ and C-5′), 129.9 (C-2′ and C-6′), 127.5 (C-9), 127.4 (q, J = 5.6 Hz, C-3′′), 126.2 (C-6), 126.0 (q, J = 272.2 Hz, CF3), 123.4 (C-1), 117.9 (C-4′), 109.3 (C-2), 104.4 (C-3), 61.6 (NCH2), 54.3 (CH2-pip), 53.7 (CH2-pip), 31.7 (CH2). Anal. calcd for C30H27F3N4: C, 71.98; H, 5.44; N, 11.19. Found: C, 72.23; H, 4.98; N, 11.02.
1-[4-(3-Fluorophenethyl)piperazinyl]-4-phenylpyrrolo[1,2-a]quinoxaline (1g). Yellow crystals (49%) mp 56 °C. 1H NMR (CDCl3) δ 9.15 (dd, 1H, J = 7.80 and 2.10 Hz, H-9), 8.01 (dd, 1H, J = 7.80 and 2.10 Hz, H-6), 7.96–7.93 (m, 2H, H-2′ and H-6′), 7.55–7.52 (m, 2H, H-3′ and H-5′), 7.48–7.43 (m, 2H, H-7 and H-8), 7.31–7.21 (m, 2H, H-5′′ and H-6′′), 7.08–6.88 (m, 2H, H-2′′ and H-4′′), 6.90 (d, 1H, J = 4.20 Hz, H-2), 6.56 (d, 1H, J = 4.20 Hz, H-3), 3.39–3.35 (m, 2H, CH2-pip), 3.14–3.07 (m, 4H, CH2-pip), 2.94–2.89 (m, 2H, CH2), 2.83–2.80 (m, 2H, CH2-), 2.64–2.61 (m, 2H, CH2-pip). Anal. calcd for C29H27FN4: C, 77.31; H, 6.04; N, 12.43. Found: C, 77.46; H, 6.15; N, 12.61.
1-[4-(4-Fluorophenethyl)piperazinyl]-4-phenylpyrrolo[1,2-a]quinoxaline (1h). Yellow crystals (55%) mp 150 °C. 1H NMR (CDCl3) δ 9.15 (dd, 1H, J = 7.70 and 2.10 Hz, H-9), 8.00 (dd, 1H, J = 7.70 and 2.10 Hz, H-6), 7.96–7.93 (m, 2H, H-2′ and H-6′), 7.55–7.51 (m, 2H, H-3′ and H-5′), 7.48–7.44 (m, 2H, H-7 and H-8), 7.30–7.21 (m, 3H, H-2′′, H-6′′ and H4′), 7.05 (t, 2H, J = 5.55 Hz, H-3′′ and H-5′′), 6.90 (d, 1H, J = 4.20 Hz, H-2), 6.56 (d, 1H, J = 4.20 Hz, H-3), 3.39–3.35 (m, 2H, CH2-pip), 3.12–3.07 (m, 4H, CH2-pip), 2.92–2.88 (m, 2H, CH2), 2.81–2.76 (m, 2H, CH2), 2.63–2.60 (m, 2H, CH2-pip). 13C NMR (CDCl3) δ 162.9 (d, J = 242.6 Hz, C-4′′), 156.2 (C-4), 144.8 (C-3a), 139.7 (C-5a), 138.8 (C-1′), 136.7 (d, J = 3.5 Hz, C-1′′), 131.5 (d, J = 7.8 Hz, C-2′′ and C-6′′), 131.0 (C-7), 130.9 (C-8), 130.1 (C-9a), 130.2 (C-3′ and C-5′), 130.0 (C-2′ and C-6′), 127.5 (C-9), 126.3 (C-6), 123.5 (C-1), 117.8 (C-4′), 116.7 (d, J = 20.9 Hz, C-3′′ and C-5′′), 109.4 (C-2), 104.5 (C-3), 61.7 (NCH2), 54.3 (CH2-pip), 53.4 (CH2-pip), 34.0 (CH2). Anal. calcd for C29H27FN4: C, 77.31; H, 6.04; N, 12.43. Found: C, 77.52; H, 6.17; N, 12.38.
1-[4-(3,4-Dichlorophenylethyl)piperazinyl]-4-phenylpyrrolo[1,2-a]quinoxaline (1i). Yellow oil (47%). 1H NMR (CDCl3) δ 9.13 (dd, 1H, J = 8.35 and 2.30 Hz, H-9), 8.00 (dd, 1H, J = 8.35 and 2.30 Hz, H-6), 7.96–7.93 (m, 2H, H-2′ and H-6′), 7.56–7.51 (m, 3H, H-2′ and H-6′), 7.56–7.51 (m, 3H, H-3′, H-4′ and H-5′), 7.49–7.43 (m, 2H, H-7 and H-8), 7.42–7.37 (m, 2H, H-3′′ and H-6′′), 7.10 (dd, 1H, J = 8.35 and 2.30 Hz, H-2′′), 6.89 (d, 1H, J = 4.30 Hz, H-2), 6.54 (d, 1H, J = 4.30 Hz, H-3), 3.37–3.33 (m, 2H, CH2), 3.08–2.98 (m, 2H, CH2-pip and CH2), 2.88–2.72 (m, 2H, CH2-pip), 2.64–2.56 (m, 2H, CH2-pip). 13C NMR (CDCl3) δ 156.2 (C-4), 144.9 (C-3a), 141.8 (C-5a), 139.8 (C-1′′), 138.8 (C-1′), 133.7 (C-3′′), 132.1 (C-5′′), 131.7 (C-7), 131.5 (C-4′′), 131.0 (C-8), 130.9 (C-2′′), 130.1 (C-9a), 130.0 (C-3′and C-5′), 129.9 (C-2′ and C-6′), 129.6 (C-6′′), 127.5 (C-9), 126.2 (C-6), 123.5 (C-1), 117.9 (C-4′), 109.3 (C-2), 104.4 (C-3), 61.2 (NCH2), 54.4 (CH2-pip), 53.7 (CH2-pip), 34.1 (CH2). Anal. calcd for C29H26Cl2N4: C, 69.46; H, 5.22; N, 11.17. Found: C, 69.41; H, 5.47; N, 10.95.
4-(4-Phenethylpiperazinyl)pyrrolo[1,2-a]quinoxaline (1j). Yellow oil (81%). 1H NMR (CDCl3) δ 7.84 (dd, 1H, J = 2.70 and 1.30 Hz, H-1), 7.76 (dd, 1H, J = 7.80 and 1.60 Hz, H-9), 7.72 (dd, 1H, J = 7.80 and 1.60 Hz, H-6), 7.37–7.25 (m, 7H, H-7, H-8, H-2′, H-3′, H-4′, H-5′ and H-6′), 6.83 (dd, 1H, J = 4.00 and 1.30 Hz, H-3), 6.78 (dd, 1H, J = 4.00 and 2.70 Hz, H-2), 3.92–3.88 (m, 4H, CH2-pip), 2.94–2.88 (m, 2H, CH2), 2.78–2.70 (m, 6H, CH2 and CH2-pip). 13C NMR (CDCl3) δ 154.0 (C-4), 141.8 (C-1′), 137.6 (C-5a), 130.2 (C-3′ and C-5′), 129.9 (C-2′ and C-6′), 128.9 (C-7), 127.6 (C-9), 127.2 (C-3a), 126.6 (C-6), 125.4 (C-4′), 121.6 (C-9a), 115.8 (C-8), 114.7 (C-1), 113.9 (C-2), 108.3 (C-3), 62.0 (NCH2), 54.7 (CH2-pip), 49.5 (CH2-pip), 35.1 (CH2). Anal. calcd for C23H24N4: C, 77.49; H, 6.78; N, 15.72. Found: C, 77.61; H, 6.92; N, 15.94.
4-[4-(3,4-Dichlorophenethyl)piperazinyl]pyrrolo[1,2-a]quinoxaline (1k). Yellow oil (75%). 1H NMR (CDCl3) δ 7.82 (dd, 1H, J = 2.70 and 1.40 Hz, H-1), 7.74 (dd, 1H, J = 7.60 and 1.40 Hz, H-9), 7.70 (dd, 1H, J = 7.60 and 1.40 Hz, H-6), 7.38–7.25 (m, 4H, H-7, H-8, H-2′ and H-5′), 7.09 (dd, 1H, J = 8.10 and 1.80 Hz, H-6′), 6.80 (dd, 1H, J = 4.00 and 1.30 Hz, H-3), 6.77 (dd, 1H, J = 4.00 and 2.70 Hz, H-2), 3.87–3.83 (m, 4H, CH2-pip), 2.85–2.80 (m, 2H, CH2), 2.76–2.64 (m, 6H, CH2 and CH2-pip). 13C NMR (CDCl3) δ 153.9 (C-4), 142.0 (C-1′), 137.5 (C-5a), 133.5 (C-3′), 132.0 (C-5′), 131.6 (C-2′), 131.4 (C-4′), 129.6 (C-6′), 128.9 (C-7), 127.2 (C-3a), 126.6 (C-9), 125.4 (C-6), 121.5 (C-9a), 115.8 (C-8), 114.7 (C-1), 113.9 (C-2), 108.2 (C-3), 61.2 (NCH2), 54.5 (CH2-pip), 49.3 (CH2-pip), 34.0 (CH2). Anal. calcd for C23H22Cl2N4: C, 64.94; H, 5.21; N, 13.17. Found: C, 65.08; H, 5.36; N, 13.35.
4-[4-(2-Trifluoromethylphenethyl)piperazinyl]pyrrolo[1,2-a]quinoxaline (1l). Yellow oil (79%). 1H NMR (CDCl3) δ 7.84 (dd, 1H, J = 2.70 and 1.30 Hz, H-1), 7.75 (dd, 1H, J = 7.80 and 1.40 Hz, H-9), 7.71 (dd, 1H, J = 7.80 and 1.40 Hz, H-6), 7.66 (d, 1H, J = 7.60 Hz, H-3′), 7.50 (t, 1H, J = 7.60 Hz, H-5′), 7.42 (d, 1H, J = 7.60 Hz, H-6′), 7.36–7.26 (m, 3H, H-7, H-8 and H-4′), 6.81 (dd, 1H, J = 4.00 and 1.30 Hz, H-3), 6.78 (dd, 1H, J = 4.00 and 2.70 Hz, H-2), 3.91–3.87 (m, 4H, CH2-pip), 3.11–3.06 (m, 2H, CH2), 2.81–2.70 (m, 6H, CH2 and CH2-pip). 13C NMR (CDCl3) δ 153.9 (C-4), 140.2 (q, J = 1.6 Hz, C-1′), 137.6 (C-5a), 133.2 (q, J = 0.8 Hz, C-5′), 132.1 (C-6′), 130.0 (q, J = 29.4 Hz, C-2′), 128.9 (C-7), 127.6 (C-4′), 127.3 (q, J = 5.6 Hz, C-3′), 127.2 (C-3a), 126.5 (C-9), 126.1 (q, J = 272.1 Hz, CF3), 125.3 (C-6), 121.6 (C-9a), 115.8 (C-8), 114.7 (C-1), 113.9 (C-2), 108.2 (C-3), 61.6 (NCH2), 54.5 (CH2-pip), 49.4 (CH2-pip), 31.4 (CH2). Anal. calcd for C24H23F3N4: C, 67.91; H, 5.46; N, 13.20. Found: C, 68.05; H, 5.22; N, 13.38.
4-[4-(2-Fluorophenethyl)piperazinyl]pyrrolo[1,2-a]quinoxaline (1m). Yellow oil (74%). 1H NMR (CDCl3) δ 7.85–7.82 (m, 1H, H-1), 7.76 (dd, 1H, J = 7.80 and 1.40 Hz, H-9), 7.70 (dd, 1H, J = 7.80 and 1.40 Hz, H-6), 7.36–7.04 (m, 6H, H-7, H-8, H-3′, H-4′, H-5′ and H-6′), 6.81–6.77 (m, 2H, H-2 and H-3), 3.97–3.94 (m, 4H, CH2-pip), 3.01–2.98 (m, 2H, CH2), 2.87–2.83 (m, 6H, CH2 and CH2-pip). 13C NMR (CDCl3) δ 162.6 (d, J = 243.3 Hz, C-2′), 153.9 (C-4), 137.6 (C-5a), 132.4 (d, J = 5 Hz, C-6′), 129.3 (d, J = 8 Hz, C-4′), 128.9 (C-7), 128.5 (d, J = 15.8 Hz, C-1′), 127.2 (C-3a), 126.5 (C-9), 125.4 (d, J = 3.2 Hz, C-5′), 125.3 (C-6), 121.6 (C-9a), 116.7 (d, J = 22.1 Hz, C-3′), 115.8 (C-8), 114.7 (C-1), 113.8 (C-2), 108.2 (C-3), 60.2 (NCH2), 54.5 (CH2-pip), 49.4 (CH2-pip), 28.2 (CH2). Anal. calcd for C23H23FN4: C, 73.77; H, 6.19; N, 14.96. Found: C, 73.85; H, 6.26; N, 15.12.
4-[4-(3-Fluorophenethyl)piperazinyl]pyrrolo[1,2-a]quinoxaline (1n). Yellow oil (81%). 1H NMR (CDCl3) δ 7.83 (dd, 1H, J = 2.70 and 1.30 Hz, H-1), 7.73 (dd, 1H, J = 8.00 and 1.50 Hz, H-9), 7.70 (dd, 1H, J = 8.00 and 1.50 Hz, H-6), 7.38–7.23 (m, 3H, H-7, H-8 and H-5′), 7.05–6.89 (m, 3H, H-2′, H-4′ and H-6′), 6.81 (dd, 1H, J = 3.90 and 1.30 Hz, H-3), 6.77 (dd, 1H, J = 3.90 and 2.70 Hz, H-2), 3.91–3.86 (m, 4H, CH2-pip), 2.93–2.87 (m, 2H, CH2), 2.78–2.69 (m, 6H, CH2 and CH2-pip). 13C NMR (CDCl3) δ 164.3 (d, J = 244.1 Hz, C-3′), 153.7 (C-4), 143.5 (d, J = 7.35 Hz, C-1′), 137.3 (C-5a), 131.3 (d, J = 8.25 Hz, C-5′), 128.9 (C-7), 127.2 (C-3a), 126.6 (C-9), 125.8 (d, J = 2.80 Hz, C-6′), 125.6 (C-6), 121.4 (C-9a), 117.0 (d, J = 20.85 Hz, C-2′), 115.9 (C-8), 114.7 (C-1), 114.5 (d, J = 21.5 Hz, C-4′), 113.9 (C-2), 108.1 (C-3), 61.2 (NCH2), 54.3 (CH2-pip), 48.8 (CH2-pip), 34.1 (CH2). Anal. calcd for C23H23FN4: C, 73.77; H, 6.19; N, 14.96. Found: C, 73.71; H, 6.33; N, 15.16.
4-[4-(4-Fluorophenethyl)piperazinyl]pyrrolo[1,2-a]quinoxaline (1o). Yellow oil (85%). 1H NMR (CDCl3) δ 7.83 (dd, 1H, J = 2.70 and 1.20 Hz, H-1), 7.75 (dd, 1H, J = 8.00 and 1.50 Hz, H-9), 7.70 (dd, 1H, J = 8.00 and 1.50 Hz, H-6), 7.37–7.26 (m, 2H, H-7 and H-8), 7.21 (dd, 2H, J = 8.70 and 5.40 Hz, H-2′ and H-6′), 7.04–6.97 (m, 2H, H-3′ and H-5′), 6.81 (dd, 1H, J = 3.90 and 1.20 Hz, H-3), 6.77 (dd, 1H, J = 3.90 and 2.70 Hz, H-2), 3.91–3.86 (m, 4H, CH2-pip), 2.89–2.84 (m, 2H, CH2), 2.77–2.66 (m, 6H, CH2 and CH2-pip). 13C NMR (CDCl3) δ 162.8 (d, J = 242.6 Hz, C-4′), 153.8 (C-4), 137.4 (C-5a), 137.0 (d, J = 3.2 Hz, C-1′), 131.5 (d, J = 7.7 Hz, C-2′ and C-6′), 128.9 (C-7), 127.2 (C-3a), 126.6 (C-9), 125.5 (C-6), 121.5 (C-9a), 116.6 (d, J = 21.2 Hz, C-3′ and C-5′), 115.8 (C-8), 114.7 (C-1), 113.9 (C-2), 108.1 (C-3), 61.8 (NCH2), 54.5 (CH2-pip), 49.1 (CH2-pip), 33.9 (CH2). Anal. calcd for C23H23FN4: C, 73.77; H, 6.19; N, 14.96. Found: C, 73.94; H, 6.05; N, 15.09.
4-[4-(3-Chlorophenyl)piperazinyl]pyrrolo[1,2-a]quinoxaline (1p). Pale-yellow oil (73%). 1H NMR (CDCl3) δ 7.90–7.88 (m, 1H, H-1), 7.82–7.76 (m, 2H, H-9 and H-6), 7.43–7.28 (m, 2H, H-7 and H-8), 7.26 (t, 1H, J = 8.10 Hz, H-5′), 7.00 (dd, 1H, J = 1.50 and 1.50 Hz, H-2′), 6.93–6.83 (m, 4H, H-4′, H-6′, H-2 and H-3), 4.01–3.97 (m, 4H, CH2-pip), 3.47–3.43 (m, 4H, CH2-pip). 13C NMR (CDCl3) δ 153.9 (C-4), 153.6 (C-1′), 137.3 (C-5a), 136.4 (C-3′), 131.5 (C-5′), 128.9 (C-7), 127.2 (C-3a), 126.7 (C-9), 125.7 (C-6), 121.5 (C-9a), 120.8 (C-8), 117.1 (C-4′), 116.0 (C-2′), 115.2 (C-6′), 114.8 (C-1), 114.0 (C-2), 108.2 (C-3), 49.9 (CH2-pip), 49.2 (CH2-pip), 33.9 (CH2). Anal. calcd for C21H19ClN4: C, 69.51; H, 5.28; N, 15.44. Found: C, 69.67; H, 5.07; N, 15.30.
4-[4-(6-Chloro-5-trifluoromethylpyridin-2-yl)piperazinyl]pyrrolo[1,2-a]quinoxaline (1q). White crystals (43%); mp 146 °C. 1H NMR (CDCl3) δ 7.88–7.86 (m, 1H, H-1), 7.77–7.73 (m, 2H, H-9 and H-6), 7.72 (d, 1H, J = 8.70 Hz, H-4′), 7.39–7.29 (m, 2H, H-7 and H-8), 6.86–6.80 (m, 2H, H-2 and H-3), 6.51 (d, 1H, J = 8.70 Hz, H-3′), 4.00–3.97 (m, 4H, CH2-pip), 3.89–3.86 (m, 4H, CH2-pip). 13C NMR (CDCl3) δ 160.6 (C-2′), 153.7 (C-4), 149.0 (q, J = 1.6 Hz, C-6′), 139.0 (q, J = 4.5 Hz, C-4′), 137.2 (C-5a), 128.9 (C-7), 127.2 (C-3a), 126.7 (C-9), 125.7 (C-6), 124.7 (q, J = 269.0 Hz, CF3), 121.3 (C-9a), 116.0 (C-8), 114.7 (C-1), 114.1 (C-2), 113.8 (q, J = 32.2 Hz, C-5′), 108.2 (C-3), 104.5 (C-3′), 48.6 (CH2-pip), 45.7 (CH2-pip). Anal. calcd for C21H17ClF3N5: C, 58.40; H, 3.97; N, 16.22. Found: C, 58.28; H, 4.07; N, 16.12.
7-Methoxy-4-[4-(2-trifluoromethylphenethyl)piperazinyl]pyrrolo[1,2-a]quinoxaline (1r). Yellow oil (79%). 1H NMR (CDCl3) δ 7.78–7.74 (m, 1H, H-1), 7.65 (d, 1H, J = 7.80 Hz, H-3′), 7.64 (d, 1H, J = 8.00 Hz, H-9), 7.51 (t, 1H, J = 7.80 Hz, H-5′), 7.43 (d, 1H, J = 7.80 Hz, H-6′), 7.33 (t, 1H, J = 7.80 Hz, H-4′), 7.20 (d, 1H, J = 2.60 Hz, H-6), 6.91 (dd, 1H, J = 8.00 and 2.60 Hz, H-8), 6.80–6.78 (m, 1H, H-3), 6.76–6.74 (m, 1H, H-2), 3.91–3.84 (m, 7H, CH3O and CH2-pip), 3.13–3.07 (m, 2H, CH2), 2.84–2.74 (m, 6H, CH2 and CH2-pip). 13C NMR (CDCl3) δ 158.7 (C-7), 154.3 (C-4), 140.3 (q, J = 3.2 Hz, C-1′), 138.7 (C-5a), 133.1 (q, J = 0.8 Hz, C-6′), 133.0 (C-5′), 130.0 (q, J = 29.4 Hz, C-2′), 127.6 (C-9), 127.3 (q, J = 5.6 Hz, C-3′), 126.0 (q, J = 272.2 Hz, CF3), 121.5 (C-3a), 121.1 (C-9a), 115.5 (C-4′), 115.4 (C-8), 113.9 (C-1), 113.5 (C-2), 110.7 (C-3), 107.9 (C-6), 61.6 (NCH2), 57.0 (CH3O), 54.5 (CH2-pip), 49.4 (CH2-pip), 31.4 (CH2). Anal. calcd for C25H25F3N4O: C, 66.07; H, 5.54; N, 12.33. Found: C, 65.94; H, 5.67; N, 12.20.
8-Methoxy-4-[4-(2-trifluoromethylphenethyl)piperazinyl]pyrrolo[1,2-a]quinoxaline (1s). Yellow oil (64%). 1H NMR (CDCl3) δ 7.73–7.71 (m, 1H, H-1), 7.66 (d, 1H, J = 8.70 Hz, H-6), 7.65 (d, 1H, J = 7.60 Hz, H-3′), 7.48 (t, 1H, J = 7.60 Hz, H-5′), 7.40 (d, 1H, J = 7.60 Hz, H-6′), 7.31 (t, 1H, J = 7.60 Hz, H-4′), 7.19 (d, 1H, J = 2.40 Hz, H-9), 6.95 (dd, 1H, J = 8.70 and 2.40 Hz, H-7), 6.79–6.77 (m, 2H, H-2 and H-3), 3.90 (s, 3H, CH3O), 3.79–3.78 (m, 4H, CH2-pip), 3.11–3.06 (m, 2H, CH2), 2.80–2.69 (m, 6H, CH2 and CH2-pip). 13C NMR (CDCl3) δ 158.2 (C-8), 152.9 (C-4), 140.2 (q, J = 1.65 Hz, C-1′), 133.2 (q, J = 0.9 Hz, C-6′), 133.1 (C-5′), 131.6 (C-5a), 130.1 (q, J = 29.3 Hz, C-2′), 130.0 (C-6), 127.8 (C-3a), 127.6 (C-4′), 127.3 (q, J = 5.6 Hz, C-3′), 126.0 (q, J = 272.1 Hz, CF3), 121.9 (C-9a), 115.4 (C-7), 114.0 (C-1), 113.6 (C-2), 107.8 (C-3), 99.3 (C-9), 61.7 (NCH2), 57.1 (CH3O), 54.5 (CH2-pip), 49.7 (CH2-pip), 31.4 (CH2). Anal. calcd for C25H25F3N4O: C, 66.07; H, 5.54; N, 12.33. Found: C, 66.20; H, 5.44; N, 12.46.
Ethyl 4-[4-(2-trifluoromethylphenethyl)piperazinyl]pyrrolo[1,2-a]quinoxaline-2-carboxylate (1t). Pale-yellow crystals (66%); mp 118 °C. 1H NMR (CDCl3) δ 8.33 (s, 1H, H-1), 7.75 (d, 1H, J = 8.00 Hz, H-9), 7.67 (d, 1H, J = 8.00 Hz, H-6), 7.64 (d, 1H, J = 7.50 Hz, H-3′), 7.49 (t, 1H, J = 7.50 Hz, H-5′), 7.43 (d, 1H, J = 7.50 Hz, H-6′), 7.41–7.26 (m, 3H, H-7, H-8 and H-4′), 7.19 (s, 1H, H-3), 4.39 (q, 2H, J = 7.20 Hz, OCH2), 3.89–3.87 (m, 4H, CH2-pip), 3.10–3.05 (m, 2H, CH2), 2.79–2.69 (m, 6H, CH2 and CH2-pip), 1.42 (t, 2H, J = 7.20 Hz, CH3). 13C NMR (CDCl3) δ 165.7 (CO), 153.6 (C-4), 140.2 (C-1′), 137.9 (C-5a), 133.1 (C-5′), 133.0 (C-7), 130.0 (q, J = 30.0 Hz, C-2′), 129.0 (C-9), 127.7 (C-6), 127.6 (C-6′), 127.3 (q, J = 5.6 Hz, C-3′), 126.3 (C-3a), 126.0 (q, J = 272.2 Hz, CF3), 125.6 (C-4′), 121.9 (C-9a), 120.5 (C-2), 119.0 (C-8), 114.9 (C-1), 109.2 (C-3), 61.9 (OCH2), 61.6 (NCH2), 54.4 (CH2-pip), 49.2 (CH2-pip), 31.4 (CH2), 15.9 (CH3). Anal. calcd for C27H27F3N4O2: C, 65.31; H, 5.48; N, 11.28. Found: C, 65.39; H, 5.29; N, 11.42.
2-Phenyl-4-[4-(2-trifluoromethylphenethyl)piperazinyl]pyrrolo[1,2-a]quinoxaline (1u). Pale-yellow crystals (67%); mp 136 °C. 1H NMR (CDCl3) δ 8.08 (s, 1H, H-1), 7.77 (d, 1H, J = 8.10 Hz, H-9), 7.74–7.68 (m, 3H, H-6, H-2′′ and H-6′′), 7.66 (d, 1H, J = 7.50 Hz, H-3′), 7.51 (t, 1H, J = 7.50 Hz, H-5′), 7.49–7.28 (m, 7H, H-4′, H-6′, H-7, H-8, H-3′′, H-4′′ and H-5′′), 7.06 (s, 1H, H-3), 3.93–3.90 (m, 4H, CH2-pip), 3.13–3.07 (m, 2H, CH2), 2.84–2.72 (m, 6H, CH2 and CH2-pip). 13C NMR (CDCl3) δ 153.4 (C-4), 140.2 (C-1′′), 137.3 (C-5a), 135.7 (C-1′), 133.4 (C-5′′), 133.3 (C-7), 130.3 (C-3′ and C-5′), 129.9 (q, J = 29.3 Hz, C-2′′), 129.1 (C-9), 128.4 (C-4′), 128.0 (C-6), 127.6 (C-3a), 127.5 (C-2′ and C-6′), 127.4 (C-6′′), 127.0 (C-2), 126.7 (C-3′′), 125.9 (C-8), 125.7 (q, J = 272.2 Hz, CF3), 125.6 (C-4′′), 122.2 (C-9a), 114.7 (C-2), 112.6 (C-1), 105.7 (C-3), 61.2 (NCH2), 53.9 (CH2-pip), 48.7 (CH2-pip), 30.7 (CH2). Anal. calcd for C30H27F3N4: C, 71.98; H, 5.44; N, 11.19. Found: C, 72.06; H, 5.53; N, 11.35.
2-Phenyl-4-[4-(3-trifluoromethylbenzyl)piperazinyl]pyrrolo[1,2-a]quinoxaline (1v). Yellow oil (54%). 1H NMR (CDCl3) δ 8.07 (s, 1H, H-1), 7.76 (d, 1H, J = 7.80 Hz, H-9), 7.73–7.69 (m, 4H, H-6, H-4′, H-2′′ and H-6′′), 7.63–7.56 (m, 2H, H-7 and H-8), 7.51–7.43 (m, 3H, H-2′, H-3′′, H-5′′), 7.37–7.28 (m, 3H, H-5′, H-6′ and H-4′′), 7.04 (s, 1H, H-3), 3.89–3.86 (m, 4H, CH2-pip), 3.67 (s, 2H, NCH2), 2.71–2.69 (m, 4H, CH2-pip). 13C NMR (CDCl3) δ 153.8 (C-4), 140.3 (C-1′′), 137.4 (C-5a), 135.8 (C-1′), 133.9 (C-6′′), 132.0 (q, J = 32.0 Hz, C-3′′), 130.3 (C-3′ and C-5′), 130.2 (C-7), 129.8 (C-3a), 128.9 (C-9), 128.3 (C-4′), 127.4 (C-2′ and C-6′), 127.2 (C-5′′), 127.1 (q, J = 3.75 Hz, C-2′′), 126.9 (C-2), 126.7 (C-6), 125.6 (C-8), 125.5 (q, J = 3.9 Hz, C-4′′), 125.4 (q, J = 271.3 Hz, CF3), 122.3 (C-9a), 114.6 (C-2), 112.4 (C-1), 105.8 (C-3), 63.8 (NCH2), 54.4 (CH2-pip), 49.3 (CH2-pip). Anal. calcd for C29H25F3N4: C, 71.59; H, 5.18; N, 11.52. Found: C, 71.75; H, 5.08; N, 11.57.
2-Phenyl-4-[4-(4-trifluoromethylbenzyl)piperazinyl]pyrrolo[1,2-a]quinoxaline (1w). Yellow oil (76%). 1H NMR (CDCl3) δ 8.08 (d, 1H, J = 0.90 Hz, H-1), 7.79 (dd, 1H, J = 7.80 and 1.80 Hz, H-9), 7.71–7.67 (m, 3H, H-6, H-2′′ and H-6′′), 7.64–7.55 (m, 2H, H-7 and H-8), 7.48–7.42 (m, 2H, H-3′′ and H-5′′), 7.35–7.29 (m, 3H, H-2′, H-6′ and H-4′′), 7.02 (d, 1H, J = 0.90 Hz, H-3), 3.88–3.85 (m, 4H, CH2-pip), 3.68 (s, 2H, NCH2), 2.72–2.69 (m, 4H, CH2-pip). 13C NMR (CDCl3) δ 153.8 (C-4), 143.8 (q, J = 1.0 Hz, C-1′′), 137.5 (C-5a), 135.8 (C-1′), 130.7 (q, J = 28.3 Hz, C-4′′), 130.6 (C-7), 130.3 (C-3′ and C-5′), 129.7 (C-3a), 129.0 (C-9), 128.3 (C-4′), 127.4 (C-2′ and C-6′), 126.7 (C-6), 126.6 (q, J = 6.3 Hz, C-3′′ and C-5′′), 126.5 (q, J = 3.5 Hz, C-2′′ and C-6′′), 125.6 (q, J = 267.8 Hz, CF3), 125.5 (C-8), 122.4 (C-9a), 114.6 (C-2), 112.4 (C-1), 105.7 (C-3), 63.9 (NCH2), 54.5 (CH2-pip), 49.4 (CH2-pip). Anal. calcd for C29H25F3N4: C, 71.59; H, 5.18; N, 11.52. Found: C, 71.72; H, 5.04; N, 11.48.
X-ray crystallography studies
Crystallographic data of compounds 1a and 1h were collected at 293 K with an Enraf-Nonius CAD-4 diffractometer with monochromatic Cu-Kα radiation (l = 1.54178 Å). The collected data were reduced using the NONIUS CAD4 software and all reflections were used for unit-cell refinement. Then the data were corrected for Lorentz and polarization effects and for empirical absorption correction.51 The structure was solved by direct methods Shelx 2013 (ref. 52) and refined using Shelx 201352 suite of programs.Colorless single crystal of 1a was obtained by slow evaporation from dichloromethane/methanol solution (70/30; v/v): triclinic, space group P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) , a = 9.5233(14) Å, b = 10.8011(19) Å, c = 11.8823(16) Å, α = 107.688(11)°, β = 110.089(10)°, γ = 100.552(11)°, V = 1035.6(3) Å3, Z = 2, δ (calcd) = 1.259 Mg m−3, FW = 392.49 for C26H24N4, F(000) = 416. Colorless single crystal of 1h was obtained by slow evaporation from methanol/dichloromethane (20/80; v/v): triclinic, space group P
, a = 9.5233(14) Å, b = 10.8011(19) Å, c = 11.8823(16) Å, α = 107.688(11)°, β = 110.089(10)°, γ = 100.552(11)°, V = 1035.6(3) Å3, Z = 2, δ (calcd) = 1.259 Mg m−3, FW = 392.49 for C26H24N4, F(000) = 416. Colorless single crystal of 1h was obtained by slow evaporation from methanol/dichloromethane (20/80; v/v): triclinic, space group P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) , a = 9.471(10) Å, b = 13.021(6) Å, c = 19.887(6) Å, α = 77.83(4)°, β = 81.76(8)°, γ = 87.25(6)°, V = 2372(3) Å3, Z = 2, δ (calcd) = 1.266 Mg m−3, FW = 904.29 for 2C29H27FN4, 0.1H2O, F(000) = 955.
, a = 9.471(10) Å, b = 13.021(6) Å, c = 19.887(6) Å, α = 77.83(4)°, β = 81.76(8)°, γ = 87.25(6)°, V = 2372(3) Å3, Z = 2, δ (calcd) = 1.266 Mg m−3, FW = 904.29 for 2C29H27FN4, 0.1H2O, F(000) = 955.
Full crystallographic results have been deposited at the Cambridge Crystallographic Data Centre (CCDC-891809, CCDC-891808, respectively), UK, as ESI.†53
Biological studies
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were analyzed in the acquisition. The analysis was performed using the CellQuest software (Becton Dickinson Immunocytometry Systems). Efflux of 100% was attributed to the cells not exposed to Nile Red and normalized with the efflux mediated via MDR transporters.
000 cells were analyzed in the acquisition. The analysis was performed using the CellQuest software (Becton Dickinson Immunocytometry Systems). Efflux of 100% was attributed to the cells not exposed to Nile Red and normalized with the efflux mediated via MDR transporters.Conflicts of interest
There are no conflicts to declare.References
- M. D. J. Richardson, Changing patterns and trends in systemic fungal infections, J. Antimicrob. Chemother., 2005, 56, i5–i11 CrossRef CAS PubMed.
- M. A. Pfaller, L. Boyken, R. J. Hollis, J. Kroeger, S. A. Messer, S. Tendolkar and D. J. Diekema, In vitro susceptibility of invasive isolates of Candida spp. to anidulafungin, caspofungin, and micafungin: six years of global surveillance, J. Clin. Microbiol., 2007, 46, 150–156 CrossRef.
- D. S. Perlin, Current perspectives on echinocandin class drugs, Future Microbiol., 2011, 6, 441–457 CrossRef CAS PubMed.
- T. C. White, K. A. Marr and R. A. Bowden, Clinical, cellular, and molecular factors that contribute to antifungal drug resistance, Clin. Microbiol. Rev., 1998, 11, 382–402 CrossRef CAS PubMed.
- R. Prasad, P. De Worgifosse, A. Goffeau and E. Balzi, Molecular cloning and characterisation of a novel gene of C. albicans, CDR1, conferring multiple resistance to drugs and antifungals, Curr. Genet., 1995, 27, 320–329 CrossRef CAS PubMed.
- D. Sanglard, F. Ischer, M. Monod and J. Bille, Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene, Microbiology, 1997, 143, 405–416 CrossRef CAS PubMed.
- R. Pasrija, D. Banerjee and R. Prasad, Structure and function analysis of CaMdr1p, a major facilitator superfamily antifungal efflux transporter protein of Candida albicans: identification of amino acid residues critical for drug/H+ transport, Eukaryot. Cell, 2007, 6, 443–453 CrossRef CAS PubMed.
- R. Prasad, S. L. Panwar and Smriti, Drug resistance in yeasts--an emerging scenario, Adv. Microb. Physiol., 2002, 46, 155–201 CrossRef CAS.
- S. Nim, P. Baghel, V. K. Tran-Nguyen, B. Peres, K. A. Nguyen, A. Di Pietro, P. Falson, R. Prasad and A. Boumendjel, Make azoles active again: chalcones as potent reversal agents of transporters-mediated resistance in Candida albicans, Future Med. Chem., 2018, 10, 2177–2186 CrossRef CAS PubMed.
- S. Nim, M. K. Rawal and R. Prasad, FK520 interacts with the discrete intrahelical amino acids of multidrug transporter Cdr1 protein and acts as antagonist to selectively chemosensitize azole-resistant clinical isolates of Candida albicans, FEMS Yeast Res., 2014, 14, 624–632 CrossRef CAS PubMed.
- M. Sharma and R. Prasad, The quorum-sensing molecule farnesol is a modulator of drug efflux mediated by ABC multidrug transporters and synergizes with drugs in Candida albicans, Antimicrob. Agents Chemother., 2011, 55, 4834–4843 CrossRef CAS.
- M. Sharma, R. Manoharlal, S. Shukla, N. Puri, T. Prasad, S. V. Ambudkar and R. Prasad, Curcumin modulates efflux mediated by yeast ABC multidrug transporters and is synergistic with antifungals, Antimicrob. Agents Chemother., 2009, 53, 3256–3265 CrossRef CAS PubMed.
- S. Shukla, Z. E. Sauna, R. Prasad and S. V. Ambudkar, Disulfiram is a potent modulator of multidrug transporter Cdr1p of Candida albicans, Biochem. Biophys. Res. Commun., 2004, 322, 520–525 CrossRef CAS PubMed.
- S. Nim, M. K. Rawal and R. Prasad, FK520 interacts with the discrete intrahelical amino acids of multidrug transporter Cdr1 protein and acts as antagonist to selectively chemosensitize azole-resistant clinical isolates of Candida albicans, FEMS Yeast Res., 2014, 14, 624–632 CrossRef CAS PubMed.
- I. Ivnitski-Steele, A. R. Holmes, E. Lamping, B. C. Monk, R. D. Cannon and L. A. Sklar, Identification of nile red as a fluorescent substrate of the Candida albicans ATP-binding cassette transporters Cdr1p and Cdr2p and the major facilitator superfamily transporter Mdr1p, Anal. Biochem., 2009, 394, 87–91 CrossRef CAS PubMed.
- L. R. Basso Jr, C. E. Gast, Y. Mao and B. Wong, Fluconazole transport into Candida albicans secretory vesicles by the membrane proteins Cdr1p, Cdr2p, and Mdr1p, Eukaryot. Cell, 2010, 9, 960–970 CrossRef PubMed.
- M. V. Keniya, E. Fleischer, A. Klinger, R. D. Cannon and B. C. Monk, Inhibitors of the Candida albicans major facilitator superfamily transporter Mdr1p responsible for fluconazole resistance, PLoS One, 2015, 10, e0126350 CrossRef PubMed.
- G. Campiani, S. Butini, C. Fattorusso, F. Trotta, S. Franceschini, M. De Angelis and K. S. Nielsen, Novel aryl piperazine derivatives with medical utility, PCT Pat., WO2006072608A2, 2006.
- G. Campiani, F. Aiello, M. Fabbrini, E. Morelli, A. Ramunno, S. Armaroli, V. Nacci, A. Garofalo, G. Greco, E. Novellino, G. Maga, S. Spadari, A. Bergamini, L. Ventura, B. Bongiovanni, M. Capozzi, F. Bolacchi, S. Marini, M. Coletta, G. Guiso and S. Caccia, Quinoxalinylethylpyridylthioureas (QXPTs) as potent non-nucleoside HIV-1 reverse transcriptase (RT) inhibitors. Further SAR studies and identification of a novel orally bioavailable hydrazine-based antiviral agent, J. Med. Chem., 2001, 44, 305–315 CrossRef CAS PubMed.
- S. Schann, S. Mayer and S. Gardan, Pyrrolo[1,2-a]quinoxaline derivatives as adenosine A3 receptor modulators and uses thereof, EP Pat., 20071798233A1, 2007.
- J. Guillon, P. Grellier, M. Labaied, P. Sonnet, J. M. Léger, R. Déprez-Poulain, I. Forfar-Bares, P. Dallemagne, N. Lemaître, F. Péhourcq, J. Rochette, C. Sergheraert and C. Jarry, Synthesis, antimalarial activity, and molecular modeling of new pyrrolo[1,2-a]quinoxalines, bispyrrolo[1,2-a]quinoxalines, bispyrido[3,2-e]pyrrolo[1,2-a]pyrazines, and bispyrrolo[1,2-a]thieno[3,2-e]pyrazines, J. Med. Chem., 2004, 47, 1997–2009 CrossRef CAS PubMed.
- J. Guillon, I. Forfar, M. Mamani-Matsuda, V. Desplat, M. Saliège, D. Thiolat, S. Massip, A. Tabourier, J. M. Léger, B. Dufaure, G. Haumont, C. Jarry and D. Mossalayi, Synthesis, analytical behaviour and biological evaluation of new 4-substituted pyrrolo[1,2-a]quinoxalines as antileishmanial agents, Bioorg. Med. Chem., 2007, 15, 194–210 CrossRef CAS PubMed.
- J. Guillon, I. Forfar, V. Desplat, S. Belisle-Fabre, D. Thiolat, S. Massip, H. Carrie, D. Mossalayi and C. Jarry, Synthesis of new 4-(E)-alkenylpyrrolo[1,2-a]quinoxalines as antileishmanial agents by Suzuki-Miyaura cross-coupling reactions, J. Enzyme Inhib. Med. Chem., 2007, 22, 541–549 CrossRef CAS PubMed.
- J. Guillon, S. Moreau, E. Mouray, V. Sinou, I. Forfar, S. Belisle-Fabre, V. Desplat, P. Millet, D. Parzy, C. Jarry and P. Grellier, New ferrocenic pyrrolo[1,2-a]quinoxaline derivatives: synthesis, and in vitro antimalarial activity, Bioorg. Med. Chem., 2008, 16, 9133–9144 CrossRef CAS PubMed.
- J. Guillon, A. Cohen, N. M. Gueddouda, R. Nath Das, S. Moreau, L. Ronga, S. Savrimoutou, L. Basmacyian, A. Monnier, M. Monget, S. Rubio, T. Garnerin, N. Azas, J. L. Mergny, C. Mullié and P. Sonnet, Design, synthesis and antimalarial activity of novel bis{N-[(pyrrolo[1,2-a]quinoxalin-4-yl)benzyl]-3-aminopropyl}amine derivatives, J. Enzyme Inhib. Med. Chem., 2017, 32, 547 CrossRef CAS PubMed.
- J. Milne, K. D. Normington and M. Milburn, Tetrahydroquinoxalinone sirtuin modulators, PCT Pat., WO2006094210A2, 2006.
- F. Grande, F. Aiello, O. De Grazia, A. Brizzi, A. Garofalo and N. Meamati, Synthesis and antitumor activities of a series of novel quinoxalinhydrazides, Bioorg. Med. Chem., 2007, 15, 288–294 CrossRef CAS PubMed.
- V. Desplat, A. Geneste, M. A. Begorre, S. Belisle Fabre, S. Brajot, S. Massip, D. Thiolat, D. Mossalayi, C. Jarry and J. Guillon, Synthesis of new pyrrolo[1,2-a]quinoxaline derivatives as potential inhibitors of Akt kinase, J. Enzyme Inhib. Med. Chem., 2008, 23, 648–658 CrossRef CAS PubMed.
- V. Desplat, S. Moreau, A. Gay, S. B. Fabre, D. Thiolat, S. Massip, G. Macky, F. Godde, D. Mossalayi, C. Jarry and J. Guillon, Synthesis and evaluation of the antiproliferative activity of novel pyrrolo[1,2-a]quinoxaline derivatives, potential inhibitors of Akt kinase. Part II, J. Enzyme Inhib. Med. Chem., 2010, 25, 204–215 CrossRef CAS PubMed.
- V. Desplat, M. Vincenzi, R. Lucas, S. Moreau, S. Savrimoutou, N. Pinaud, J. Lesbordes, E. Peyrilles, M. Marchivie, S. Routier, P. Sonnet, F. Rossi, L. Ronga and J. Guillon, Synthesis and evaluation of the cytotoxic activity of novel ethyl 4-[4-(4-substituted piperidin-1-yl)]benzyl-phenylpyrrolo[1,2-a]quinoxaline-carboxylate derivatives in myeloid and lymphoid leukemia cell lines, Eur. J. Med. Chem., 2016, 113, 214–227 CrossRef CAS PubMed.
- J. Guillon, M. Le Borgne, C. Rimbault, S. Moreau, S. Savrimoutou, N. Pinaud, S. Baratin, M. Marchivie, S. Roche, A. Bollacke, A. Pecci, L. Alvarez, V. Desplat and J. Jose, Eur. J. Med. Chem., 2013, 65, 205–222 CrossRef CAS PubMed.
- C. Vidaillac, J. Guillon, C. Arpin, I. Forfar-Bares, J. Grellet, S. Moreau, D. H. Caignard, C. Jarry and C. Quentin-Noury, Synthesis of omeprazole analogues and evaluation of these as potential inhibitors of the multidrug efflux pump NorA of Staphylococcus aureus, Antimicrob. Agents Chemother., 2007, 51, 831–838 CrossRef CAS PubMed.
- C. Vidaillac, J. Guillon, S. Moreau, C. Arpin, A. Lagardère, S. Larrouture, P. Dallemagne, D. H. Caignard, C. Quentin-Noury and C. Jarry, Synthesis of new 4-[2-(alkylamino)ethylthio]pyrrolo[1,2-a]quinoxaline and 5-[2-(alkylamino)ethylthio]pyrrolo[1,2-a]thieno[3,2-e]pyrazine derivatives, as potential bacterial multidrug resistance pump inhibitors, J. Enzyme Inhib. Med. Chem., 2007, 22, 620–631 CrossRef CAS PubMed.
- S. Wang, A. Folkes, I. Chuckowree, X. Cockcroft, S. Sohal, W. Miller, J. Milton, S. P. Wren, N. Vicker, P. Depledge, J. Scott, L. Smith, H. Jones, P. Mistry, R. Faint, D. Thompson and S. Cocks, Studies on pyrrolopyrimidines as selective inhibitors of multidrug-resistance-associated protein in multidrug resistance, J. Med. Chem., 2004, 47, 1329–1338 CrossRef CAS PubMed.
- S. Wang, N. C. Wan, J. Harrison, W. Miller, I. Chuckowree, S. Sohal, T. C. Hancox, S. Baker, A. Folkes, F. Wilson, D. Thompson, S. Cocks, H. Farmer, A. Boyce, C. Freathy, J. Broadbridge, J. Scott, P. Depledge, R. Faint, P. Mistry and P. Charlton, Design and synthesis of new templates derived from pyrrolopyrimidine as selective multidrug-resistance-associated protein inhibitors in multidrug resistance, J. Med. Chem., 2004, 47, 1339–1350 CrossRef CAS PubMed.
- J. Robert and C. Jarry, Multidrug resistance reversal agents, J. Med. Chem., 2003, 46, 4805–4817 CrossRef CAS.
- T. Suzuki, N. Fukazawa, K. San-nohe, W. Sato, O. Yano and T. Tsuruo, Structure-activity relationship of newly synthesized quinoline derivatives for reversal of multidrug resistance in cancer, J. Med. Chem., 1997, 40, 2047–2052 CrossRef CAS PubMed.
- P. D. Eckford and F. J. Sharom, ABC efflux pump-based resistance to chemotherapy drugs, Chem. Rev., 2009, 109, 2989–3011 CrossRef CAS PubMed.
- A. S. Guram, R. A. Rennels and S. L. Buchwald, A simple catalytic method for the conversion of aryl bromides to arylamines, Angew Chem. Int. Ed. Engl., 1995, 34, 1348–1350 CrossRef CAS.
- I. Castellote, J. J. Vaquero, J. Fernandez-Gadea and J. Alvarez-Builla, Pyrrolodiazines. 6. Palladium-catalyzed arylation, heteroarylation, and amination of 3,4-dihydropyrrolo[1,2-a]pyrazines, J. Org. Chem., 2004, 69, 8668–8675 CrossRef CAS PubMed.
- I. Castellote, J. J. Vaquero and J. Alvarez-Builla, Palladium-catalysed amination of 2-acyl-1-alkyl-5-bromopyrroles, Tetrahedron Lett., 2004, 45, 769–772 CrossRef CAS.
- J. Guillon, S. Moreau, L. Ronga, L. Basmacyian, A. Cohen, S. Rubio, G. Bentzinger, S. Savrimouto, N. Azas, C. Mullié and P. Sonnet, Design, synthesis and antimalarial activity of some new aminoalcoholpyrrolo[1,2-a]quinoxaline derivatives, Lett. Drug Des. Discov., 2016, 13, 932–942 CrossRef CAS.
- R. L. White, D. S. Burgess, M. Mandruru and J. A. Bosso, Comparison of three different in vitro methods of detecting synergy: time-kill, checkerboard, and E test, Antimicrob. Agents Chemother., 1996, 40, 1914–1918 CrossRef CAS PubMed.
- F. C. Odds, Synergy, antagonism, and what the chequerboard puts between them, J. Antimicrob. Chemother., 2003, 52, 1 CrossRef CAS PubMed.
- R. Franz, S. L. Kelly, D. C. Lamb, D. E. Kelly, M. Ruhnke and J. Morschhauser, Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains, Antimicrob. Agents Chemother., 1998, 42, 3065–3072 CrossRef CAS.
- R. Franz, M. Ruhnke and J. Morschhauser, Molecular aspects of fluconazole resistance development in Candida albicans, Mycoses, 1999, 42, 453–458 CrossRef CAS PubMed.
- M. M. Masood, M. Irfan, P. Khan, M. F. Alajmi, A. Hussain, J. Garrison, Md. T. Rehman and M. Abid, 1,2,3-Triazole-quinazolin-4(3H)-one conjugates: evolution of ergosterol inhibitor as anticandidal agent, RSC Adv., 2018, 8, 39611–39625 RSC.
- F. Pagniez, N. Lebouvier, Y. M. Na, I. Ourliac-Garnier, C. Picot, M. Le Borgne and P. Le Pape, Biological exploration of a novel 1,2,4-triazole-indole hybrid molecule as antifungal agent, J. Enzyme Inhib. Med. Chem., 2020, 35, 398–403 CrossRef CAS PubMed.
- A. K. Redhu, A. H. Shah and R. Prasad, MFS transporters of Candida species and their role in clinical drug resistance, FEMS Yeast Res., 2016, 16, fow043 CrossRef PubMed.
- V. K. Tran-Nguyen, R. Prasad, P. Falson and A. Boumendjel, Modulators of the efflux pump CaCdr1p of Candida albicans: mechanisms of action and chemical features, Curr. Med. Chem., 2017, 24, 3242–3253 CrossRef CAS PubMed.
- G. M. Sheldrick, SADABS, University of Göttingen, Germany, Standard software references, 1996 Search PubMed.
- G. M. Sheldrick, A short history of SHELX, Acta Crystallogr., Sect. A: Found. Crystallogr., 2008, 64, 112–122 CrossRef CAS PubMed.
- Supplementary X-ray crystallographic data.†.
- K. Nakamura, M. Niimi, K. Niimi, A. R. Holmes, J. E. Yates, A. Decottignies, B. C. Monk, A. Goffeau and R. D. Cannon, Functional expression of Candida albicans drug efflux pump Cdr1p in a Saccharomyces cerevisiae strain deficient in membrane transporters, Antimicrob. Agents Chemother., 2001, 45, 3366–3374 CrossRef CAS PubMed.
- S. Shukla, P. Saini, Smriti, S. Jha, S. V. Ambudkar and R. Prasad, Functional characterization of Candida albicans ABC transporter Cdr1p, Eukaryot. Cell, 2003, 2, 1361–1375 CrossRef CAS PubMed.
- D. Sanglard and F. C. Odds, Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences, Lancet Infect. Dis., 2002, 2, 73–85 CrossRef CAS PubMed.
- D. Hiller, D. Sanglard and J. Morschhauser, Overexpression of the MDR1 gene is sufficient to confer increased resistance to toxic compounds in Candida albicans, Antimicrob. Agents Chemother., 2006, 50, 1365–1371 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. CCDC 891808 and 891809. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9ra09348f |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |